Abstract
Chemical openers for KCNQ potassium channels are useful probes both for understanding channel gating and for developing therapeutics. The five KCNQ isoforms (KCNQ1 to KCNQ5, or Kv7.1 to Kv7.5) are differentially localized. Therefore, the molecular specificity of chemical openers is an important subject of investigation. Native KCNQ1 normally exists in complex with auxiliary subunits known as KCNE. In cardiac myocytes, the KCNQ1-KCNE1 (IsK or minK) channel is thought to underlie the IKs current, a component critical for membrane repolarization during cardiac action potential. Hence, the molecular and pharmacological differences between KCNQ1 and KCNQ1-KCNE1 channels have been important topics. Zinc pyrithione (ZnPy) is a newly identified KCNQ channel opener, which potently activates KCNQ2, KCNQ4, and KCNQ5. However, the ZnPy effects on cardiac KCNQ1 potassium channels remain largely unknown. Here we show that ZnPy effectively augments the KCNQ1 current, exhibiting an increase in current amplitude, reduction of inactivation, and slowing of both activation and deactivation. Some of these are reminiscent of effects by KCNE1. In addition, neither the heteromultimeric KCNQ1-KCNE1 channels nor native IKs current displayed any sensitivity to ZnPy, indicating that the static occupancy by a KCNE subunit desensitizes the reversible effects by a chemical opener. Site-directed mutagenesis of KCNQ1 reveals that residues critical for the potentiation effects by either ZnPy or KCNE are clustered together in the S6 region overlapping with the critical gating determinants. Thus, the convergence of potentiation effects and molecular determinants critical for both an auxiliary subunit and a chemical opener argue for a mechanistic overlap in causing potentiation.
Voltage-gated potassium channels are critical for membrane excitability. In cardiac tissue, potassium currents are important elements responsible for the repolarization of action potential. Among the different potassium current components, IKs and IKr are two key determinants for the duration of cardiac action potential (1). Molecular and genetic studies have shown that the IKs component is likely formed by the heteromultimeric assembly of subunits encoded by KCNQ1 (Kv7.1) and KCNE1 (IsK or minK) (2, 3), whereas the current encoded by hERG (human ether-ago-go related gene) is responsible for IKr (4, 5). Genetic mutations of genes encoding these subunits commonly resulted in reduction of channel expression or conductance that causes congenital long QT syndrome (1, 6–8). In addition, IKr and, to some extent, IKs are common targets of unintended block by non-cardiac drugs causing acquired long QT syndrome (9, 10). Both forms of long QT syndrome are life-threatening cardiac conditions with a shared feature of current reduction (9–11). Unlike channel blockers, the chemical activators for hERG and KCNQ1 are rare but valuable probes. Understanding of channel activation and investigation of channel openers are of considerable interest both in terms of gating mechanisms and in terms of developing therapeutic intervention.
Voltage-gated potassium channels consist of pore-forming α subunits and auxiliary regulatory β subunits that contribute to diverse physiological functions, e.g. Kvβ subunits (12). KCNE proteins are auxiliary subunits of at least five members, KCNE1 to KCNE5 (13, 14). Each KCNE subunit consists of a single transmembrane segment. All five KCNE members (KCNE1 to KCNE5) are capable of coassembly with KCNQ1 (15, 16). In the case of KCNQ1 with KCNE1, the resultant heteromultimeric current is similar to IKs in cardiac tissue. Both KCNE1 and KCNE3 increase the maximum conductance of KCNQ1, whereas the association with KCNE2, KCNE4, and KCNE5 results in inhibition (2, 3, 16, 17). The effects of KCNE1 on KCNQ1 include increasing overall current, slowing the activation and deactivation kinetics, and removal of inactivation (2, 3). There is also evidence suggesting an increase in the single channel conductance of KCNQ1 (18–20). KCNE3 stabilizes KCNQ1 in the open state and augments current amplitude to a level comparable with that by KCNE1 (17). Several residues in the KCNQ1 S6 domain (e.g. Ser338, Phe339, and Phe340) critical for the augmentation by KCNE1 and KCNE3 have been identified (21–23), although the interaction mechanism still remains elusive.
Recently, several KCNQ-activating compounds have been identified, some of which are in clinical trials for anti-convulsive applications (24–28). These compounds are interesting in several ways. First, their structures are sufficiently distinct and appear to affect different aspects of channel properties that lead to more active channels (29, 30). Second, mutagenesis studies revealed that they indeed recognize different “agonistic” sites on KCNQ channels (31, 32). Furthermore, one KCNQ channel complex is capable of interacting with more than one class of chemical openers. As a result, the tripartite complex displays a hybrid response, tunable by different concentrations and/or ratios of the chemical openers (33). Specificity for either isoform or subunit composition is a topic critical for the molecular understanding of these cation channel openers. Because the KCNQ2–5 subunits are more commonly found in the nervous system, whereas KCNQ1 is predominantly localized in cardiac and other non-excitable tissues where they are in complex with KCNE subunits, it is particularly relevant to investigate subunit specificity among KCNQ1, KCNQ1-KCNE, and KCNQ2–5 channels.
We have reported that bis(1-hydroxy-2(1H)-pyridineselonato-O,S) zinc, commonly known as zinc pyrithione (ZnPy),2 is a potent activator of KCNQ 1, 2, 4, and 5 channels but not KCNQ3 (34). The effects of ZnPy on neuronal KCNQ channels include both a hyperpolarizing shift in the voltage dependence of activation and an increase in current amplitude. Of particular interest, mutagenesis studies have revealed that KCNQ2(A306T) in the S6 segment significantly reduced current augmentation of ZnPy (34). The corresponding region of KCNQ2(A306T) in KCNQ1 was implicated for interactions with KCNE1 (21–23). We thus hypothesized that the coassembly of KCNQ1 with KCNE1 subunits may lead to a distinct molecular specificity or pharmacological response, which would be relevant to finding compounds specific for either neuronal or cardiac KCNQ channels. Using a combination of electrophysiological analyses and site-directed mutagenesis, we have evaluated channel properties affected by ZnPy, tested whether KCNE auxiliary subunits affect ZnPy sensitivity, and investigated the molecular determinants critical for KCNE modulation and KCNQ1 pharmacology to chemical openers.
EXPERIMENTAL PROCEDURES
cDNAs and Mutagenesis—KCNQ1 and KCNE1 cDNA were gifts from Dr. M. C. Sanguinetti (University of Utah). KCNE3 cDNA was a gift from Dr. T. V. McDonald (Albert Einstein School of Medicine). Point mutations in the KCNQ1 channel were introduced by using the QuikChange II site-directed mutagenesis kit (Stratagene) and verified by DNA sequencing.
Cell Culture and Transfection—Chinese hamster ovary (CHO) cells were grown in 50/50 Dulbecco's modified Eagle's medium/F-12 (Cellgro, Manassas, VA) with 10% fetal bovine serum and 2 mm l-glutamine (Invitrogen). To express KCNQ1 and KCNQ1-KCNE1, cells were split at 24 h before transfection, plated in 60-mm dishes, and transfected with Lipofectamine2000™. After transfection, cells were split and replated onto coverslips coated with poly-l-lysine (Sigma-Aldrich). Plasmid expressing CD4 as a marker was cotransfected. Prior to recording, anti-CD4 Dynabeads (Invitrogen) were added to the medium to allow for identification of the transfected cells. To coexpress KCNQ1 and KCNE3, cells were electroporated with a Nucleofector™ kit for CHO-K1 cells (Amaxa, Gaithersburg, MD) according to the manufacturer's instruction. A green fluorescent protein cDNA (Amaxa) was cotransfected to allow identification of the transfected cells with the fluorescent microscope. The cDNA concentration of KCNQ1 or KCNE was 200 ng/μl, and the molar ratio of KCNE/KCNQ1 was 1:1.
Electrophysiological Recording in CHO Cells—Whole-cell voltage clamp recording was carried out using cultured CHO cells at room temperature by an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA). The electrodes were pulled from borosilicate glass capillaries (World Precision Instruments, Sarasota, FL). When filled with the intracellular solution, the electrodes have resistances of 3–5 MΩ. Pipette solution contained (in mm) 145 KCl, 1 MgCl2, 5 EGTA, 10 HEPES, and 5 MgATP (pH 7.3 with KOH). During the recording, constant perfusion of extracellular solution was maintained using a B-channel valve BPS-8 model perfusion system (ALA Scientific Instruments, Westburg, NY). Extracellular solution contained (in mm) 140 NaCl, 3 KCl, 2 CaCl2, 1.5 MgCl2, 10 HEPES, and 10 glucose (pH 7.4 with NaOH). Signals were filtered at 1 KHz and digitized using a DigiData 1322A with pClamp 9.2 software (Molecular Devices). Series resistance was compensated by 60–80%. In the present study, three types of voltage protocols were used. The holding potential was -80 mV in all voltage protocols. To record the KCNQ1 current, the cells were stimulated by a series of 1,500-ms depolarizing steps from -70 mV to +50 mV in 10-mV increments. A testing step to -120 mV was applied to obtain tail currents. To record KCNQ1-KCNE1 currents, longer depolarizing steps (3,000 ms) were used to elicit the currents, and a -50-mV testing step was used to obtain tail currents. To record the KCNQ1-KCNE3 current, slightly different voltage protocols were used. The currents were elicited by a series of depolarizing steps from -100 to +80 mV in 20-mV increments. The tail current testing step was -50 mV.
Isolation of Cardiac Myocytes and Native IKs Recording—Single myocytes were isolated from the left ventricle of adult guinea pig in a Langerdorff perfusion system as described previously (35). Briefly, the hearts were removed quickly via midline thoracotomy and perfused with a Ca2+-free Tyrode's solution containing collagenase (6 mg/ml) and protease (0.1 mg/ml) for ∼5–6 min. Then the hearts were switched to Kraft-Bruhe solution perfusion for 5 min, and the ventricles were minced and gently triturated to single cells. The cells were stored at 4 °C in Kraft-Bruhe solution until use. Kraft-Bruhe solution contained the following (in mm): 50 l-glutamic acid, 80 KOH, 40 KCl, 3 MgSO4, 25 KH2PO4, 10 HEPES, 1 EGTA, 20 taurine, and 10 glucose (pH 7.4).
Before recording, the myocytes were transferred to a recording chamber perfused with Tyrode's solution. Both Itotal and IKs were recorded with conventional configuration of patch-clamp technique using an Axopatch 200B amplifier (Molecular Devices). Pipettes were pulled from borosilicate glass capillaries (World Precision Instruments). When filled with the intracellular solution containing (in mm) 120 KCl, 10 KH2PO4, 1 MgSO4, 5 EGTA, and 5 HEPES (pH 7.2 with KOH), the pipettes have resistances of 3–5 MΩ. Itotal was elicited by 3-s depolarizing pulses from a holding potential of -80mV to various test potentials between -70 and +70 mV in 10-mV increments in Tyrode's solution perfusion. Tyrode's solution contained (in mm) 135 NaCl, 5.4 KCl, 1 MgCl2, 0.33 NaH2PO4, 5 HEPES, and 5 glucose (pH 7.4 with NaOH) and was oxygenated with 100% O2. To isolate for IKs, the external solution was switched to Na+-free solution containing (in mm) 132 N-methyl-d-glucamine (for sodium ion replacement), 1.0 CaCl2, 1.0 MgCl2, 10 HEPES, 5 glucose, 0.05 lanthanum chloride to block IKr, and 0.005 nifedipine to block L-type calcium current (pH 7.4 with HCl).
Modeling—Three-dimensional structural models for the KCNQ1 S5 and S6 domains were generated using the solved crystal structure of Kv1.2 (Protein Data Bank code 2A79) as a template. The corresponding domains between KCNQ1 and Kv1.2 were aligned with the DNASTAR MegAlign program using standard parameters. The KCNQ1 models were constructed using DeepView/SWISS-PDBViewer (36). The structural representation was performed with the POV-Ray program.
Data and Statistical Analysis—Patch-clamp data were processed using Clampfit 9.2 (Molecular Devices) and then analyzed in GraphPad Prism 4 (GraphPad, San Diego, CA). The activation curve was fitted by the Boltzmann Sigmoidal equation: G = Gmin+ (Gmax - Gmin)/(1 + exp(V - V½)/S)), where Gmax is the maximum conductance, Gmin is the minimum conductance, V½ is the voltage for reaching 50% of maximum conductance, and S is the slope factor. The dose-response curve was fitted by the Hill equation: E = Emax/(1 + (EC50/C)P), where EC50 is the drug concentration producing half of the maximum response, and P is the Hill coefficient. The activation and deactivation trace were fitted by the standard (or power) exponential equation using Clampfit 9.2. Data are presented as means ± S.E. Significance was estimated using a paired two-tailed Student's t test.
RESULTS
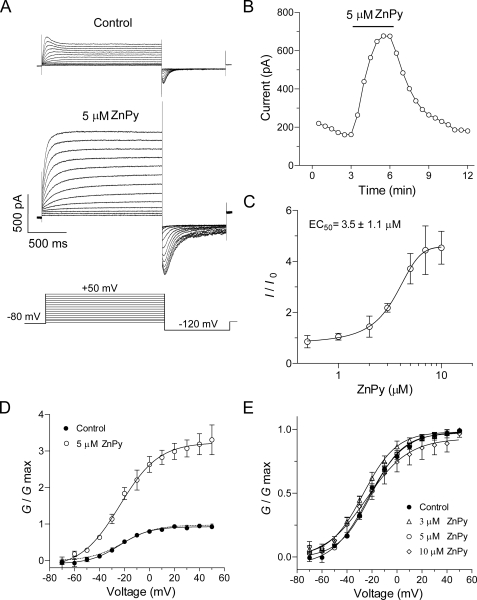
Augmentation of KCNQ1 Channels by ZnPy—To examine ZnPy modulation on the KCNQ1 current, we first expressed the KCNQ1 cDNA in CHO cells and recorded the channel activity with a whole-cell voltage clamp. KCNQ1 displayed a characteristic outward current with visible inactivation (Fig. 1A). In the presence of 5 μm ZnPy, steady-state currents at different depolarizing voltages were greatly potentiated, and inactivation was no longer readily detectable during the depolarizing phase (Fig. 1A). The ZnPy-mediated potentiation was fully reversible upon removal of ZnPy (Fig. 1B), consistent with the idea of modulating channel activity instead of protein density on cell surface. Examination of current amplitude increase using steady-state currents at different concentrations of ZnPy revealed a half-maximal value (EC50) of 3.5 ± 1.1 μm (n > 3) (Fig. 1C).
FIGURE 1.
ZnPy potentiates KCNQ1 homomultimers expressed in CHO cells. A, the representative traces of the KCNQ1 currents before and after application of 5 μm ZnPy. The cell was held at -80 mV. The KCNQ1 currents were elicited by a series of voltage steps from -70 mV to +50 mV in 10-mV increments (inset). Scale bars and voltage-step protocol are as indicated. B, time course of a representative KCNQ1 peak current in the presence of 5 μm ZnPy. C, concentration-potentiation curve of ZnPy (n > 3). D, conductance-voltage curves of KCNQ1 in the absence or presence of ZnPy (5 μm). The conductance at each depolarized voltage (from -70 to +50 mV) was normalized to the conductance at +50 mV in the control (n > 3). The dashed line is a fit curve in the presence of ZnPy after rescaling Gmax to 1. E, conductance-voltage curves after application of the indicated concentrations of ZnPy (n > 3).
Previous work has shown that ZnPy-mediated potentiation of neuronal KCNQ2 channels involves both a hyperpolarizing shift of half-maximal activation voltage (V½) and an increase in overall conductance (Gmax) (34). To determine the ZnPy effects on the KCNQ1 homomultimer, we examined the G-V curve in the presence or absence of 5 μm ZnPy, which causes an ∼80% potentiation. In the absence of ZnPy, the V½ value was -23.2 ± 1.1 mV, similar to the value of the earlier reports (3, 37, 38) (Fig. 1D). Further experiments with a range of ZnPy concentrations revealed no significant change of V½ either (Fig. 1E). Hence, ZnPy increases overall conductance of KCNQ1 without affecting voltage dependence.
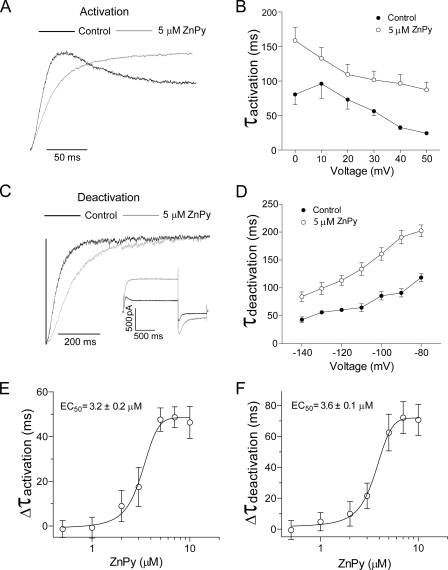
Modulation of Kinetic Properties of KCNQ1 by ZnPy—To investigate ZnPy-mediated modulation on the close-open transition of the KCNQ1 channel, effects on both activation and deactivation were examined. The currents induced by depolarizing from -70 mV to +50 mV displayed a rapid activation followed by characteristic inactivation. In the presence of 5 μm ZnPy, the time constant of activation was slowed from 23.4 ± 2.4 ms to 74.0 ± 12.5 ms (n = 3; p < 0.01) (Fig. 2A). This effect was seen in a range of depolarizing voltages (Fig. 2B). In addition, ZnPy also induced slowing of deactivation in hyperpolarizing voltages (Fig. 2, C and D). The reduction of deactivation rate is consistent with the overall increase of current amplitude or Gmax.
FIGURE 2.
ZnPy effects on channel kinetics of KCNQ1 homomultimers. A, the normalized upstroke phases from the full traces (see inset in C) in the control (black line) and after application of 5 μm ZnPy (gray line) are shown. B, the time constant of activation was fit to standard (or power) exponential function and was plotted against the different voltages. C, the normalized tail currents from full traces in the absence (black line) and presence (gray line) of 5 μm ZnPy are shown. D, the deactivation rate was fit to standard exponential function, and the time constant was plotted against the different voltages. E and F show the dose-dependent relationship of ZnPy concentration to changes in either activation rate (E) or deactivation rate (F).
If ZnPy affects both activation and deactivation through the same interaction that causes the increase of current amplitude, one should see similar EC50 values. Indeed, when plotting the change of time constants for activation or deactivation against concentrations of ZnPy, we found EC50 values of 3.2 ± 0.2 μm (n > 3) for activation and 3.6 ± 0.1 μm (n > 3) for deactivation (Fig. 2, E and F). Thus, the EC50 values measured for current amplitude, activation, and deactivation time constants are essentially the same, providing evidence for the notion that one class of interaction (or binding site) is responsible for the activation and deactivation changes and for the current potentiation (Fig. 1C).
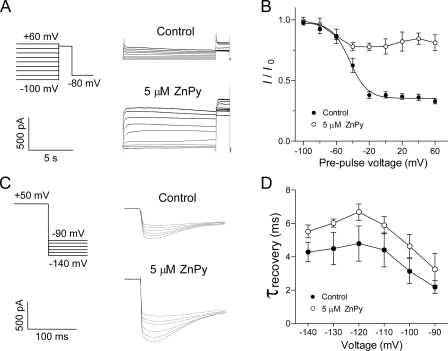
Among different KCNQ channels, inactivation is a distinctive feature for KCNQ1. The overall increase in current amplitude could be contributed in part through inhibition of inactivation (Fig. 1A). Using the voltage pulse protocols outlined in Fig. 3A, the inactivation kinetics in the presence or absence of ZnPy was evaluated. In the absence of drug treatment, noticeable inactivation was observed (Fig. 3, A and B). However, in the presence of 5 μm of ZnPy, the inactivation was largely diminished. At a higher concentration of 10 μm, a similar effect was observed (data not shown). When examining the recovery from inactivation, the effect was rather minimal (n > 4; p > 0.1) (Fig. 3, C and D). Thus, the inhibition of inactivation by ZnPy is a contributing factor for an overall increase of current amplitude.
FIGURE 3.
ZnPy reduces the voltage-dependent inactivation. A, the steady-state inactivation of the KCNQ1 channel was investigated by the protocol shown in the left panel, a series of 10-s-long pre-pulses from -100 mV to +60 mV in 20-mV increments followed by a test pulse at +50 mV. The right panel shows the currents recorded before and after application of 5 μm ZnPy. B, the steady-state inactivation curves with or without 5 μm ZnPy are shown. C, the protocol to study recovery from inactivation is shown in the left panel: inward KCNQ1 tail currents were recorded from -140 mV to -90 mV in 10-mV increments after a depolarization step to +50 mV. The initial phases of the tail currents at these potentials are shown (right panel). D, the initial rising phases preceding slower deactivation of the tail currents were fit to a single exponential function and plotted against the corresponding voltage.
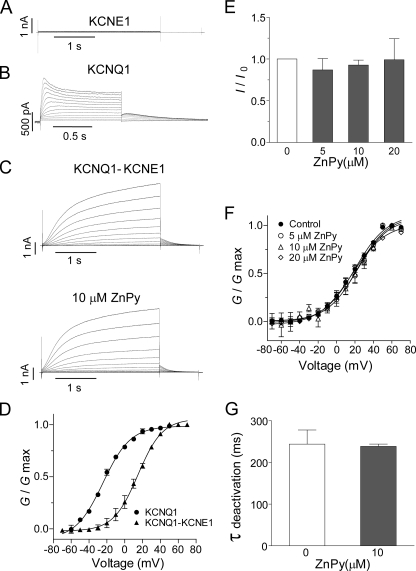
Coassembly with KCNE1 Desensitizes ZnPy-mediated Augmentation—Coassembly of KCNQ1 with KCNE1 is thought to form the native IKs (2, 3). The modulation of KCNQ1 by KCNE1 displays considerable similarity to ZnPy-induced effects. Both KCNE1 and ZnPy increase current amplitude, decrease inactivation, and slow activation and deactivation (Table 1), although their effects on the G-V curve are different. To test any potential alteration of sensitivity to ZnPy, we coexpressed KCNQ1 and KCNE1 (Fig. 4C). In the presence of 10 μm ZnPy, there were no obvious effects of drug-induced further potentiation (Fig. 4, C and E). It is known that the heteromultimeric KCNQ1-KCNE1 channels have a depolarizing V½ shift of 38.5 ± 1.5 mV compared with homomultimeric KCNQ1 channels (Fig. 4D). In the presence of different concentrations of ZnPy, the V½ values remain unchanged (Fig. 4F). In addition, no changes were detected for deactivation in the presence of ZnPy (Fig. 4G).
TABLE 1.
Comparision of ZnPy, KCNE1, and KCNE3 effects on KCNQ1 G is the channel conductance in the presence of either ZnPy or auxiliary subunits, whereas G0 is the conductance of KCNQ1 without ZnPy or any auxiliary subunits (KCNE1 or KCNE3). Both ZnPy and KCNE increase the conductance ∼4-fold; ZnPy and KCNE1 slow the activation and deactivation, whereas KCNE3 accelerates the deactivation. In addition, ZnPy inhibits KCNQ1 inactivation, whereas KCNE completely abolishes the inactivation. Significance was estimated by using a paired t-test; n > 4. NA, not applicable.
| KCNQ1a | ZnPy (5 μm)a | KCNQ1-KCNE1a | KCNQ1-KCNE3b | |
|---|---|---|---|---|
| G/G0 | 1.0 | 4.2 ± 0.8c | 4.3 ± 0.9c | 4.4 ± 1.5c |
| τ activation (ms) | 23.4 ± 4.9 | 73.7 ± 12.5c | τ1, 1297.6 ± 432.5c; τ2, 218.0 ± 46.8 | 25.9 ± 2.8 |
| τ deactivation (ms) | 60.6 ± 6.2 | 113.20 ± 15.41c | 243.8 ± 47.9c | 21.6 ± 3.4c |
| Inactivation (%) | 67.2 ± 3.7 | 14.2 ± 12.5c | NA | NA |
The KCNQ1 and KCNQ1-KCNE1 currents were elicited by depolarization to +50 mV
The KCNQ1-KCNE3 currents were elicited by depolarization to +40 mV
*, p < 0.05 compared with KCNQ1
FIGURE 4.
The heteromultimeric KCNQ1-KCNE1 channels lack ZnPy sensitivity. A and B show the currents induced from CHO cells transiently transfected by KCNE1 and KCNQ1, respectively. C, the currents recorded in the cells cotransfected by KCNQ1 and KCNE1 in the absence or presence of 10 μm ZnPy are shown. D, comparison of voltage-dependent activation of KCNQ1 homomultimers and KCNQ1-KCNE1 heteromultimeric channels is shown. E, the histogram shows steady current of KCNQ1-KCNE1 heteromultimers in the presence of ZnPy at the indicated concentrations. F, G-V curves of KCNQ1-KCNE1 in the presence of different concentrations of ZnPy are shown. G, the histogram shows the effects of 10 μm ZnPy on the deactivation rate of KCNQ1-KCNE1.
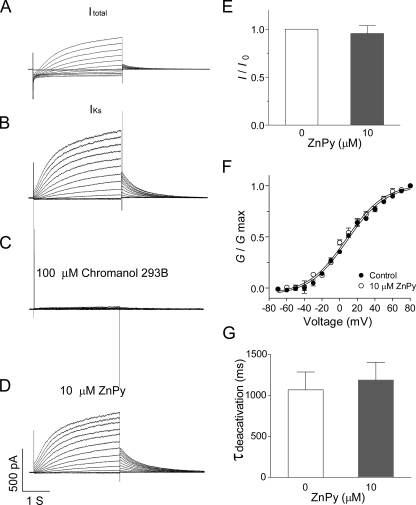
It is believed that coassembly of KCNE1 and KCNQ1 forms the native IKs channels. Therefore, if KCNE1 indeed abolishes the KCNQ1 sensitivity to ZnPy, native IKs should display no sensitivity. To examine this possibility, ventricular cardiac myocytes from guinea pig were acutely isolated. Using the previously reported conditions (see “Experimental Procedures”), the IKs component was isolated and showed a characteristic inhibition by 100 μm chromanol 293B (Fig. 5, A–C) (39). In the presence of ZnPy, the IKs displayed no detectable change in macroscopic currents, voltage sensitivity, or deactivation kinetics (Fig. 5, D–G). Hence, neither recombinant KCNQ1-KCNE1 nor native IKs displays the sensitivity to ZnPy. This is in further agreement with KCNE1-mediated desensitization.
FIGURE 5.
Native IKs recorded from guinea pig cardiac myocytes lacks ZnPy sensitivity. A, the total current (Itotal) elicited by a series of depolarization steps from -70 mV to +70 mV in 10-mV increments in normal Tyrode's solution, which includes 140 mm Na+, 5.4 mm K+, and 1.8 mm Ca2+, is shown. B, the current recorded in a modified Na+-free external solution supplemented with 50 μm La3+ to block IKr and 5 μm nifedipine to block ICa is shown. C, the currents recorded in the modified external solution were inhibited by 100 μm IKs selective inhibitor chromanol 293B. D, the currents recorded in the same cells in B after application of 10 μm ZnPy are shown. E, the histogram shows that 10 μm ZnPy did not increase the native IKs amplitude. F, G-V curves of native IKs in the absence or presence of 10 μm ZnPy are shown. G, the histogram shows that 10 μm ZnPy did not affect the deactivation rate of the native IKs.
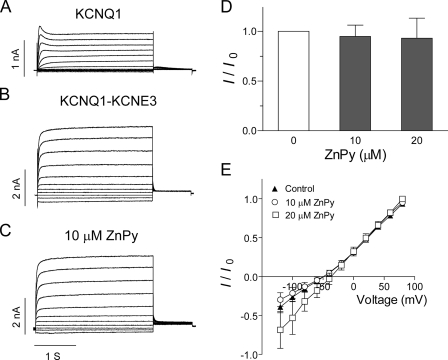
The five known KCNE subunits may be divided according to their effects. Both KCNE1 and KCNE3 display potentiation when coassembled with pore-forming α subunits. In contrast, the other KCNE subunits display inhibitory effects. KCNE3 has a 35% identity with KCNE1. Coassembly of KCNE3 with KCNQ1 produces a constitutive and potentiated current (17). Because the critical KCNQ1 residues for KCNE3 lie in proximity to those for KCNE1, and KCNE1 abolished ZnPy potentiation, we sought to examine whether KCNE-mediated desensitization is generalizable. Indeed, we found that ZnPy caused no detectable potentiation on the KCNQ1-KCNE3 heteromultimeric channel in CHO cells (Fig. 6, A–D). In addition, the I-V relationship has no significant change in presence of either 10 or 20 μm ZnPy (Fig. 6E). Thus, coassembly of KCNQ1 with KCNE3 also causes desensitization to ZnPy.
FIGURE 6.
ZnPy effects on the heteromultimeric KCNQ1-KCNE3 channels. A and B show the currents induced by KCNQ1 and heteromultimeric KCNQ1-KCNE3. Compared with the KCNQ1 homomultimer currents, coexpression with KCNE3 caused a constitutively active current. C, the currents recorded in the same cells in B after application of 10 μm ZnPy are shown. D, the histogram shows the effects of 10 and 20 μm ZnPy on KCNQ1-KCNE3 heteromultimers. E, the I-V relationship of KCNQ1-KCNE3 in the absence or presence of 10 or 20 μm ZnPy is shown.
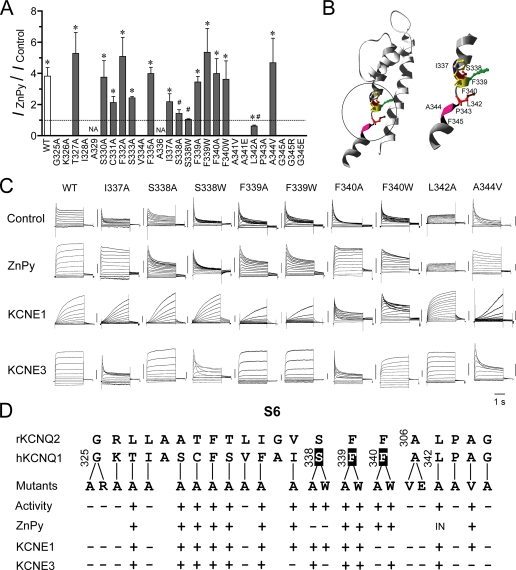
Molecular Determinants for KCNE1 and ZnPy Modulation—Earlier work has identified residues in KCNQ1 critical for KCNE1 modulation (21–23). Of particular interest, KCNQ1 with mutations of Phe339 or Phe340 displays much reduced effects by KCNE1. Residues in the corresponding area of KCNQ2 also affect the ZnPy sensitivity (34). We thus constructed point mutations in this region and tested for sensitivity to KCNE1, KCNE3, and ZnPy. Fig. 7A illustrates a segment of the KCNQ1 S6 domain, from Gly325 to Gly345, and the individually mutated residues are as indicated. Mutants with functional expression were tested for sensitivity to 10 μm ZnPy. Although most functional KCNQ1 mutants displayed the ZnPy-mediated potentiation, three mutants show reduction of ZnPy potentiation (Fig. 7, A and C). The S338A mutation dramatically reduced ZnPy potentiation from 4.5-fold to 1.4-fold, whereas S338W completely abolished ZnPy potentiation. Furthermore, ZnPy has an inhibitory effect on the L342A mutation. Because the mutant itself displays no inactivation (Fig. 7C), one possibility would be that the mutation does not affect the binding but causes ZnPy to preferably bind the close state. Investigation of the mutants for functional coexpression with KCNE1 indicated that most mutants, including S338A, S338W, and L342A, are sensitive to KCNE1 modulation. An immediately adjacent residue, Phe340, when changed to either alanine or tryptophan, renders a complete loss of modulation by KCNE1 (Fig. 7, B and D). The Phe339 residue was shown previously to be important for the modulation by KCNE1 (21, 22). Mutations of Phe339 to either alanine or tryptophan preserve sensitivity to ZnPy modulation. Interestingly, we began to find more cells no longer display full sensitivity to KCNE1 modulation under the same 1:1 transfection ratio of KCNQ1 and KCNE1 (Fig. 7C and data not shown). This may represent an intermediate effect by the mutation in perturbing coassembly. The loss or disruption of KCNE1 modulation could be identified by lacking potentiation of overall conductance or a noticeable inactivation phase. Together, the evidence from point mutation sensitivity studies reveals that critical residues for either ZnPy or KCNE1 modulation are clustered in the S6 domain.
FIGURE 7.
Point mutants reveal critical residues for ZnPy and KCNE modulation. A, the histogram shows the potentiation effect of 10 μm ZnPy on KCNQ1 S6 point mutants (at +50 mV) (n > 3). Each mutation site was indicated based on the predicted transmembrane regions. The dashed line indicates a potentiation level of 1 (no effect). Significance was estimated using analysis of variance; n > 3. *, p < 0.05 compared with the control level (dashed line); #, p < 0.05 compared with the ZnPy potentiation effect on the wild type (WT). B, the modeled structure of an isolated KCNQ1 S5-S6 domain is shown. Left panel, residues essential for ZnPy, R-L3, KCNE1, and KCNE3 are colored differently. Right panel, enlarged structure of the S6 domain. C, the representative traces of mutants alone or coexpression with auxiliary subunits (KCNE1 or KCNE3) are shown. D, summary of mutagenesis results is shown. The inset IN indicates an inhibitory effect.
When tested with KCNE3, most mutants displayed sensitivity as indicated in Fig. 7D. Intriguingly, the residue Ser338, when mutated to tryptophan, lost the sensitivity to KCNE3, whereas a mutation to alanine was sensitive to KCNE3. The adjacent residue, Phe340, when mutated to alanine, lost the sensitivity, whereas a mutation to tryptophan was sensitive to KCNE3. This indicates a strict dependence of side-chain size, as only conserved changes are tolerable. Mutations of I337A and A344V led to the loss of KCNE3 modulation. Mutations of Phe339, which did not display full sensitivity to KCNE1, were sensitive to KCNE3. These experiments provide evidence that the Ile337 to Ala344 region is critical for KCNE3 modulation and that the modulation desensitizes the ZnPy potentiation. Using homology modeling and the Kv1.2 crystal structure, we constructed a KCNQ1 model, and the location of critical residues for KCNE1, KCNE3, and ZnPy were assigned (Fig. 7B). A short segment between Ile337 and Val344 harbors the key determinants, either overlapping or in immediate proximity to the gating hinge or PAG motif (40). This supports that one restrictive region in S6 is involved in augmentation for both auxiliary subunits and ZnPy.
DISCUSSION
There are several measurable changes that contribute to the overall current potentiation of KCNQ1 by ZnPy. The simultaneous slowing of both activation and deactivation is an intriguing behavior of ZnPy, seen in KCNQ1 (Fig. 2) and KCNQ2 (34). This behavior bares some resemblance to that by KCNE1. A similar effect was also reported for L-364,373 ((3-R)-1, 3-dihydro-5-(2-fluorophenyl)-3-(1H-indol-3-ylmethyl)-1-methyl-2H-1,4-benzodiazepine-2) (also known as R-L3), a benzodiazepine-like compound with lower potency compared with that of ZnPy (41). Indeed, the slowing of deactivation is in agreement with the overall current potentiation. But the slowing of activation rate would have an opposite effect on potentiation. The potentiation of neuronal KCNQ channels by ZnPy causes a significant hyperpolarization shift of voltage dependence, whereas ZnPy has no effects on the activation dependence curve of KCNQ1 (Fig. 1). Hypothetically, the ZnPy effect on activation kinetics could simply elongate the first latency of channel opening, a notion that requires further investigation.
KCNQ1 channels display prominent inactivation upon depolarization (Figs. 1A and 3). Several voltage-gated channels openers (e.g. sodium channel activator DPI 201-106 and hERG activator RPR260243) have been reported to confer potentiation through inhibiting inactivation (42, 43). Noticeably, in addition to KCNQ1, inactivation was recently reported for KCNQ4 and KCNQ5. BMS-204352, an activator of neuronal KCNQ channels, inhibits the KCNQ4 inactivation (44). In the presence of 5 μm ZnPy, the inactivation of KCNQ1 was almost completely removed at the depolarizing voltages with no significant change of recovery from inactivation. For those mutants that possess significant inactivation, the inhibition of inactivation by ZnPy was also observed (Fig. 7C and data not shown). In addition, KCNE1 and KCNE3 abolish KCNQ1 inactivation, which contributes to the amplitude potentiation of the heteromultimeric channels (45–47). Thus, inhibition of KCNQ1 inactivation may be a key potentiation mechanism by both auxiliary subunits and chemical openers of KCNQ1. However, the quantitative analysis and kinetic modeling are required to quantify the degree of the kinetics changes caused by ZnPy and to what extent the changes could account for the increase in amplitude.
Earlier reports have provided the evidence that alteration of signal channel kinetics could contribute to the overall potentiation of KCNQ1 by KCNE subunits (19, 20, 48). The potentiation by ZnPy on KCNQ2 and KCNQ4 appears to have no effect on the single conductance (34). Given the considerable similarity between ZnPy and KCNE1 shown in the present study, it would be interesting to determine whether ZnPy could alter the single channel conductance of KCNQ1.
Although ZnPy strongly potentiates KCNQ1, it does not have an effect on the heteromultimeric KCNQ1-KCNE1 and KCNQ1-KCNE3 channels. The loss of sensitivity may be a result of auxiliary subunit assembly that competes, masks, or decouples the binding and/or allosteric effects by ZnPy. S338A showed a lower sensitivity to ZnPy compared with the wild type, whereas S338W does not have sensitivity to ZnPy. For the mutation L342A, ZnPy inhibited the amplitude. This suggests that Ser338 and Leu342 are critical for ZnPy potentiation. Interestingly, an adjacent residue, Ile337, was reported to be critical for R-L3 effects (49). In the KCNQ1 homology model (Fig. 7B), all three residues locate in the lower portion of S6, which suggests this region is essential for augmentation of KCNQ1 by chemical openers. Voltage-gated potassium channel activation is proposed to be the result of simultaneous bending of all S6 domains to open the pore (50–54). For KCNQ1, a flexible motif PAG (residues 343–345) is important for channel activation (40). Although the precise interaction sites for KCNE1 and KCNE3 modulations are still being actively investigated, the critical role of this S6 region is clearly evident. Regardless of the precise roles of KCNE subunits in abolishing the ZnPy effect, either via competition for binding or by allosteric changes, the convergence of molecular determinants critical for argumentation of KCNQ1 channels argues for the possibility of a mechanistic overlap in endowing the potentiation. Indeed, the differences between ZnPy and KCNE1 effects are also noticeable, including differential effects on the G-V curve, and mutations of Ser338 and Phe340 have a different sensitivity to ZnPy and KCNE1. Further studies are required to clarify whether and to what extent their mechanistic actions overlap.
Alteration of pharmacology for an ion channel by an accessory subunit has been reported in a number of cases, e.g. Refs. 41 and 55–58. Complete desensitization of an agonistic ligand for voltage-gated ion channels is rare, perhaps in part due to few openers that are available. For KCNQ channels, a known case so far is differential potentiation by R-L3 for KCNQ1 and KCNQ1-KCNE1, in which case there is a noticeable reduction of R-L3 effects for the heteromultimeric channels when titrated with amounts of the injected cRNA in the Xenopus oocyte expression system (41). This is intriguing especially in light of a recent paper arguing for KCNQ1-KCNE1 with 4:2 stoichiometry (59) and the evidence that one KCNQ1 channel complex may recruit more than one type of KCNE subunits (60). Indeed, ZnPy and retigabine are two KCNQ openers and interact with different sites (34). When applied together, they cause a hybrid effect indicative of simultaneous binding to one channel complex (33). In the proposed “4:2” complex, even if KCNE physically blocks a ZnPy site, a simple model would expect there are two “vacant” sites. Our present data indicate no ZnPy effect at all in the presence of KCNE1 or KCNE3, although these results could not distinguish whether the zinc pyrithione actually binds to the heteromultimeric channel. This is an interesting direction for future studies.
Acknowledgments
We thank Drs. M. Sanguinetti and T. McDonald for gifts of cDNA. We thank Drs. E. Marban and K. Chang for gifts of isolated cardiac myocytes and thank our colleagues and members of the Li laboratory for valuable discussions and comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM070959 and GM078579 (to M. L.). This work was also supported by a postdoctoral fellowship award from the American Heart Association (to Z. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ZnPy, zinc pyrithione; CHO, Chinese hamster ovary.
References
- 1.Zeng, J., Laurita, K. R., Rosenbaum, D. S., and Rudy, Y. (1995) Circ. Res. 77 140-152 [DOI] [PubMed] [Google Scholar]
- 2.Barhanin, J., Lesage, F., Guillemare, E., Fink, M., Lazdunski, M., and Romey, G. (1996) Nature 384 78-80 [DOI] [PubMed] [Google Scholar]
- 3.Sanguinetti, M. C., Curran, M. E., Zou, A., Shen, J., Spector, P. S., Atkinson, D. L., and Keating, M. T. (1996) Nature 384 80-83 [DOI] [PubMed] [Google Scholar]
- 4.Sanguinetti, M. C., Jiang, C., Curran, M. E., and Keating, M. T. (1995) Cell 81 299-307 [DOI] [PubMed] [Google Scholar]
- 5.Trudeau, M. C., Warmke, J. W., Ganetzky, B., and Robertson, G. A. (1995) Science 269 92-95 [DOI] [PubMed] [Google Scholar]
- 6.Duggal, P., Vesely, M. R., Wattanasirichaigoon, D., Villafane, J., Kaushik, V., and Beggs, A. H. (1998) Circulation 97 142-146 [DOI] [PubMed] [Google Scholar]
- 7.Splawski, I., Tristani-Firouzi, M., Lehmann, M. H., Sanguinetti, M. C., and Keating, M. T. (1997) Nat. Genet. 17 338-340 [DOI] [PubMed] [Google Scholar]
- 8.Tyson, J., Tranebjaerg, L., Bellman, S., Wren, C., Taylor, J. F., Bathen, J., Aslaksen, B., Sorland, S. J., Lund, O., Malcolm, S., Pembrey, M., Bhattacharya, S., and Bitner-Glindzicz, M. (1997) Hum. Mol. Genet. 6 2179-2185 [DOI] [PubMed] [Google Scholar]
- 9.Roden, D. M., Lazzara, R., Rosen, M., Schwartz, P. J., Towbin, J., and Vincent, G. M. (1996) Circulation 94 1996-2012 [DOI] [PubMed] [Google Scholar]
- 10.Napolitano, C., Schwartz, P. J., Brown, A. M., Ronchetti, E., Bianchi, L., Pinnavaia, A., Acquaro, G., and Priori, S. G. (2000) J. Cardiovasc. Electrophysiol. 11 691-696 [DOI] [PubMed] [Google Scholar]
- 11.Sanguinetti, M. C. (1999) Ann. N. Y. Acad. Sci. 868 406-413 [DOI] [PubMed] [Google Scholar]
- 12.Xu, J., and Li, M. (1998) Trends Cardiovasc. Med. 8 229-234 [DOI] [PubMed] [Google Scholar]
- 13.Abbott, G. W., Sesti, F., Splawski, I., Buck, M. E., Lehmann, M. H., Timothy, K. W., Keating, M. T., and Goldstein, S. A. (1999) Cell 97 175-187 [DOI] [PubMed] [Google Scholar]
- 14.Piccini, M., Vitelli, F., Seri, M., Galietta, L. J., Moran, O., Bulfone, A., Banfi, S., Pober, B., and Renieri, A. (1999) Genomics 60 251-257 [DOI] [PubMed] [Google Scholar]
- 15.Abbott, G. W., and Goldstein, S. A. (2001) Mol. Interv. 1 95-107 [PubMed] [Google Scholar]
- 16.Bendahhou, S., Marionneau, C., Haurogne, K., Larroque, M. M., Derand, R., Szuts, V., Escande, D., Demolombe, S., and Barhanin, J. (2005) Cardiovasc. Res. 67 529-538 [DOI] [PubMed] [Google Scholar]
- 17.Schroeder, B. C., Waldegger, S., Fehr, S., Bleich, M., Warth, R., Greger, R., and Jentsch, T. J. (2000) Nature 403 196-199 [DOI] [PubMed] [Google Scholar]
- 18.Pusch, M. (1998) Pflugers Arch. 437 172-174 [DOI] [PubMed] [Google Scholar]
- 19.Sesti, F., and Goldstein, S. A. (1998) J. Gen. Physiol. 112 651-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang, Y., and Sigworth, F. J. (1998) J. Gen. Physiol. 112 665-678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panaghie, G., Tai, K. K., and Abbott, G. W. (2006) J. Physiol. 570 455-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melman, Y. F., Um, S. Y., Krumerman, A., Kagan, A., and McDonald, T. V. (2004) Neuron 42 927-937 [DOI] [PubMed] [Google Scholar]
- 23.Tapper, A. R., and George, A. L., Jr. (2001) J. Biol. Chem. 276 38249-38254 [DOI] [PubMed] [Google Scholar]
- 24.Blackburn-Munro, G., Dalby-Brown, W., Mirza, N. R., Mikkelsen, J. D., and Blackburn-Munro, R. E. (2005) CNS Drug Rev. 11 1-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fatope, M. O. (2001) Drugs 4 93-98 [PubMed] [Google Scholar]
- 26.Miceli, F., Soldovieri, M. V., Martire, M., and Taglialatela, M. (2008) Curr. Opin. Pharmacol. 8 65-74 [DOI] [PubMed] [Google Scholar]
- 27.Porter, R. J., Nohria, V., and Rundfeldt, C. (2007) Neurotherapeutics 4 149-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porter, R. J., Partiot, A., Sachdeo, R., Nohria, V., and Alves, W. M. (2007) Neurology 68 1197-1204 [DOI] [PubMed] [Google Scholar]
- 29.Peretz, A., Degani, N., Nachman, R., Uziyel, Y., Gibor, G., Shabat, D., and Attali, B. (2005) Mol. Pharmacol. 67 1053-1066 [DOI] [PubMed] [Google Scholar]
- 30.Xiong, Q., Gao, Z., Wang, W., and Li, M. (2008) Trends Pharmacol. Sci. 29 99-107 [DOI] [PubMed] [Google Scholar]
- 31.Schenzer, A., Friedrich, T., Pusch, M., Saftig, P., Jentsch, T. J., Grotzinger, J., and Schwake, M. (2005) J. Neurosci. 25 5051-5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wuttke, T. V., Seebohm, G., Bail, S., Maljevic, S., and Lerche, H. (2005) Mol. Pharmacol. 67 1009-1017 [DOI] [PubMed] [Google Scholar]
- 33.Xiong, Q., Sun, H., Zhang, Y., Nan, F., and Li, M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3128-3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong, Q., Sun, H., and Li, M. (2007) Nat. Chem. Biol. 3 287-296 [DOI] [PubMed] [Google Scholar]
- 35.Akao, M., Ohler, A., O'Rourke, B., and Marban, E. (2001) Circ. Res. 88 1267-1275 [DOI] [PubMed] [Google Scholar]
- 36.Guex, N., and Peitsch, M. C. (1997) Electrophoresis 18 2714-2723 [DOI] [PubMed] [Google Scholar]
- 37.Chouabe, C., Neyroud, N., Richard, P., Denjoy, I., Hainque, B., Romey, G., Drici, M. D., Guicheney, P., and Barhanin, J. (2000) Cardiovasc. Res. 45 971-980 [DOI] [PubMed] [Google Scholar]
- 38.Melman, Y. F., Domenech, A., de la Luna, S., and McDonald, T. V. (2001) J. Biol. Chem. 276 6439-6444 [DOI] [PubMed] [Google Scholar]
- 39.Busch, A. E., Suessbrich, H., Waldegger, S., Sailer, E., Greger, R., Lang, H., Lang, F., Gibson, K. J., and Maylie, J. G. (1996) Pflugers Arch. 432 1094-1096 [DOI] [PubMed] [Google Scholar]
- 40.Seebohm, G., Strutz-Seebohm, N., Ureche, O. N., Baltaev, R., Lampert, A., Kornichuk, G., Kamiya, K., Wuttke, T. V., Lerche, H., Sanguinetti, M. C., and Lang, F. (2006) Biophys. J. 90 2235-2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salata, J. J., Jurkiewicz, N. K., Wang, J., Evans, B. E., Orme, H. T., and Sanguinetti, M. C. (1998) Mol. Pharmacol. 54 220-230 [DOI] [PubMed] [Google Scholar]
- 42.Nilius, B., Benndorf, K., Markwardt, F., and Franke, T. (1987) Gen. Physiol. Biophys. 6 409-424 [PubMed] [Google Scholar]
- 43.Kang, J., Chen, X. L., Wang, H., Ji, J., Cheng, H., Incardona, J., Reynolds, W., Viviani, F., Tabart, M., and Rampe, D. (2005) Mol. Pharmacol. 67 827-836 [DOI] [PubMed] [Google Scholar]
- 44.Jensen, H. S., Grunnet, M., and Olesen, S. P. (2007) Biophys. J. 92 2747-2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grunnet, M., Olesen, S. P., Klaerke, D. A., and Jespersen, T. (2005) Biochem. Biophys. Res. Commun. 328 1146-1153 [DOI] [PubMed] [Google Scholar]
- 46.Seebohm, G., Scherer, C. R., Busch, A. E., and Lerche, C. (2001) J. Biol. Chem. 276 13600-13605 [DOI] [PubMed] [Google Scholar]
- 47.Tristani-Firouzi, M., and Sanguinetti, M. C. (1998) J. Physiol. 510 37-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romey, G., Attali, B., Chouabe, C., Abitbol, I., Guillemare, E., Barhanin, J., and Lazdunski, M. (1997) J. Biol. Chem. 272 16713-16716 [DOI] [PubMed] [Google Scholar]
- 49.Seebohm, G., Pusch, M., Chen, J., and Sanguinetti, M. C. (2003) Circ. Res. 93 941-947 [DOI] [PubMed] [Google Scholar]
- 50.del Camino, D., Holmgren, M., Liu, Y., and Yellen, G. (2000) Nature 403 321-325 [DOI] [PubMed] [Google Scholar]
- 51.Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B. T., and MacKinnon, R. (2002) Nature 417 523-526 [DOI] [PubMed] [Google Scholar]
- 52.Kelly, B. L., and Gross, A. (2003) Nat. Struct. Biol. 10 280-284 [DOI] [PubMed] [Google Scholar]
- 53.Tikhonov, D. B., and Zhorov, B. S. (2004) Biophys. J. 87 1526-1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding, S., Ingleby, L., Ahern, C. A., and Horn, R. (2005) J. Gen. Physiol. 126 213-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bett, G. C., Morales, M. J., Beahm, D. L., Duffey, M. E., and Rasmusson, R. L. (2006) J. Physiol. 576 755-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menuz, K., Stroud, R. M., Nicoll, R. A., and Hays, F. A. (2007) Science 318 815-817 [DOI] [PubMed] [Google Scholar]
- 57.Wang, H. S., Brown, B. S., McKinnon, D., and Cohen, I. S. (2000) Mol. Pharmacol. 57 1218-1223 [PubMed] [Google Scholar]
- 58.Kerst, G., Brousos, H., Schreiber, R., Nitschke, R., Hug, M. J., Greger, R., and Bleich, M. (2002) J. Membr. Biol. 186 89-100 [DOI] [PubMed] [Google Scholar]
- 59.Morin, T. J., and Kobertz, W. R. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1478-1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manderfield, L. J., and George, A. L., Jr. (2008) FEBS J. 275 1336-1349 [DOI] [PubMed] [Google Scholar]