Abstract
The pathophysiology underlying mitochondrial dysfunction in insulin-resistant skeletal muscle is incompletely characterized. To further delineate this we investigated the interaction between insulin signaling, mitochondrial regulation, and function in C2C12 myotubes and in skeletal muscle. In myotubes elevated insulin and glucose disrupt insulin signaling, mitochondrial biogenesis, and mitochondrial bioenergetics. The insulin-sensitizing thiazolidinedione pioglitazone restores these perturbations in parallel with induction of the mitochondrial biogenesis regulator PGC-1α. Overexpression of PGC-1α rescues insulin signaling and mitochondrial bioenergetics, and its silencing concordantly disrupts insulin signaling and mitochondrial bioenergetics. In primary skeletal myoblasts pioglitazone also up-regulates PGC-1α expression and restores the insulin-resistant mitochondrial bioenergetic profile. In parallel, pioglitazone up-regulates PGC-1α in db/db mouse skeletal muscle. Interestingly, the small interfering RNA knockdown of the insulin receptor in C2C12 myotubes down-regulates PGC-1α and attenuates mitochondrial bioenergetics. Concordantly, mitochondrial bioenergetics are blunted in insulin receptor knock-out mouse-derived skeletal myoblasts. Taken together these data demonstrate that elevated glucose and insulin impairs and pioglitazone restores skeletal myotube insulin signaling, mitochondrial regulation, and bioenergetics. Pioglitazone functions in part via the induction of PGC-1α. Moreover, PGC-1α is identified as a bidirectional regulatory link integrating insulin-signaling and mitochondrial homeostasis in skeletal muscle.
Understanding the pathophysiology initiating the development of insulin resistance should augment our capacity to identify novel therapeutic targets for the prevention and treatment of type 2 diabetes. This biology remains incompletely characterized in part due to the complexity of the interaction of multiple organ systems and the multiplicity of intracellular perturbations within these organs governing the development of insulin resistance. The major peripheral organ systems implicated in insulin resistance include skeletal muscle, adipose tissue, liver, and the immune system. The enhancement of our understanding of this biology will require the combination of reductionist and systems biological approaches.
The complexity of the effects of insulin resistance in a single organ is exemplified in skeletal muscle where disruption in glucose uptake (1), insulin signaling (2, 3), glycogen synthesis (4) and in mitochondrial biology (5–7) are evident in insulin-resistant subjects up to two decades before their developing diabetes. A fundamental question arising is whether disruption of skeletal muscle mitochondria is a primary component in this disease pathophysiology or whether it is a consequence of reduced aerobic activity in response to alternate metabolic perturbations associated with insulin resistance and diabetes (8). Although this question has not been definitively answered, increasing mitochondrial electron flux via skeletal muscle-restricted overexpression of uncoupling protein homologues blunts diet-induced insulin resistance (9) and reverses diabetes (10). In skeletal myotubes this modest uncoupling has been shown to augment mitochondrial fatty acid oxidation (11). Taken together these data suggest that enhancing skeletal muscle mitochondrial metabolic activity would be an attractive strategy to prevent or reverse insulin resistance.
Interestingly, the thiazolidinedione class of insulin sensitizers do induce mitochondrial oxygen consumption and up-regulate the mitochondrial biogenesis program in adipose tissue (12, 13). Whether these effects are operational in skeletal muscle are less certain (13–15), and if present, whether these changes are linked to insulin signaling is unknown. The objectives of this study were to determine whether the thiazolidinedione pioglitazone modulates the mitochondrial biogenesis program and mitochondrial respiration and to delineate the link between insulin signaling and mitochondrial biology in insulin-resistant skeletal muscle.
Insulin resistance was evoked by exposing murine C2C12 myotubes to high glucose and insulin levels. These cells showed disruption in insulin-mediated signaling in the mitochondrial regulatory programs and in mitochondrial bioenergetics. Pioglitazone restored insulin signaling and the mitochondrial regulatory and bioenergetic phenotype. Affymetrix gene array analysis shows a disproportionate pioglitazone-mediated induction of genes encoding for mitochondrion-functioning proteins. Furthermore, the master regulator of mitochondrial biogenesis, peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α)2 was up-regulated by pioglitazone. We explored the role of PGC-1α in the interaction between insulin signaling and mitochondrial biology. PGC-1α overexpression significantly augments insulin-mediated signaling in parallel with rescuing mitochondrial biology in insulin-resistant myotubes. Depletion of PGC-1α attenuates insulin signal transduction under control conditions. Moreover, the direct genetic disruption of the insulin receptor results in the down-regulation of PGC-1α and reduced mitochondrial bioenergetics. These data show that PGC-1α is required to maintain insulin-mediated signal transduction in C2C12 myotubes and that its induction is sufficient to restore the integrity of insulin-mediated signaling and mitochondrial function under insulin-resistant conditions. In primary myoblasts pioglitazone restores the insulin resistance-mediated blunting of mitochondrial bioenergetics. Furthermore, the integration of insulin signaling and mitochondrial functioning is further supported where insulin receptor knock-out mouse primary skeletal myoblasts show diminished mitochondrial bioenergetic capacity. Together, these data show a direct link between insulin signaling and mitochondrial integrity and implicate PGC-1α as a bidirectional regulator coordinating signal transduction with organelle homeostasis in the pathophysiology of insulin resistance. Pioglitazone functions in part through the concordant restoration of insulin signaling and of the associated mitochondrial deficits.
EXPERIMENTAL PROCEDURES
Cell Culture—C2C12 cells (ATCC, Manassas, VA) were differentiated into myotubes by serum depletion. Insulin receptor knock-out and wild type skeletal myoblasts were harvested from 1–3-day-old neonatal pups using heterozygous B6:129S4-Insrtm1Dac/J mice from The Jackson Laboratory, Bar Harbor, ME. The pups were phenotyped by measuring their glucose levels and genotyped according to protocol (16). Primary neonatal muscle cells were harvested and cultured as previously described (17). The insulin-resistant phenotype was evoked by exposure to high levels of glucose (40 mm) and insulin (100 nm, Sigma) for 2–3 days. 50 μm Pioglitazone (Axxora, San Diego, CA) or vehicle control was added 24 h before harvesting in selected studies.
Mouse Studies—Seven-week-old male db/db mice (C57BL/6J strain; The Jackson Laboratory) were treated with pioglitazone (30 mg/kg body weight, once daily, n = 8) or 0.5% carboxymethyl cellulose (0.1 ml/10 g body weight as vehicle, n = 8) by oral gavage for 2 weeks. Blood glucose levels were measured after overnight fasting before and at the end of the treatment period. After 14 days of treatment mice were sacrificed, and skeletal muscle was harvested from quadriceps muscle and snap-frozen in liquid nitrogen. Protein was extracted using radioimmune precipitation assay buffer, and Western blot analysis was performed as described below using an anti-PGC-1α antibody (a gift from D. Kelly, Washington University, St. Louis, MO). Age, strain, and diet-matched C57BL/6J controls were euthanized at the same time point for a comparison of skeletal muscle PGC-1α levels. Animal and primary muscle culture experiments were approved by the NHLBI Animal Care and Use Committee.
Protein Analysis—Protein samples were analyzed by Western blot analysis. Membranes were incubated with specific antibodies directed against total protein or phosphorylated protein. Proteins of interest included subunits I and III of cytochrome oxidase (COX I, MitoSciences, Eugene, OR; COX III, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), ND6 (MitoSciences), myogenin (BD Biosciences), cytochrome c, Akt, p44/p42 mitogen-activated protein kinase (ERK), insulin receptor substrate 1 (IRS-1; Cell Signaling, Danvers, MA), voltage-dependent anion channel (Molecular Probes, Eugene, OR/Invitrogen), insulin receptor subunit β (InsRβ), SIRT1 (Upstate/Millipore, Charlottesville, VA), PGC-1α (a gift from D. Kelly), and β-actin (Ambion, Austin, TX). Blots were developed using SuperSignal West Chemiluminescence kit (Pierce).
Mitochondrial DNA Copy Measurement—Mitochondrial DNA copy number was assessed as described previously (14). In brief, DNA was collected from whole cell lysates. Real-time PCR was performed for COX I and for the nuclear encoded 18 S. The ratio of COX I DNA copies to 18 S represents the relative mitochondrial copy number.
Gene Expression Analysis—After isolation and quantification of RNA from C2C12 myotubes and wild type primary skeletal myoblasts, the reverse transcription reaction was performed using SuperScript III reverse transcriptase kit (Invitrogen). Real time quantitative-PCR was performed using SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA) and MJ Research DNA Engine Opticon 2 fluorescence detection system (Bio-Rad). Sequences of specific primers are listed in supplemental Table 1. The mRNA concentration was calculated from the cycle threshold values using a standard curve and normalized to the expression levels of 18 S ribosomal RNA.
Gene Array—After RNA isolation (RNeasy kit, Qiagen, Valencia, CA), RNA amplification was performed using the RiboAmp OA 1 Round kit (Arcturus, Sunnyvale, CA) followed by in vitro transcription and transcript labeling (ENZO Bioarray RNA transcript labeling kit, ENZO Life Sciences, Farmingdale, NY). cRNA purification and fragmentation of the purified cRNA were performed using GeneChip Sample Module (Qiagen). Microarray chips (MOE430A, Affymetrix, Santa Clara, CA) were incubated with the fragmented RNA samples. Images were processed using Micro Array Suite software (Affymetrix). Data were analyzed using Ingenuity software.
Flow Cytometry—Mitochondrial size and inner mitochondrial membrane potential were determined using MitoTracker green FM (100 nm) and the mitochondria specific dye tetramethylrhodamine methyl ester (25 nm), respectively (Molecular Probes). Briefly, C2C12 myotubes or primary myoblasts were harvested and incubated with the dye for 30 min at 37 °C and analyzed using the FACSCalibur (BD Biosciences) and Cellquest Software (Becton Dickinson Immunocytometry Systems).
Oxygraphy—C2C12 myotubes were harvested and resuspended in complete medium. Whole cell respiration was measured using the BD Oxygen Biosensor System (BD Biosciences). Total oxygen consumption was measured concurrently comparing the three groups, measured as relative change in fluorescence as described previously (12).
ATP Content—Cells were incubated in ice-cold lysis buffer (20 mm Tris pH 7.0, 0.5% Nonidet P-40, 25 mm NaCl, 2.5 mm EDTA). Cellular ATP content was measured spectrophotometrically in cell lysates with a specific luminescence kit using firefly luciferase and D-luciferin (Molecular Probes).
Adenoviral Transduction—Adenoviral vectors encoding for full-length mouse PGC-1α Ad-Track-FLAG-HA-PGC-1α) and SIRT1 (Ad-Track-FLAG-HA-SIRT1), respectively, were described previously and kindly provided by P. Puigserver (Harvard Medical School, Boston, MA). Adeno-GFP that was used as control for adenoviral infection experiments was kindly provided by K. Walsh (Boston University School of Medicine, Boston, MA).
siRNA Transfection Experiments—C2C12 myotubes were transfected with indicated amounts of SMARTpool® siRNA targeting PGC-1α or InsRβ (Dharmacon, Chicago, IL) using Oligofectamine (Invitrogen) following the manufacturers' protocol and harvested 3 days after transfection.
Acute Insulin Stimulation and Immunoprecipitation—Cells were stimulated with 100 nm insulin for 5 or 10 min at 37 °C, and protein was extracted using radioimmune precipitation assay buffer containing protease inhibitor mix (Roche Diagnostics) and NaF (1 mm), Na3VO4 (0.1 mm), β-glycerophosphate (100 μm, Sigma). 500 μg of protein was incubated with 4 μl of anti-IRS antibody (Cell signaling), and 1 mg of protein was incubated with anti-InsRβ antibody (H-70, Santa Cruz) in radioimmune precipitation assay buffer overnight. 20 μl of protein A/G Plus Agarose (Santa Cruz Biotechnology) was added for 2 h, and agarose beads were washed in radioimmune precipitation assay buffer and boiled in sample loading buffer. The supernatant was used for electrophoresis and Western blot. Phosphorylated IRS-1 and phosphorylated InsRβ were detected using an anti-phosphotyrosine antibody (Upstate/Millipore) and horseradish peroxidase-conjugated secondary anti-mouse antibody, and bands were visualized using SuperSignal West chemiluminescence substrate kit (Pierce).
Promoter Analysis—The murine 2-kilobase PGC-1α promoter-luciferase reporter construct was purchased from Add-gene (Cambridge, MA). C2C12 myoblasts were transfected with PGC-1α-promoter/reporter plasmids using FuGENE 6 transfection reagent (Roche Diagnostics). pRL-SV40/control plasmid (Promega, Madison, WI) was co-transfected as a control for the transfection efficiency. Cells were incubated overnight and were further incubated for 24 h with 50 μm pioglitazone or vehicle. PGC-1α promoter activity was determined using Dual Luciferase Reporter assay system (Promega) as relative firefly luciferase luminescence normalized to the renilla luciferase luminescence according to the manufacturer's instruction.
Statistical Analysis—Differences between data groups were evaluated for significance using Student t test or one-way analysis of variance. p < 0.05 was considered statistically significant, and data are expressed as mean ± S.E. The asterisk denotes p < 0.05 versus control. Gene array expression levels were analyzed after segregation to exclude genes probesets employing a false discovery rate of >10% and a -fold change <1.5. A Fisher's exact test was employed to evaluate whether significantly regulated genes encoding mitochondrial proteins are disproportionately represented relative to the total number of probesets on the microarray chips. n ≥ 4 repeated experiments were performed in all studies.
RESULTS
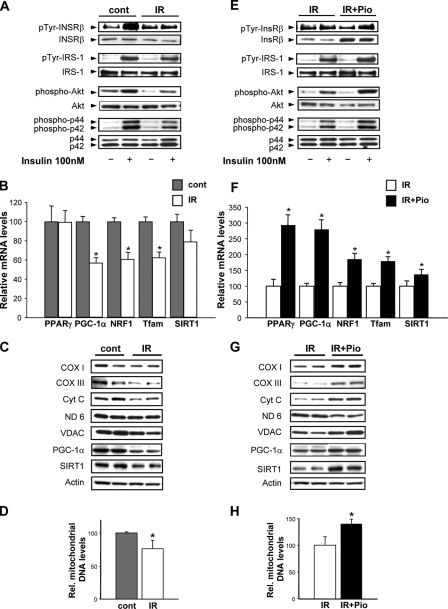
C2C12 myotubes exposed to elevated glucose and insulin were used to characterize the interaction between insulin-signaling and mitochondrial regulation and function. To validate insulin resistance, insulin-activated phosphorylation of the InsRβ and of downstream kinases including tyrosine phosphorylation of IRS-1 and phosphorylation of Akt and of ERK isoforms were compared with non-insulin-stimulated controls. The insulin receptor and the downstream kinases were induced >4-fold in control cells, and this induction is significantly blunted in the insulin resistance cells (Fig. 1A). To delineate whether elevated glucose and insulin perturb mitochondrial regulation, we examined the mitochondrial biogenesis regulatory program. The transcriptional regulators of the intergenomic control of mitochondrial biogenesis, including nuclear respiratory factor 1 and transcription factor A of mitochondria, were down-regulated ∼40% in insulin-resistant cells. This was mirrored by the down-regulation of PGC-1α (Fig. 1B). We measured the transcript and protein levels of SIRT1, the NAD+-dependent deacetylase known to activate PGC-1α in skeletal muscle (18). SIRT1 gene expression is unchanged, but SIRT1 protein levels is reduced ∼38% in insulin-resistant cells (Fig. 1, B and C, respectively). In parallel, the steady-state levels of mitochondrial electron transfer chain proteins including the nuclear-encoded cytochrome c and the mitochondrial-encoded cytochrome oxidase subunits I and III proteins are significantly diminished in parallel with the reduction in PGC-1α under insulin-resistant conditions (Fig. 1C). Interestingly, the NADH dehydrogenase subunit 6 (ND6 mitochondrial-encoded protein) is not modulated at the protein level in insulin-resistant myotubes. The expression of the outer mitochondrial membrane voltage-dependent anion channel is also significantly reduced in insulin-resistant cells (Fig. 1C). To establish whether this down-regulation is reflected at the mitochondrial genomic level, we quantified the relative mitochondrial versus genomic DNA copy number. In insulin-resistant myotubes the relative mitochondrial DNA copy number is reduced ∼24% (Fig. 1D). These data are supported by the diminution in the mitochondrial mass of ∼29% (Fig. 2A).
FIGURE 1.
Insulin signaling and mitochondrial phenotype in response to insulin resistance and pioglitazone. Control myotubes were exposed to 25 mm glucose, and the insulin resistant myotubes were exposed to 40 mm glucose and 100 nm insulin for 48 h. A represents insulin-responsive signaling as shown by the representative Western blots (pTyr, phosphotyrosine; cont, control; IR, insulin resistance). B, transcript levels, expressed as % control, of genes encoding for mitochondrial regulatory proteins (PPARγ, peroxisome proliferator-activated receptor γ; NRF1, nuclear respiratory factor 1; Tfam, mitochondrial transcription factor A and SIRT1. C, steady-state mitochondrial electron transfer chain protein levels (COX I and III, cytochrome c oxidase subunits I and III; Cyt c, cytochrome c; ND 6, NADH dehydrogenase, subunit 6; VDAC, voltage-dependent anion channel) as well as levels of voltage-activated anion channel, SIRT1, and actin as a housekeeping protein; D, relative mitochondrial genomic copy number (% control). E shows insulin-responsive signaling comparing insulin-resistant to pioglitazone (Pio)-treated insulin-resistant myotubes. F, transcript levels, expressed as % control, of genes encoding for mitochondrial regulatory proteins. G, steady-state mitochondrial electron transfer chain protein levels comparing insulin-resistant to insulin-resistant myotubes exposed to pioglitazone. H, relative mitochondrial genomic copy number (% control). The asterisk represents a p < 0.05 versus the respective control in this and all subsequent figures (unless otherwise stated).
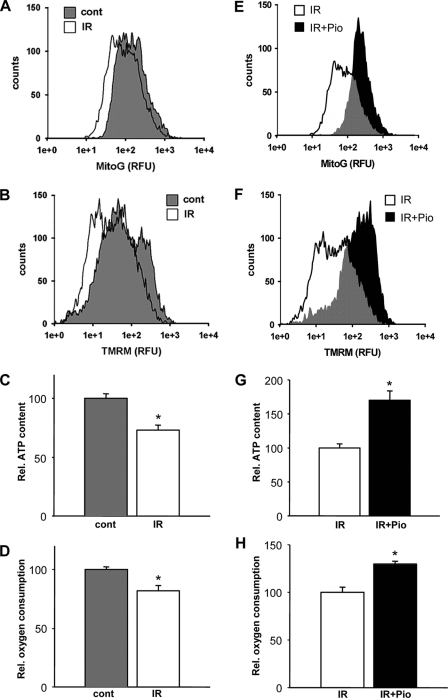
FIGURE 2.
Insulin resistance and pioglitazone modulation of mitochondrial biology. The mitochondrial bioenergetic profile was compared between control and insulin-resistant myotubes and to the administration of pioglitazone. Results are representative cytometric profile of MitoTracker green, which represents that relative mitochondrial size (A), and tetramethylrhodamine methyl ester (TMRM), which represents the relative mitochondrial membrane potential (% control) (B) comparing control to insulin-resistant cells. C, total cellular ATP levels. D, the relative rate of oxygen consumption (% control). E, comparing insulin resistant cells to those exposed to pioglitazone (Pio) showing a representative cytometric profile of MitoTracker green fluorescence depicting the relative mitochondrial size. F, tetramethylrhodamine methyl ester represents the relative mitochondrial membrane potential (% insulin-resistant controls). G, total cellular ATP levels. H, relative rate of oxygen consumption (% insulin-resistant controls). RFU, relative fluorescent units.
The thiazolidinedione class of anti-diabetic agents function as peroxisome proliferator-activated receptor ligands and improves overall systemic insulin sensitivity. We determined whether pioglitazone would concurrently modulate insulin signaling and the mitochondrial regulatory program. Pioglitazone restores InsRβ, IRS-1, Akt, and ERK phosphorylation in the insulin-resistant myotubes and increases InsRβ levels (Fig. 1E). In parallel, pioglitazone augments the expression levels of genes encoding for mitochondrial biogenesis regulatory proteins (Fig. 1F), the steady-state electron transfer chain protein levels (Fig. 1G), the mitochondrial genomic copy number (Fig. 1H), and mitochondrial mass (Fig. 2E).
To test the functional consequences of these regulatory events during insulin resistance and the following the administration of pioglitazone, we measured the bioenergetic profile in these myotubes. Insulin-resistant myotubes exhibit an ∼26% reduction in the mitochondrial energetic capacity as measured by the relative mitochondrial membrane potential (Fig. 2B), a ∼28% reduction in total cellular ATP content (Fig. 2C), and a ∼33% reduction in the rate of cellular oxygen consumption (Fig. 2D) compared with control myotubes. Pioglitazone enhances the relative mitochondrial membrane potential, oxygen consumption, and ATP production in insulin-resistant cells (Figs. 2, F–H). Interestingly, the induction of this mitochondrial regulatory program by pioglitazone is not exclusively dependent on the insulin-resistant milieu in that the induction of the mitochondrial biogenesis program and its sequelae was also seen, albeit to varying degrees in the control cells (supplemental Fig. 1). Pioglitazone did not enhance insulin-induced signaling in the control cells (data not shown).
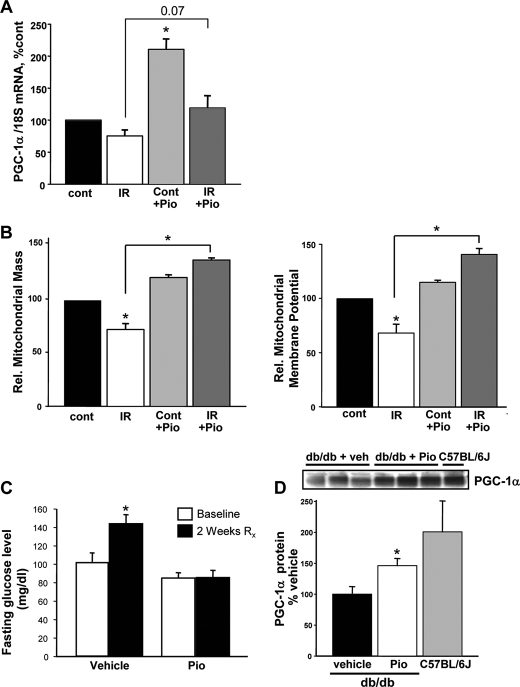
Murine skeletal myoblasts were then employed to confirm whether pioglitazone replicates this ameliorative mitochondrial profile in primary cells. Under basal conditions, pioglitazone significantly up-regulates PGC-1α gene expression (Fig. 3A) in parallel with the induction seen in C2C12 myotubes. In response to insulin resistance, the mRNA expression of PGC-1α is modestly induced above base line in response to pioglitazone (Fig. 3A). The measurement of mitochondrial mass and membrane potential directly parallels the findings shown in C2C12 myotubes with a diminution in response to insulin resistance and complete restoration after pioglitazone administration (Fig. 3B). The ability of pioglitazone to induce PGC-1α in skeletal muscle was additionally confirmed in db/db mice. These mice are deficient in the leptin receptor and develop progressive obesity and diabetes with the onset of elevated glucose levels at around 8 weeks of age. Seven-week-old mice were treated with pioglitazone or vehicle control for 2 weeks and then sacrificed for skeletal muscle extraction. The vehicle-treated mice showed a significant rise in blood glucose over this period. In contrast, the pioglitazone-treated db/db mice maintained their base-line fasting blood sugar levels (Fig. 3C). In parallel, the steady-state PGC-1α protein levels is significantly elevated in the pioglitazone mice compared with the vehicle-treated control mice (Fig. 3D). It is of interest that the db/db mice developing hyperglycemia have lower skeletal muscle PGC-1α levels compared with strain-matched C57BL/6J control mice.
FIGURE 3.
Pioglitazone restores the mitochondrial bioenergetic profile in primary myocytes and in the db/db mouse. A, pioglitazone (Pio) up-regulates the expression of the gene encoding for PGC-1α in primary myoblasts. B, pioglitazone reverses the diminution in mitochondrial size and membrane potential evoked by insulin resistance in primary skeletal myoblasts. C, pioglitazone prevents the development of fasting hyperglycemia in db/db mice. D, pioglitazone up-regulates PGC-1α steady-state protein levels in db/db mice. The levels of PGC-1α were diminished in untreated db/db mice compared with strain-matched C57BL/6J control mice.
To explore the regulatory role of pioglitazone in modulating these bioenergetic changes in insulin-resistant C2C12 cells, we compared the expression profiles of insulin-resistant and pioglitazone-treated insulin-resistant myotubes using Affymetrix-gene array analysis. Probeset expression of genes designated to have mitochondrial functions, as defined by the terms mitochondria, mitochondrion, or mitochondrial in the gene ontology data base of Entrez Gene elicited 921 mitochondrial functioning probesets of a total of 22,689, with 253 probesets associated with mitochondrial biogenesis. Using a 10% false discovery rate and a cutoff of 1.5-fold induction elucidated a total of 436 pioglitazone regulated probesets of which 50 were mitochondrial-related and 21 associated with mitochondrial biogenesis. This results in a highly significant and disproportionate up-regulation of genes supporting mitochondrial function by pioglitazone. The pioglitazone-regulated genes are shown in supplemental Table 2.
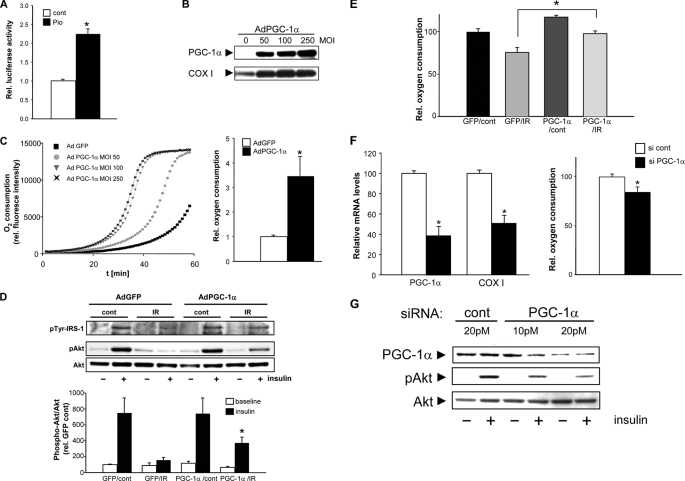
As PGC-1α was down-regulated in insulin-resistant myotubes and induced by pioglitazone, we determined whether pioglitazone directly transactivates PGC-1α. Using the mouse PGC-1α promoter-luciferase construct, we show that pioglitazone significantly transactivates PGC-1α in C2C12 cells (Fig. 4A). To evaluate whether this up-regulation of PGC-1α could modulate insulin signal transduction in parallel to the up-regulation of the mitochondrial biogenesis program, we employed an adenoviral expression vector harboring the murine PGC-1α cDNA. Infection efficiency was in excess of 80%, and the infection with the PGC-1α construct resulted in a significant induction of the PGC-1α protein levels (Fig. 4B). This augmentation resulted in an increase of mitochondrial respiration in insulin-resistant myotubes in a multiplicity of infection dose-dependent manner as shown by oxygen consumption (Fig. 4C). To test the effect of this induction on insulin signaling, the threonine phosphorylation of Akt was measured in response to GFP versus PGC-1α infection. GFP vector infection had no restorative effect on Akt phosphorylation, but PGC-1α overexpression significantly increases phosphorylation under insulin-resistant conditions (Figs. 4D) and concomitantly restored the rate of oxygen consumption (Fig. 4E). To test the converse effect of PGC-1α, we knocked down PGC-1α levels in the C2C12 cells using siRNA and assessed the insulin responsiveness of Akt. PGC-1α knockdown led to a significant reduction of the mRNA levels of mitochondrial-encoded cytochrome c oxidase subunit I levels and lowered oxygen consumption rate (Fig. 4F). In addition, progressive PGC-1α knockdown resulted in the incremental diminution of insulin-stimulated Akt phosphorylation (Fig. 4G). Together these data support a role for PGC-1α in the integrated regulation of both mitochondrial biogenesis and insulin signal transduction that appears to be operational under basal and insulin-resistant conditions. Interestingly, overexpression of SIRT1, a known post-translational activator of PGC-1α, partially normalizes the blunted insulin response in insulin-resistant myotubes, and this ameliorative effect is further enhanced with the concurrent transduction of PGC-1α (supplemental Fig. 2).
FIGURE 4.
PGC-1α modulates mitochondrial biology and insulin signaling. A shows murine PGC-1α promoter activation in response to pioglitazone (cont, control; Pio, pioglitazone). B, infection of PGC-1α up-regulates PGC-1α and COX I protein levels. MOI, multiplicity of infection. C, augmentation of relative rate of oxygen consumption in cells with increasing levels of PGC-1α and the relative O2 consumption comparing GFP to PGC-1α vector at a multiplicity of infection of 100. D shows the restoration of insulin-mediated signaling (% GFP/cont without insulin) by PGC-1α overexpression (pTyr, phosphotyrosine; cont, control). E, induction of oxygen consumption by PGC1α overexpression in, insulin resistance myotubes. F, effect of PGC-1α silencing on transcript levels and on oxygen consumption (% si cont); G, the diminution of Akt threonine phosphorylation by insulin after partial silencing of PGC-1α.
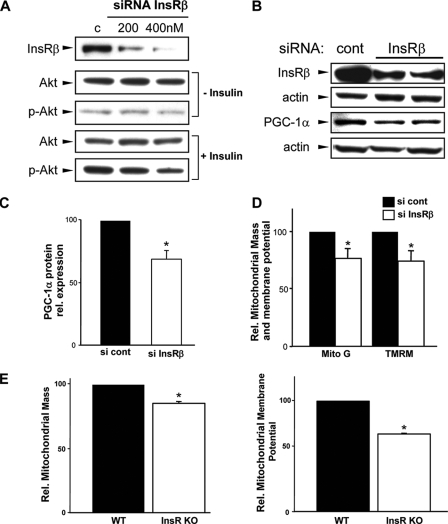
To further analyze the specificity of PGC-1α in linking the aberrations in insulin signaling and mitochondrial dysfunction, we investigated PGC-1α and mitochondrial bioenergetics in response to the knockdown of the insulin receptor. The siRNA knockdown of InsRβ resulted in desensitization to insulin-mediated activation of Akt (Fig. 5A). This blunting of insulin signal transduction is associated with a significant reduction in the steady-state protein levels of PGC-1α and of mitochondrial bioenergetic function (Fig. 5, B–D). Finally, as the siRNA analysis in C2C12 myotubes demonstrates a direct link between disruption in insulin receptor signaling and mitochondrial function, we determined whether this mitochondrial bioenergetic profile is perturbed in skeletal myoblasts harvested from insulin receptor knock-out mice. The lack of insulin receptor leads to neonatal lethality in the first 5 days of life (16). To preempt this, neonatal mice were sacrificed on day 2 to prepare primary skeletal myoblast cultures. At the time of euthanasia the blood glucose levels averaged 51 ± 12 mg/dl in the wild type control pups and 469 ± 38 mg/dl in the knock-out animals (p = 0.0005). As shown in Fig. 5E, the mitochondrial mass and membrane potential in these insulin receptor knock-out myoblasts are significantly reduced compared with the wild type controls and show the same profile evident in insulin-resistant C2C12 myotubes.
FIGURE 5.
Disruption of insulin signaling and mitochondrial respiration. A, effect of knockdown of InsRβ on Akt phosphorylation (p-Akt). B, PGC-1α levels in response to knockdown of InsRβ. C, quantitative reduction of PGC-1α protein after InsRβ knockdown. D, attenuation in mitochondrial size and membrane potential in insulin receptor knockdown cells. TMRM, tetramethylrhodamine methyl ester; Mito G, MitoTracker green. E, in insulin receptor knock-out (KO) skeletal myoblasts the mitochondrial membrane potential and mitochondrial mass is diminished compared with wild type (WT) control cells.
DISCUSSION
Pioglitazone and rosiglitazone, the two clinically used thiazolidinediones, activate mitochondrial biogenesis in adipocytes (12, 19). This is proposed to function, in part, through the direct induction of PGC-1α via a peroxisome proliferator-activated receptor γ response element within the PGC-1α promoter region (20). In contrast, in skeletal muscle the capacity of rosiglitazone to restore insulin signaling and mitochondrial biogenesis has been questioned (14, 21). This may be due in part to the inability of rosiglitazone to activate the PGC-1α promoter-luciferase construct in skeletal myotubes (22). As pioglitazone has distinct metabolic effects compared with rosiglitazone (23), we investigated the role of this thiazolidinedione on skeletal muscle mitochondrial biogenesis and on insulin sensitivity. We show that pioglitazone activates the murine PGC-1α promoter-reporter construct in C2C12 myotubes, up-regulates the gene encoding for PGC-1α in C2C12 and in primary skeletal myoblasts, and increases PGC-1α protein levels in quadriceps muscle of db/db mice. Furthermore, we demonstrate that the administration of pioglitazone restores insulin signaling, augments the mitochondrial biogenesis program, and restores the mitochondrial bioenergetic capacity in insulin-resistant myotubes. In parallel with these changes, pioglitazone modulates genes encoding for additional mitochondrial and cellular functioning proteins in insulin-resistant myotubes.
We focused our further investigations on the role of PGC-1α in integrating mitochondrial biology and insulin signaling. As background, prior in vivo studies have been undertaken to explore the role of PGC-1α in skeletal muscle. Skeletal muscle-targeted deletion of PGC-1α results in fiber-type switching with a reduction in slow-twitch oxidative fibers and in exercise intolerance (24). In keeping with the dominant role of skeletal muscle for glucose uptake, these mice exhibit impaired post-prandial glucose uptake (25). Surprisingly, despite this tissue-targeted PGC-1α depletion, systemic consequences are manifest in that peripheral insulin sensitivity was maintained and this skeletal muscle perturbation initiated cross-talk with the pancreatic β-cells, resulting in the development of β-cell hyperplasia (25). Conversely, short-term induction of PGC-1α in an inducible system results in enhanced glucose uptake, glycogen storage, and in the induction of genes encoding for mitochondrial oxidative phosphorylation (26). In contrast, chronic robust skeletal muscle PGC-1α activation to prevent or reverse insulin resistance is unlikely in that the skeletal muscle-specific overexpression of PGC-1α resulted in impaired glucose tolerance and a reduction in skeletal muscle GLUT4 expression (27). Collectively, these in vivo studies do establish a primary role of PGC-1α in maintaining mitochondrial homeostasis and glucose tolerance but also demonstrate that the robust activation of this regulatory protein is unlikely to be an optimal therapeutic strategy. Our data show that the pharmacologic and modest induction of PGC-1α is feasible and concordantly restores insulin-mediated signal transduction and mitochondrial regulatory and bioenergetic function. Of note, the concept that modest PGC-1α induction is beneficial has recently also been confirmed where modest induction of PGC-1α (∼25%) in an isolated rat muscle enhanced both insulin sensitivity and palmitate oxidation (28).
Of further interest in this study we find that PGC-1α functions as a bidirectional regulatory link between insulin signaling and mitochondrial homeostasis. We demonstrate that siRNA knockdown of PGC-1α disrupts insulin-mediated signaling and mitochondrial function and that the overexpression of this transcriptional coactivator under insulin-resistant conditions is sufficient to restore both mitochondrial respiratory function and to partially rescue insulin-mediated signaling. The bidirectional control of PGC-1α between insulin signaling and mitochondrial biology is further supported by the down-regulation of PGC-1α protein and a diminution of mitochondrial bioenergetics after siRNA knockdown of the insulin receptor β subunit. This diminution in mitochondrial mass and membrane potential is also shown in primary skeletal myoblasts lacking the insulin receptor. Together these data implicate a central role for PGC-1α in integrating insulin-responsive signaling and mitochondrial homeostasis in skeletal muscle. Our data further support the strong association between the disruption in skeletal muscle mitochondrial dysfunction and the development of insulin resistance (6, 29) and the association with concordant down-regulation of PGC-1α-responsive genes in type 2 diabetes (30) and of PGC-1α itself in skeletal muscle from subjects with polycystic ovary syndrome-associated insulin resistance (31).
As shown by the adverse phenotype following robust overexpression of PGC-1α in skeletal muscle (27), a modest induction of this regulatory protein would be optimal for therapeutic efficacy to enhance skeletal muscle insulin sensitivity. Although pioglitazone does modestly induce PGC-1α, the long term clinical use of the thiazolidinediones is questionable as their therapeutic potential has recently been assuaged by evidence linking these compounds to systemic adverse effects (32, 33). Our study shows that pioglitazone activates SIRT1, and this functions in concert with PGC-1α to restore mitochondrial biology and insulin signaling. Hence, the activation or up-regulation of SIRT1 may be a more amenable target for therapeutic manipulation. The therapeutic potential for SIRT1 activation may soon be realized in that numerous small molecule activators of SIRT1 have been identified, and these compounds have been shown to improve glucose tolerance in rodent models of diabetes in parallel with the induction of skeletal muscle mitochondrial content as measured by their capacity to up-regulate citrate synthase activity (34). Likewise, resveratrol, which activates SIRT1, AMP kinase, and mitochondrial biogenesis have been shown to attenuate age and high fat diet-induced insulin resistance (35, 36). From a mechanistic perspective SIRT1 has also been shown to enhance insulin sensitivity in insulin-resistant myotubes via repression of the protein-tyrosine phosphatase 1B (37). Recently, small molecule activators of PGC-1α itself has been described (38). These compounds need to be explored as alternative agents to modify this program in skeletal muscle and to evaluate whether they too can restore insulin signaling.
The interrelationship between perturbed insulin signaling and mitochondrial dysfunction in skeletal muscle insulin resistance has been shown in insulin-resistant off-spring of diabetic parents (3). However, the regulatory link between these two processes has not been extensively explored. Our data support that PGC-1α functions to integrate the insulin signaling pathway and the regulation of mitochondrial biology. The mechanism whereby the diminution in insulin signaling down-regulates PGC-1α does not appear to have been explored. In contrast, the mechanism whereby PGC-1α activation restores insulin signaling may speculatively result from the induction in mitochondrial metabolism with a reduction in diacylglycerol accumulation. Diacylglycerol has been shown to inactivate Akt via diacylglycerol-activated protein kinase serine phosphorylation of Akt (39). Insulin signaling is also perturbed by excess reactive oxygen species (40). PGC-1α has been shown to up-regulate anti-oxidant defense systems in skeletal myotubes (41), and a second possible mechanism whereby the reduction in PGC-1α may disrupt insulin signaling may be via regulation of reactive oxygen species homeostasis. The investigation of both of these mechanisms warrants investigation.
In summary, in this study we show that elevated insulin and glucose levels in the absence of exogenous manipulation of free fatty acid levels disrupts insulin signal transduction, the mitochondrial biogenesis regulatory program, and mitochondrial respiratory function. The thiazolidinedione pioglitazone can restore these programs and is shown to directly transactivate PGC-1α and increase the steady-state protein levels of SIRT1. The necessity and requirement of PGC-1α to integrate insulin signaling and mitochondrial biogenesis and function in skeletal myotubes is evident whereby overexpression of PGC-1α can restore insulin signaling and the mitochondrial bioenergetic phenotype, and its genetic knockdown has the opposite phenotype. Furthermore, knockdown of the insulin receptor itself is sufficient to down-regulate PGC-1α and disrupt mitochondrial bioenergetic function. This study supports a central role of PGC-1α in the pathophysiology of insulin resistance and suggests that this transcriptional coactivator has a novel role in the bidirectional integration of insulin signaling and mitochondrial homeostasis.
Supplementary Material
Acknowledgments
We thank J. Philip McCoy of the Flow cytometry core for assistance and expertise and the NHLBI Laboratory of Animal Medicine and Surgery for expertise and support.
This work was supported, in whole or in part, by the National Institutes of Health (NHLBI Division of Intramural Research). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Tables 1 and 2.
Footnotes
The abbreviations used are: PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; siRNA, small interfering RNA; COX, cytochrome oxidase; ERK, extracellular signal-regulated kinase; IR, insulin resistance; IRS-1, insulin receptor substrate 1; InsRβ, insulin receptor subunit β; GFP, green fluorescent protein.
References
- 1.Karlsson, H. K., Ahlsen, M., Zierath, J. R., Wallberg-Henriksson, H., and Koistinen, H. A. (2006) Diabetes 55 1283–1288 [DOI] [PubMed] [Google Scholar]
- 2.Storgaard, H., Song, X. M., Jensen, C. B., Madsbad, S., Bjornholm, M., Vaag, A., and Zierath, J. R. (2001) Diabetes 50 2770–2778 [DOI] [PubMed] [Google Scholar]
- 3.Morino, K., Petersen, K. F., Dufour, S., Befroy, D., Frattini, J., Shatzkes, N., Neschen, S., White, M. F., Bilz, S., Sono, S., Pypaert, M., and Shulman, G. I. (2005) J. Clin. Investig. 115 3587–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen, K. F., Dufour, S., Savage, D. B., Bilz, S., Solomon, G., Yonemitsu, S., Cline, G. W., Befroy, D., Zemany, L., Kahn, B. B., Papademetris, X., Rothman, D. L., and Shulman, G. I. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12587–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patti, M. E., Butte, A. J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., Miyazaki, Y., Kohane, I., Costello, M., Saccone, R., Landaker, E. J., Goldfine, A. B., Mun, E., DeFronzo, R., Finlayson, J., Kahn, C. R., and Mandarino, L. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen, K. F., Dufour, S., Befroy, D., Garcia, R., and Shulman, G. I. (2004) N. Engl. J. Med. 350 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Befroy, D. E., Petersen, K. F., Dufour, S., Mason, G. F., de Graaf, R. A., Rothman, D. L., and Shulman, G. I. (2007) Diabetes 56 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei, M., Gibbons, L. W., Mitchell, T. L., Kampert, J. B., Lee, C. D., and Blair, S. N. (1999) Ann. Intern. Med. 130 89–96 [DOI] [PubMed] [Google Scholar]
- 9.Choi, C. S., Fillmore, J. J., Kim, J. K., Liu, Z. X., Kim, S., Collier, E. F., Kulkarni, A., Distefano, A., Hwang, Y. J., Kahn, M., Chen, Y., Yu, C., Moore, I. K., Reznick, R. M., Higashimori, T., and Shulman, G. I. (2007) J. Clin. Investig. 117 1995–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gates, A. C., Bernal-Mizrachi, C., Chinault, S. L., Feng, C., Schneider, J. G., Coleman, T., Malone, J. P., Townsend, R. R., Chakravarthy, M. V., and Semenkovich, C. F. (2007) Cell Metab. 6 497–505 [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Martinez, C., Sibille, B., Solanes, G., Darimont, C., Mace, K., Villarroya, F., and Gomez-Foix, A. M. (2001) FASEB J. 15 2033–2035 [DOI] [PubMed] [Google Scholar]
- 12.Wilson-Fritch, L., Nicoloro, S., Chouinard, M., Lazar, M. A., Chui, P. C., Leszyk, J., Straubhaar, J., Czech, M. P., and Corvera, S. (2004) J. Clin. Investig. 114 1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boden, G., Homko, C., Mozzoli, M., Showe, L. C., Nichols, C., and Cheung, P. (2005) Diabetes 54 880–885 [DOI] [PubMed] [Google Scholar]
- 14.Pagel-Langenickel, I., Schwartz, D. R., Arena, R. A., Minerbi, D. C., Johnson, D. T., Waclawiw, M. A., Cannon, R. O., III, Balaban, R. S., Tripodi, D. J., and Sack, M. N. (2007) Am. J. Physiol. Heart Circ. Physiol. 293 2659–2666 [DOI] [PubMed] [Google Scholar]
- 15.Mensink, M., Hesselink, M. K., Russell, A. P., Schaart, G., Sels, J. P., and Schrauwen, P. (2007) Int. J. Obes. (Lond) 31 1302–1310 [DOI] [PubMed] [Google Scholar]
- 16.Accili, D., Drago, J., Lee, E. J., Johnson, M. D., Cool, M. H., Salvatore, P., Asico, L. D., Jose, P. A., Taylor, S. I., and Westphal, H. (1996) Nat. Genet. 12 106–109 [DOI] [PubMed] [Google Scholar]
- 17.Rando, T. A., and Blau, H. M. (1994) J. Cell Biol. 125 1275–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhart-Hines, Z., Rodgers, J. T., Bare, O., Lerin, C., Kim, S. H., Mostoslavsky, R., Alt, F. W., Wu, Z., and Puigserver, P. (2007) EMBO J. 26 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bogacka, I., Xie, H., Bray, G. A., and Smith, S. R. (2005) Diabetes 54 1392–1399 [DOI] [PubMed] [Google Scholar]
- 20.Hondares, E., Mora, O., Yubero, P., de la Concepcion, M. R., Iglesias, R., Giralt, M., and Villarroya, F. (2006) Endocrinology 147 2829–2838 [DOI] [PubMed] [Google Scholar]
- 21.Yaspelkis, B. B., III, Lessard, S. J., Reeder, D. W., Limon, J. J., Saito, M., Rivas, D. A., Kvasha, I., and Hawley, J. A. (2007) Am. J. Physiol. Endocrinol. Metab. 293 941–949 [DOI] [PubMed] [Google Scholar]
- 22.Schuler, M., Ali, F., Chambon, C., Duteil, D., Bornert, J. M., Tardivel, A., Desvergne, B., Wahli, W., Chambon, P., and Metzger, D. (2006) Cell Metab. 4 407–414 [DOI] [PubMed] [Google Scholar]
- 23.Deeg, M. A., Buse, J. B., Goldberg, R. B., Kendall, D. M., Zagar, A. J., Jacober, S. J., Khan, M. A., Perez, A. T., and Tan, M. H. (2007) Diabetes Care 30 2458–2464 [DOI] [PubMed] [Google Scholar]
- 24.Handschin, C., Chin, S., Li, P., Liu, F., Maratos-Flier, E., Lebrasseur, N. K., Yan, Z., and Spiegelman, B. M. (2007) J. Biol. Chem. 282 30014–30021 [DOI] [PubMed] [Google Scholar]
- 25.Handschin, C., Choi, C. S., Chin, S., Kim, S., Kawamori, D., Kurpad, A. J., Neubauer, N., Hu, J., Mootha, V. K., Kim, Y. B., Kulkarni, R. N., Shulman, G. I., and Spiegelman, B. M. (2007) J. Clin. Investig. 117 3463–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wende, A. R., Schaeffer, P. J., Parker, G. J., Zechner, C., Han, D. H., Chen, M. M., Hancock, C. R., Lehman, J. J., Huss, J. M., McClain, D. A., Holloszy, J. O., and Kelly, D. P. (2007) J. Biol. Chem. 282 36642–36651 [DOI] [PubMed] [Google Scholar]
- 27.Miura, S., Kai, Y., Ono, M., and Ezaki, O. (2003) J. Biol. Chem. 278 31385–31390 [DOI] [PubMed] [Google Scholar]
- 28.Benton, C. R., Nickerson, J. G., Lally, J., Han, X. X., Holloway, G. P., Glatz, J. F., Luiken, J. J., Graham, T. E., Heikkila, J. J., and Bonen, A. (2008) J. Biol. Chem. 283 4228–4240 [DOI] [PubMed] [Google Scholar]
- 29.Martin, B. C., Warram, J. H., Krolewski, A. S., Bergman, R. N., Soeldner, J. S., and Kahn, C. R. (1992) Lancet 340 925–929 [DOI] [PubMed] [Google Scholar]
- 30.Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., Houstis, N., Daly, M. J., Patterson, N., Mesirov, J. P., Golub, T. R., Tamayo, P., Spiegelman, B., Lander, E. S., Hirschhorn, J. N., Altshuler, D., and Groop, L. C. (2003) Nat. Genet. 34 267–273 [DOI] [PubMed] [Google Scholar]
- 31.Skov, V., Glintborg, D., Knudsen, S., Jensen, T., Kruse, T. A., Tan, Q., Brusgaard, K., Beck-Nielsen, H., and Hojlund, K. (2007) Diabetes 56 2349–2355 [DOI] [PubMed] [Google Scholar]
- 32.Nissen, S. E., and Wolski, K. (2007) N. Engl. J. Med. 356 2457–2471 [DOI] [PubMed] [Google Scholar]
- 33.Wan, Y., Chong, L. W., and Evans, R. M. (2007) Nat. Med. 13 1496–1503 [DOI] [PubMed] [Google Scholar]
- 34.Milne, J. C., Lambert, P. D., Schenk, S., Carney, D. P., Smith, J. J., Gagne, D. J., Jin, L., Boss, O., Perni, R. B., Vu, C. B., Bemis, J. E., Xie, R., Disch, J. S., Ng, P. Y., Nunes, J. J., Lynch, A. V., Yang, H., Galonek, H., Israelian, K., Choy, W., Iffland, A., Lavu, S., Medvedik, O., Sinclair, D. A., Olefsky, J. M., Jirousek, M. R., Elliott, P. J., and Westphal, C. H. (2007) Nature 450 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baur, J. A., Pearson, K. J., Price, N. L., Jamieson, H. A., Lerin, C., Kalra, A., Prabhu, V. V., Allard, J. S., Lopez-Lluch, G., Lewis, K., Pistell, P. J., Poosala, S., Becker, K. G., Boss, O., Gwinn, D., Wang, M., Ramaswamy, S., Fishbein, K. W., Spencer, R. G., Lakatta, E. G., Le Couteur, D., Shaw, R. J., Navas, P., Puigserver, P., Ingram, D. K., de Cabo, R., and Sinclair, D. A. (2006) Nature 444 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagouge, M., Argmann, C., Gerhart-Hines, Z., Meziane, H., Lerin, C., Daussin, F., Messadeq, N., Milne, J., Lambert, P., Elliott, P., Geny, B., Laakso, M., Puigserver, P., and Auwerx, J. (2006) Cell 127 1109–1122 [DOI] [PubMed] [Google Scholar]
- 37.Sun, C., Zhang, F., Ge, X., Yan, T., Chen, X., Shi, X., and Zhai, Q. (2007) Cell Metab. 6 307–319 [DOI] [PubMed] [Google Scholar]
- 38.Arany, Z., Wagner, B. K., Ma, Y., Chinsomboon, J., Laznik, D., and Spiegelman, B. M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 4721–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, L., Zhang, Y., Chen, N., Shi, X., Tsang, B., and Yu, Y. H. (2007) J. Clin. Investig. 117 1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blendea, M. C., Jacobs, D., Stump, C. S., McFarlane, S. I., Ogrin, C., Bahtyiar, G., Stas, S., Kumar, P., Sha, Q., Ferrario, C. M., and Sowers, J. R. (2005) Am. J. Physiol. Endocrinol. Metab. 288 353–359 [DOI] [PubMed] [Google Scholar]
- 41.St-Pierre, J., Lin, J., Krauss, S., Tarr, P. T., Yang, R., Newgard, C. B., and Spiegelman, B. M. (2003) J. Biol. Chem. 278 26597–26603 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.