Abstract
Trafficking of Smad proteins between the cytoplasm and nucleus is a critical component of transforming growth factor β (TGF-β) signal transduction. Smad4 translocates into the nucleus either in response to TGF-β stimulation or when its nuclear export is blocked by leptomycin B (LMB). We demonstrate that both TGF-β-induced and basal state spontaneous nuclear import of Smad4 require importin 7 and 8 (Imp7,8). Our data suggest that in the nuclear import of Smad4, the role of Imp8 is irreplaceable by Imp7, and that Smads preferentially bind Imp8. Interestingly, in contrast to its mammalian counterpart Smad4, Drosophila Medea appears to utilize different mechanisms for TGF-β-induced or basal state nuclear accumulation, with the latter independent of Msk (Drosophila Imp7/8) function. In addition, overexpression of Imp8 alone was sufficient to cause an increased concentration of Smad1, 3 and 4 in the nucleus, but had very limited effects on Smad2. These observations suggest selective involvement of Imp8/Msk in nuclear import of different Smads under different conditions.
Cytokines in the transforming growth factor β (TGF-β)2 family are important regulators of embryonic development and tissue homeostasis (1). From Drosophila to mammals, the TGF-β signaling network centers around the Smad proteins, which are categorized into R-Smads (receptor-activated Smads, i.e. Smads1/2/3/5/8), Co-Smads (i.e. Smad4), and I-Smads (inhibitory Smads, i.e. Smads6/7). The R- and Co-Smads are essential mediators of TGF-β signaling, while the I-Smads served to dampen TGF-β signaling in a feedback manner (2-4). R-Smads and Smad4 are detected primarily in the cytoplasm or evenly throughout the cell, but accumulate in the nucleus upon TGF-β stimulation coincident with R-Smads phosphorylation at their C termini (5, 6). Such intracellular movement of Smads underlies the inducibility of transcriptional regulation by Smads. In contrast, the I-Smad, Smad7 is largely present in the nucleus at the basal state and reportedly undergoes nucleus-to-cytoplasm translocation in response to TGF-β. Therefore, deciphering the molecular mechanisms mediating nuclear import and export of Smads is crucial for understanding how the TGF-β signal is transduced into the nucleus and how this process is regulated.
R-Smads and Smad4 can enter the nucleus in both the basal state and upon TGF-β stimulation, regardless of the phosphorylation state at the C termini of R-Smads (7, 8). Recent RNA interference (RNAi) studies in both Drosophila and human cells provided strong evidence that Msk and its human orthologs Imp7 and Imp8 are indispensable for nuclear accumulation of activated phospho-R-Smads (9). However, Msk and Imp7/8 are not as essential for basal state R-Smad nuclear import, because knockdown of Msk and Imp7/8 did not affect distribution of Mad and Smad2/3 in unstimulated cells (9). These observations suggested that basal state nuclear import of R-Smads might entail additional pathways, including the importin-independent or the importin-β1-mediated mechanism suggested by previous in vitro studies (10-13).
It is not immediately obvious whether Msk/Imp7/8 would be involved in nuclear import of the other Smads including Smad4 and Smad7. Smad7 resides preferentially in the nucleus in unstimulated cells, and Smad4 could become predominantly nuclear when the export factor CRM-1 is blocked by LMB, without TGF-β signaling and completely separate from R-Smads (14, 15).
In this study, we used RNAi to evaluate the roles of Imp7 and Imp8 in the nuclear import of Smad4 and Smad7 either at the basal state or upon TGF-β stimulation. In addition, we overexpressed Imp7 and 8 to comprehensively examine their impact on the distribution of R-Smads, Smad4, and Smad7. These experiments verified the roles of Imp7/8 in nuclear import of Smad4 and ruled out the involvement of either Imp7 or Imp8 in regulating subcellular distribution of Smad7. Interestingly, our data also suggest that despite sequence similarities to other R- and Co-Smads, Smad2 and Medea (Drosophila Smad4) are not bona fide import cargoes of Imp8/Msk. These observations suggested specificity in Smad nuclear import through Imp8.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Cytokine Treatment—293T and HeLa cell culture were maintained in Dulbecco's Modified Eagle's media (DME) supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 units/ml) (all from Invitrogen). Cells were grown at 37 °C in 5% CO2. Mammalian cells were transfected using Lipofectamine 2000 following the manufacturer's protocol (Invitrogen). Drosophila S2R+ cells were grown at 25 °C in Schneider's Drosophila Medium with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 units/ml, all from Invitrogen). Effectene was used to transfect S2R+ cells using the manufacturer's recommended procedures (Qiagen). For TGF-β treatment, human TGF-β1 (R&D Systems) was used at a final concentration of 100 pm for 1 h. Leptomycin B (LMB, Sigma) was used at 10 ng/ml for 30 min.
RNAi in Mammalian and Drosophila Cells—For mammalian cells, synthetic siRNA duplexes (Dharmacon) were transfected at 40 nm using HiperFect following the vendor's instructions (Qiagen). The sequence for the Imp8 siRNAs was described in our previous publication (9). Sequences for the other siRNAs: Imp7a, CAAUUGCAGCUUUG-UAUUAuu; Imp7, UAAGCAGGCUGGU-GAA GAUuu; Imp7 + 8, GAUGGAGCCCUGCAUAUGAdTT. For Drosophila cells, the procedure for RNAi, as well as the dsRNA targeting msk was described previously (9).
Antibodies—The anti-Imp7/8 antibody was raised against recombinant amino acids 885-1038 of human Imp8. The Smad4 antibody has been described previously (16). Other antibodies were purchased from commercial sources (polyclonal and monoclonal anti-Flag: Sigma; anti-HA: Cell Signaling; antilamin A/C: Cell Signaling; Alexa 488-conjugated anti-mouse IgG: Invitrogen).
Subcellular Fractionation—Cells were treated with trypsin and harvested into ice-cold phosphate-buffered saline. The cell pellet was resuspended in ice-cold buffer containing 20 mm HEPES pH 7.4, 250 mm sucrose, 5 mm MgCl2, 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 100 μg/ml digitonin (Calbiochem), 2 mm dithiothreitol, and supplemented with protease inhibitors. After 5 min, the nuclei were pelleted by centrifugation at 800 × g for 7 min at 4 °C. The supernatant was used as the cytoplasmic fraction. The nuclear pellet was further extracted for 15 min on ice in 20 mm TrisCl, pH 7.4, 250 mm NaCl, 5 mm MgCl2, and 0.5% Nonidet P-40. After centrifugation (16,000 × g for 10 min at 4 °C), the supernatant was collected as the nuclear fraction.
Immunofluorescence Staining and Quantification—The procedures for immunofluorescence staining were described before (9). Fluorescent microscopy was done using a Nikon TE2000-S inverted microscope, and the images were captured by a Spot RT-KE digital camera (Diagnostic Instruments, Inc.). For confocal microscopy, a DMIRE2 Inverted Microscope and a TCS scanning system from Leica were used (Leica). In one method to quantify nuclear versus cytoplasmic localization, cells were categorized based on whether the immunofluorescence signal was predominantly nuclear, even or predominantly cytoplasm. Cells falling into each category were counted in a blind fashion (n > 150). In a second method, fluorescence microscopy images captured under the same exposure conditions were analyzed using Image J to quantify per unit area staining intensity in the cytoplasm and nucleus (∼20 positively stained cells from multiple fields).
Protein-Protein Interaction—293T cells with or without prior TGF-β treatment (100 pm, 1 h) were harvested and lysed for 20 min at 4 °C in a buffer containing 20 mm HEPES, pH 7.4, 200 mm NaCl, 1% Triton X-100, 5% glycerol, 2 mm dithiothreitol, and protease inhibitors. After centrifugation, the supernatant was diluted 1:2 with 20 mm HEPES and used for immunoprecipitation using anti-Flag-conjugated-agarose beads (Sigma) or anti-Imp7/8 plus protein A beads (Roche Applied Science). After incubation for 6 h, the beads were pelleted and washed 3× in the same buffer used during immunoprecipitation.
RESULTS
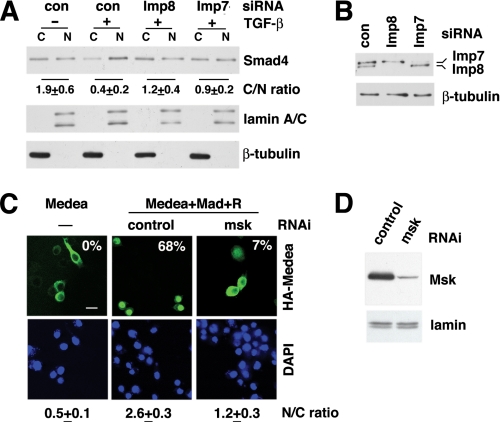
Impact of Imp7 and 8 Knockdown on Nuclear Translocation of Smad4—In unstimulated HeLa cells, endogenous Smad4 was detected in both cytoplasmic and nuclear fractions by immunoblotting. TGF-β treatment led to a decrease in the cytoplasm/nucleus ratio of Smad4 concentration reflecting signal-induced nuclear accumulation of Smad4 (Fig. 1A). When Imp7 or Imp8 was individually depleted by siRNAs, the cytoplasm-to-nucleus translocation of Smad4 in response to TGF-β was substantially reduced, suggesting a requirement for Imp7 and Imp8 in nuclear import (Fig. 1, A and B).
FIGURE 1.
Imp7 and Imp8 are required for signal-induced nuclear accumulation of Smad4 in mammalian and Drosophila cells. A, HeLa cells were transfected with siRNAs and stimulated with TGF-β as indicated. Cytosolic (C) and nuclear (N) fractions were analyzed by indicated immunoblotting. Smad4 signals were quantified by Image J, and the cytoplasmic/nuclear ratios were calculated (C/N ratio, mean ± S.D., n = 3). Lamin A/C and β-tubulin were used as markers for nuclear and cytoplasmic fractions, respectively. B, whole cell extracts from HeLa cells transfected with indicated siRNAs were examined by immunoblotting to detect Imp7/8 and β-tubulin (loading control). C, CuSO4-inducible expression vector encoding HA-Medea was transfected into Drosophila S2R+ cells alone, or together with Mad and the receptor kinases Punt and Tkv (R) to activate the Dpp pathway. Double-stranded RNAs were co-transfected for RNAi as indicated. After CuSO4 induction for 3 h, cells were stained with an anti-HA antibody and DAPI. Representative fluorescence micrographs are shown, and the percentages of cells having HA-Medea predominantly in the nucleus are shown in each case (n > 150). The nuclear/cytoplasmic ratio of HA-Medea staining was measured by Image J as described under “Experimental Procedures” (N/C ratio, mean ± S.D., n = 20). Scale bar: 10 μm. D, cell extracts from C were examined by immunoblotting to detect Msk and lamins (loading control).
In the Drosophila cell line S2R+, the Smad4 ortholog Medea was largely excluded from the nucleus when overexpressed (Fig. 1C). Because immunostaining patterns for overexpressed proteins are usually heterogeneous, we categorized cells based on Medea staining patterns as predominantly cytoplasmic, even, or predominantly nuclear. The percentage of cells in each category provides quantitative measurement of nuclear translocation. Using this method, we found, as reported previously, that upon co-expression of Mad and the TGF-β receptor kinases Punt and Thickvien (Tkv), Medea was present predominantly in the nucleus of 68% of transfected cells (n > 150, Fig. 1C) (17). In contrast, when the Imp7/8 ortholog Msk was knocked down by RNAi, only 7% of the transfected cells contain Medea mostly in the nucleus (Fig. 1, C and D). As a second method to quantify nuclear import, we analyzed multiple confocal microscopic images (taken under the same conditions) using Image J to measure HA-Medea staining intensity per unit area in cytoplasm and nucleus. The calculated nucleus/cytoplasmic ratio (n = 20 in each case) also verified that Msk depletion impaired nuclear accumulation of Medea in cells activated by Punt/Tkv (Fig. 1C, N/C ratio). Therefore, in both Drosophila and mammalian cells, Imp7 and 8 are important for signal-induced nuclear accumulation of the Co-Smad, Smad4.
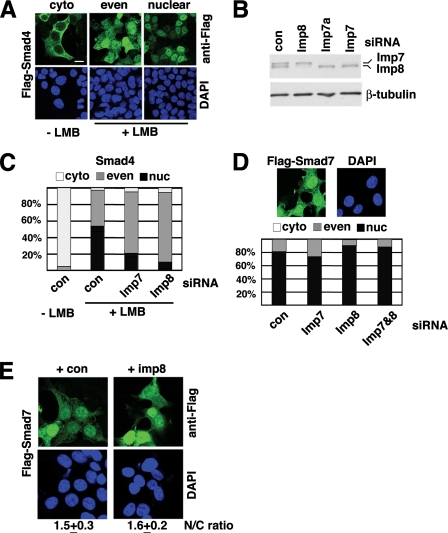
We next tested whether the spontaneous, TGF-β-independent nuclear import of Smad4 also relies on Imp7 and 8. In 293T cells transfected with Flag-Smad4-encoding plasmids, Smad4 was predominantly in the cytoplasm in 95% of the cells (n > 300, see Fig. 2A for representative patterns). After treatment with LMB, in 57% of the cells Flag-Smad4 was predominantly in the nucleus; in 42% of them Flag-Smad4 concentration was equal in the cytoplasm and nucleoplasm, and only 1% of the cells still had Flag-Smad4 mostly in the cytoplasm. This clearly indicated a shift of Smad4 into the nucleus in the presence of LMB.
FIGURE 2.
Roles of Imp7/8 in basal state nuclear import of Smad4 and Smad7. A, three representative patterns of Flag-Smad4 distribution are shown, cyto, predominantly cytoplasmic; even, equal distribution inside and outside of the nucleus; nuclear, predominantly nuclear. 293T cells were treated with LMB as indicated and immunostained with an anti-Flag antibody. Scale bar: 15 μm. B, 293T cells transfected with indicated siRNA duplexes were analyzed by anti-Imp7/8 immunoblotting to verify knockdown efficacy and specificity. β-Tubulin served as a loading control. C, Flag-Smad4 expression vector was co-transfected into 293T cells with indicated siRNAs and treated with LMB. Cells exhibiting the three Flag-Smad4 distribution patterns were counted, and the percentages are shown (n > 250 in each case). D, 293T cells were transfected with a Flag-Smad7 expression vector and stained with anti-Flag antibodies. Cells were scored for the Flag-Smad7 distribution pattern and counted (n > 250). A representative Flag-Smad7 distribution is shown. E, 293T cells were transfected with Flag-Smad7, with or without co-transfection of Imp8 as indicated. The nuclear/cytoplasmic ratio of Flag-Smad7 staining (N/C ratio: mean ± S.D.) was measured as described under “Experimental Procedures.”
Using a polyclonal antibody that recognizes both Imp7 and 8, we verified that Imp7 or Imp8 was knocked down individually (Fig. 2B). More importantly, co-transfecting Flag-Smad4 with siRNA against either Imp7 or 8 led to substantially decreased percentage of cells exhibiting nuclear accumulation of Flag-Smad4 in response to LMB (Fig. 2C). The direct measurement of nuclear/cytoplasmic ratio of Flag-Smad4 staining also indicated that LMB-induced nuclear import of Smad4 was decreased upon Imp7 or Imp8 knockdown (supplemental Table S1). Thus, the TGF-β-independent nuclear import of Smad4 also relied on Imp7 and 8.
Subcellular Distribution of Smad7 Is Not Affected by Levels of Imp7 and 8—When overexpressed in 293T and HeLa cells, Smad7 was present in both the cytoplasm and nucleus with a higher concentration in the nucleus, consistent with previous reports (Fig. 2D and data not shown) (18, 19). We wished to ascertain whether Imp7 or Imp8 might also take part in the nuclear import of Smad7. Co-transfection with siRNAs against Imp7 and/or 8 did not noticeably change the distribution pattern of Smad7, suggesting that Imp7 and 8 are not required for Smad7 import into the nucleus (Fig. 2D and supplemental Table S2). Moreover, we overexpressed Imp8 together with Smad7, and again we did not observe any changes in the nuclear/cytoplasmic ratio of Smad7 distribution (Fig. 2E). Therefore, we concluded that Imp7 and 8 were not critical regulators of Smad7 nuclear import.
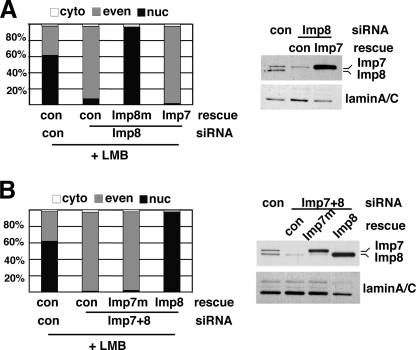
Imp8 Can Replace Imp7 in Mediating the Nuclear Import of Smad4—To verify that the siRNA effects were not due to off-target artifacts, we tested whether the import defects could be rescued by expressing silent mutants of Imp7 or 8 that are resistant to the siRNAs. This also allowed us to determine if the functions of Imp7 and 8 are interchangeable. We focused on nuclear import of Smad4 in LMB-treated cells. In Imp8 knockdown cells, overexpressing the siRNA-resistant Imp8 readily restored the nuclear import of Flag-Smad4, confirming the specificity of the RNAi result (Fig. 3A and supplemental Table S3.). In contrast, when Imp7 was overexpressed, the import defect due to Imp8 depletion could not be rescued (Fig. 3A and supplemental Table S3). Immunoblotting confirmed that overexpression of Imp7 was successful (Fig. 3A). Therefore, by itself Imp7 could not replace Imp8 to import Smad4.
FIGURE 3.
Imp8 cannot be replaced by Imp7 in mediating nuclear import of Smad4. A and B, 293T cells were transfected with indicated siRNA duplexes and rescuing plasmids, together with Flag-Smad4. After LMB treatment, cells were stained with anti-Flag. Quantification of cells exhibiting various Smad4 distribution patterns is shown (n > 250). The efficacy of knockdown and expression levels of rescue constructs was verified in the immunoblots (right panels). LaminA/C served as the loading control.
Furthermore, in cells with combined knockdown of Imp7 and Imp8, introducing siRNA-resistant Imp8, but not Imp7, was sufficient to reinstate nuclear accumulation of Smad4 upon LMB treatment (Fig. 3B and supplemental Table S4). It is important to note that overexpressing Imp8 did not increase the Imp7 protein level (Fig. 3B). Taken together, these knockdown and rescue experiments suggested that in basal state nuclear import of Smad4, Imp8 plays an irreplaceable role while Imp7 can be substituted by Imp8.
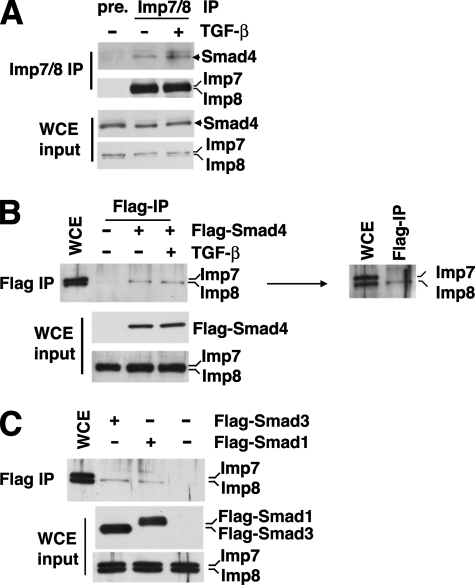
Smads Preferentially Interact with Imp8—To evaluate the interaction between endogenous Smad4 and Imp7/8, we first carried out co-immunoprecipitation experiment using an antibody recognizing both Imp7 and 8. Indeed, Smad4 was co-immunoprecipitated from 293T cell extract using the anti-Imp7/8 antibody (Fig. 4A). Treatment with TGF-β did not affect the Smad4-Imp7/8 interaction to a significant degree, consistent with the notion that Imp8 mediates both the basal state and TGF-β-induced nuclear import of Smad4 (Fig. 4A).
FIGURE 4.
Interaction between Smads and Imp8. A, 293T cell extract with or without prior TGF-β stimulation were immunoprecipitated with polyclonal anti-Imp7/8 antibodies. The bound proteins were analyzed by anti-Smad4 immunoblotting. The expression levels of indicated proteins are also shown (WCE input). WCE, whole cell extract. Pre-immune anti-sera were used in the control immunoprecipitation (pre.). B, 293T cells were transfected with Flag-Smad4 and treated with TGF-β as indicated. The cell extract was then subjected to anti-Flag immunoprecipitation. The bound proteins were analyzed by anti-Imp7/8 immunoblotting. The immunoblot on the right shows a better resolved gel which verifies that only Imp8 (lower band) was detected in the anti-Flag-Smad4 immunoprecipitate. C, same immunoprecipitation experiments as in B except that the 293T cells were transfected with Flag-Smad1 or -Smad3.
To examine if Smad4 has any preference in interacting with either Imp7 or Imp8, we transfected 293T cells with Flag-Smad4 and carried out anti-Flag immunoprecipitation. Interestingly, we detected only Imp8 in the immunoprecipitate (Fig. 4B, see the right panel for a better resolved immunoblot). Again, TGF-β treatment did not affect Imp8-Smad4 association (Fig. 4B). For comparison we tested Flag-Smad1 and Flag-Smad3, and indeed these two R-Smads also bound more strongly with endogenous Imp8 (Fig. 4C). These data suggested that the roles of Imp7 and Imp8 in Smad nuclear import are not equal, which also corroborate with the above observation that Imp8 was functionally irreplaceable by Imp7 in nuclear import of Smad4.
Overexpression of Imp8 Is Sufficient to Drive Smad4 into the Nucleus Independent of TGF-β—When expressed in HeLa or 293T cells, Smad4 mostly resided in the cytoplasm, presumably because the CRM-1-mediated nuclear export dominated over the nuclear import (Fig. 5A). Upon co-transfection with Imp7, more cells now exhibited even distribution of Smad4 in- and outside of the nucleus, indicating enhanced nuclear import, but few cells could qualify as having Smad4 predominantly in the nucleus (Fig. 5A and supplemental Fig. S1A). In contrast, overexpression of Imp8 had a much stronger effect, and in ∼70% of the cells, now Smad4 was predominantly in the nucleus (Fig. 5A and supplemental Fig. S1A). Measuring the nuclear/cytoplasmic ratio of Flag-Smad4 staining also confirmed that Imp8 is much more potent in driving Smad4 into the nucleus (Fig. 5A, N/C ratio). Imp7 and Imp8 were expressed at comparable levels in these experiments judging by immunostaining and immunoblotting (Fig. 5A and supplemental Fig. S1C). Therefore, these experiments once again suggested that Imp8 is a more potent nuclear import factor for Smad4 than Imp7.
FIGURE 5.
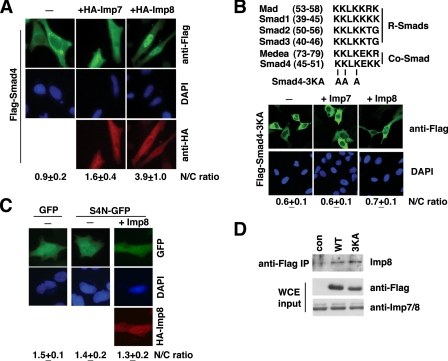
Imp8 is sufficient to re-distribute Smad4 into the nucleus without TGF-β. A, HeLa cells were transfected with Flag-Smad4 in combination with empty vector, HA-Imp7, or HA-Imp8 as indicated. Shown are anti-Flag (green), -HA (red) double immunostaining, and DAPI staining. The nuclear/cytoplasmic ratio (N/C ratio: mean ± S.D.) of anti-Flag staining was measured as described. B, upper, alignment of the KKLK motifs in Smads, and the 3KA mutations. Lower, Flag-Smad4-3KA mutant was transfected into HeLa cells together with Imp7 or Imp8 as indicated, and fluorescence micrographs of anti-Flag staining are shown. The measurement of N/C ratio (mean ± S.D.) of anti-Flag staining was done as in A. C, HeLa cells were transfected with indicated combination of plasmids (S4N-GFP: aa1-63 of Smad4 fused to the N-terminal of EGFP), and fluorescence microscopic images of GFP, anti-HA, and DAPI stainings are shown. Nuclear/cytoplasmic ratios of GFP signals were quantified as in A. D, 293T cells were transfected with Flag-tagged wild type (WT) or 3KA mutant Smad4 constructs as indicated, together with non-tagged Imp8. Anti-Flag immunoprecipitation was carried out, and the bound proteins were analyzed with anti-Imp7/8 immunoblotting (top panel). The expression levels of Flag-Smad4, Flag-Smad4-3KA, and Imp7/8 in whole cell extract (WCE) were verified by the indicated immunoblotting.
A Smad4 Motif Required but Not Sufficient for Imp8-mediated Nuclear Import—Previous studies have identified a Lysrich motif in the MH1 domain of Smad4 (45KKLKEKK51) that was necessary for Smad4 to accumulate in the nucleus upon treatment with LMB (15). This motif is highly conserved in R- and Co-Smads in Drosophila and mammals (Fig. 5B). Despite its similarity to a classic nuclear localization signal (cNLS), previous studies have clearly indicated that this Lys-rich sequence in Smads is not a cNLS, which by definition should bind importin α (10-13). When the first three Lys residues in this motif were mutated to Ala, overexpression of Imp8 or Imp7 could no longer drive this Smad4 mutant into the nucleus (Fig. 5B and supplemental Fig. S1B). However, a Smad4 fragment (amino acids 1-63) containing this Lys-rich motif but not the nuclear export sequence (NES) was unable to target a heterologous protein EGFP into the nucleus, even with Imp8 overexpression (Fig. 5C), suggesting that this Lys-rich motif in Smad4 is not sufficient to mediate nuclear import through Imp8. Interestingly, the Lys-to-Ala mutation did not affect interaction between Smad4 and Imp8 when examined by co-immunoprecipitation (Fig. 5D). This raised a possibility that the KKLK motif may serve a critical role in Smad4 nuclear import that is separate from interaction with Imp8.
Msk Is Not Required for Basal State Nuclear Import of Medea in Drosophila Cells—The MH1 domain including the KKLK motif in Medea is highly homologous to that of its mammalian counterpart Smad4, but there is considerable divergence in the linker region, and there is an additional 26 amino acids at the N terminus of Medea. Even though data in Fig. 1 clearly suggested that Medea import in stimulated cells was dependent on the Imp7/8 ortholog Msk, we were interested in testing whether at basal state, without the complication of possible piggyback import together with phospho-Mad, nuclear import of Medea was also mediated by Msk.
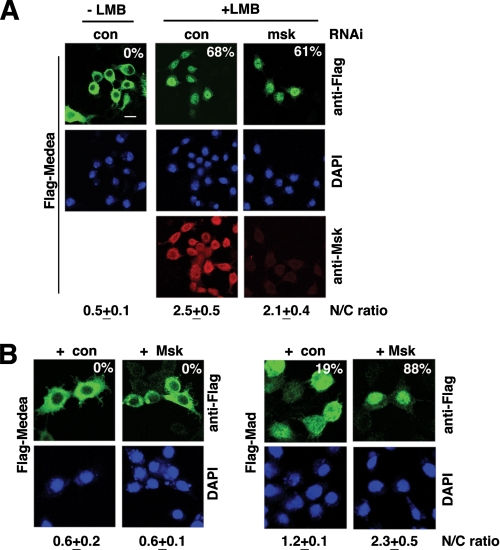
Like Smad4, Medea changed from mostly cytoplasmic to predominantly nuclear when cells were treated with LMB, indicating that both Smad4 and Medea are exported by CRM-1 (Fig. 6A). However, LMB-induced nuclear accumulation of Medea was entirely normal when Msk was depleted by RNAi (Fig. 6A). The efficacy of Msk RNAi was verified by immunostaining and that the nuclear import of Mad was severely affected (Fig. 6A and data not shown). Furthermore, we tested RNAi against all of the known karyopherin family members in Drosophila, and none appeared to be required for LMB-induced Medea import into the nucleus (data not shown). Moreover, when we overexpressed Msk, Medea still remained entirely cytoplasmic while in the control experiment nuclear import of Mad was clearly enhanced by Msk, further suggesting that Msk was not a critical factor in the basal state nuclear import of Medea (Fig. 6B).
FIGURE 6.
Nuclear import of Medea at basal state is independent of Msk in Drosophila cells. A, S2R+ cells conditionally expressing Flag-Medea were subjected to the indicated RNAi. The cells were then treated with CuSO4 to induce Flag-Medea expression, and also with LMB. Cells were co-stained with anti-Flag (green), anti-Msk (red), and DAPI. The percentages of cells having Flag-Medea predominantly in the nucleus are indicated (n > 200). In addition, the nuclear/cytoplasmic ratio (N/C ratio: mean ± S.D.) of anti-Flag staining intensity was measured as above. Scale bar: 10 μm. B, S2R+ cells were transfected with vectors for Flag-Medea together with Msk or empty vectors (con). Cells transfected with Flag-Mad were used for comparisons. Representative anti-Flag micrographs are shown. The percentages of cells having anti-Flag staining predominantly in the nucleus are shown. The nuclear/cytoplasmic ratio (N/C ratio: mean ± S.D.) of anti-Flag signal was measured as in A.
Therefore, although Medea and Smad4 are considered orthologs and both are exported by the same factor CRM-1, the spontaneous nuclear import at the basal state appeared to be mediated by completely different mechanisms. Given the data in Figs. 1C and 6 and the established fact that Medea complexes with Mad upon Mad phosphorylation, we reasoned that in stimulated Drosophila cells, phospho-Mad piggybacked Medea into the nucleus via Msk.
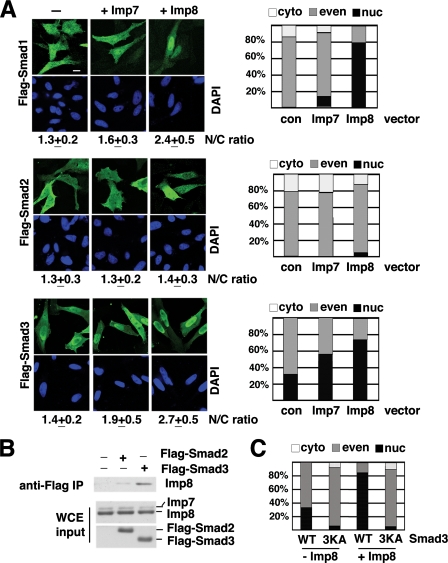
Roles of Imp7 and 8 in Nuclear Import of R-Smads at Basal State—We next investigated whether overexpression of Imp7 or 8 would also alter R-Smad distribution in the absence of TGF-β. Flag-tagged Smad1, 2, and 3 were transfected into HeLa cells, with or without co-transfection of Imp7 or Imp8, and then examined by anti-Flag immunofluorescence staining. As in the above experiments, the nuclear versus cytoplasmic distribution was quantified with two different methods (Fig. 7A). Co-transfection with Imp7 had very limited effects on the distribution patterns of Smad1 and 3 (Fig. 7A). In contrast, Imp8 clearly enhanced the nuclear accumulation of Smad1 and Smad3 (Fig. 7A). Importantly, the overexpression level of Imp7 was actually higher than that of Imp8 in these experiments, suggesting that the differences in R-Smad import efficiency was not due to differential expression of Imp7 and Imp8 (supplemental Fig. S1C). In contrast to both Smad1 and Smad3, the pattern of Smad2 changed very little upon Imp8 overexpression, and most remained evenly distributed in the cytoplasm and nucleus (Fig. 7A). The same was also observed in 293T cells (data not shown). Therefore, among R-Smads, Imp8 is effective in importing Smad1 and Smad3, but much weaker for Smad2. Consistent with this, Smad2 did appear to interact more weakly with Imp8 in comparison to Smad3 (Fig. 7B).
FIGURE 7.
Impact of Imp7 or Imp8 overexpression on cellular distribution of R-Smads. A, HeLa cells were transfected with Flag-tagged Smad1, 2, or 3, in combination with control vector, Imp7, or Imp8 as indicated. Cells were immunostained with an anti-Flag antibody (green), and the cells exhibiting different Flag-immunostaining patterns were counted, and the percentages are presented in the charts (panels on the right). Micrographs of representative fields are also shown, and the nuclear/cytoplasmic ratios (N/C ratio: mean ± S.D.) of anti-Flag signal were measured as described. Scale bar: 15 μm. B, 293T cells were transfected with Flag-Smad3 or Flag-Smad2, together with non-tagged Imp8. For control, cells transfected with only Imp8 was used. Anti-Flag immunoprecipitation was carried out, and the bound proteins were examined with anti-Imp7/8 immunoblotting. The levels of overexpressed proteins in whole cell extract (WCE) were also analyzed by anti-Imp7/8 or anti-Flag immunoblotting (lower panels). C, wild type or 3KA mutant Smad3 (40KKLK → 40AALA) was transfected into HeLa cells with or without Imp8. Cells were stained with anti-Flag, and the percentages of cells exhibiting different staining patterns are shown (n > 200).
Smad1, 2, 3, and 4 have highly homologous MH1 domains, including the KKLK motif at relatively the same positions (Fig. 5B). As in Smad4, changing the KKLK sequence to AALA in Smad3 also prevented Imp8 from importing this mutant Smad3 into the nucleus (Fig. 7C and supplemental Table S5).
Combining the data in Figs. 5, 6 and 7, we reached the conclusion that the KKLK motif is required, but not sufficient, for Imp8-mediated nuclear import of Smads at the basal state. Thus, there must be additional structural requirements in Smads that enable nuclear import via Imp8, and such structural elements are probably not present in Smad2 and Medea.
DISCUSSION
We recently demonstrated that TGF-β-induced nuclear accumulation of R-Smads depended on Imp7/8 in both Drosophila and mammalian cells. Here we expanded the study to Co-Smad and I-Smad. Both gain- and loss-of-function analyses suggested a critical role of Imp8 in basal state and TGF-β-induced nuclear import of Smad4, while the subcellular distribution of Smad7 appeared to be independent of Imp7 and Imp8 regulation. Our data have now revealed that in both Drosophila and mammalian cells, the signal-induced nuclear accumulation of all R- and Co-Smads clearly require Msk/Imp7/8. On the other hand, for nuclear import in unstimulated cells, Msk/Imp7/8 is dispensable except in the case of Smad4. In addition, overexpression of Msk/Imp7/8 could enhance basal state nuclear import of some, but not all R- and Co-Smads. These observations support the idea that only selected members of the Smad family are direct import cargoes of Msk/Imp8, and different Smads utilize multiple import pathways for nuclear accumulation under different circumstances.
Imp7 versus Imp8 in Smad Import—Msk is the only Imp7/8 ortholog in Drosophila and its evolution into Imp7 and Imp8 may reflect demand for cargo specification (20). Imp7 and Imp8 are nearly 80% homologous. The sequence similarity extends throughout the proteins, with the lowest homology in the last ∼100 amino acid residues (21). Only two previous studies examined Imp7 and Imp8 functions side-by-side. In the case of glucocorticoid receptor (GR), both Imp7 and Imp8 exhibited similar interaction with GR, but only Imp7 was able to import GR into the nucleus in vitro (22). In another study, the signal recognition particle protein 19 (SRP19) was shown to bind a number of importins with similar affinity including Imp7 and 8, and yet in the in vitro reconstituted assay, only Imp8 and transportin showed ability to import SRP19 (23).
Here, we provide evidence arguing that the functions of Imp7 and Imp8 in Smad nuclear import are not exchangeable. This conclusion derives from three observations. In RNAi rescue experiments, overexpression of Imp7 could not overcome the deficiency in Imp8; whereas overexpression of Imp8 readily restored Smad4 nuclear import in Imp7 knockdown cells. Secondly, even though endogenous levels of Imp7 and 8 are similar, in co-immunoprecipitation experiments, Smad1, Smad3, and Smad4 all interacted much more strongly with endogenous Imp8 than with Imp7. Thirdly, increased expression of Imp8 caused a dramatic redistribution of Smad1, Smad3, and Smad4 into the nucleus, but overexpression of Imp7 to a comparable level had a much weaker effect.
Although RNAi experiments suggested that both Imp7 and Imp8 are required for nuclear accumulation of Smads to a similar degree, Imp7 and 8 may serve different roles in this process. Our data are most consistent with the notion that Imp7 and 8 have non-redundant roles and may participate in different aspects of Smad nuclear import.
Structural Elements in Smads Required for Nuclear Import by Imp8—The KKLK sequence is conserved in Drosophila and mammalian R-Smads and Smad4, and mutation to AALA abrogated nuclear import of Smad4 and Smad3 (this report and Ref. 15). However, the KKLK motif is not sufficient to facilitate nuclear import of a heterologous protein. In addition, while the KKLK motif is present in Smad2 and Medea, these two Smads are not directly imported by Imp8/Msk.
The precise function of the KKLK motif is still not clear. Mutating KKLK to AALA did not affect Imp8 interaction with Smad4. One interpretation is that the KKLK motif is critical for Smad4 import at the steps after Imp8 association. Alternatively, the Imp8-Smad4 interaction involves the KKLK and other domains, but the KKLK sequence is required for an interaction with Imp8 that would generate an import-competent configuration. Indeed there are many examples in which binding to an importin does not result in actual nuclear import (22, 24, 25). It is also possible that another factor binding the KKLK motif is crucial for nuclear import of Smads.
For Smad2, the interaction with Imp8 was also significantly weaker when compared with Smad3. The C-terminal MH2 domains of Smad2 and Smad3 are almost identical, so the differences between the two in terms of Imp8 interaction and nuclear import is likely to reside within the MH1 and the linker region. This is also the region in R-Smads where interaction with Imp8 was mapped (9). The MH1 domains are relatively conserved among all Smads but the linkers are much more divergent. Further investigation is needed to determine precisely what structural elements distinguish Smad2 and Medea from the rest of R- and Co-Smads in their ability to be imported by Imp8 or Msk.
In the meantime, questions also arise about how Smad2 and Medea are imported into the nucleus. It is very possible that under TGF-β stimulation, since R-Smads and Co-Smad form complexes, they could be co-imported. Indeed, RNAi experiments clearly demonstrated that Smad2 and Medea import in response to TGF-β activation was dependent on Imp8 and Msk, respectively (9). At the basal state, Smad2 may be able to enter the nucleus through a previously described importin-independent mechanism that depends on interaction with nucleoporins (7, 10). It is reasonable to propose that Medea can be imported through a similar mechanism because Smad4 was shown to bind nucleoporins as well (11). At present, we favor the notion that there are multiple parallel pathways for importing R-Smads and Co-Smads into the nucleus at the basal state; on the other hand upon TGF-β stimulation, Imp8/Msk is the rate-limiting factor in nuclear import of all R- and Co-Smads. A possible exception is that phospho-Smad2 homotrimers may enter the nucleus through different mechanisms in response to TGF-β, without the involvement of Imp8.
The Balance of Smad Nuclear Import and Export Forces—When overexpressed in HeLa or 293T cells, Smad1, Smad3, and Smad4 display different distribution patterns, probably reflecting the sum of counteracting nuclear import and export forces. In all three cases, overexpression of Imp8 was sufficient to cause redistribution of these Smads into the nucleus. It is interesting to note that the level of endogenous Imp8 is apparently sufficient for TGF-β-induced nuclear import of R- and Co-Smads, but clearly not enough to counteract the export factors without TGF-β stimulation. Although it remains to be determined if Imp8 activity can be enhanced by TGF-β, current data seem to support the hypothesis that TGF-β facilitates nuclear accumulation of R-Smads and Smad4 mainly by suppressing their nuclear export activities (8, 26).
Collectively, our observations suggest an important role for Imp8 in nuclear translocation of Smad4, in addition to R-Smads. It will be interesting to further investigate how the functions of Imp8 in Smad import could be regulated.
Supplementary Material
Acknowledgments
We thank Dr. R. Padgett (Rutgers University) for providing the original Medea-expressing plasmid, and Dr. Q. Xu for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA108509. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1-S5.
Footnotes
The abbreviations used are: TGF-β, transforming growth factor β; HA, hemagglutinin; LMB, leptomycin B; RNAi, RNA interference; DAPI, 4′,6-diamidino-2-phenylindole; siRNA, short interfering RNA; R-Smad, receptor-activated Smad; I-Smad, inhibitory Smad.
References
- 1.Massagué, J., Blain, S. W., and Lo, R. S. (2000) Cell 103 295-309 [DOI] [PubMed] [Google Scholar]
- 2.Shi, Y., and Massagué, J. (2003) Cell 113 685-700 [DOI] [PubMed] [Google Scholar]
- 3.Massague, J., Seoane, J., and Wotton, D. (2005) Genes Dev. 19 2783-2810 [DOI] [PubMed] [Google Scholar]
- 4.Derynck, R., and Zhang, Y. E. (2003) Nature 425 577-584 [DOI] [PubMed] [Google Scholar]
- 5.Xu, L., and Massague, J. (2004) Nat Rev Mol. Cell. Biol. 5 209-219 [DOI] [PubMed] [Google Scholar]
- 6.Reguly, T., and Wrana, J. L. (2003) Trends Cell Biol. 13 216-220 [DOI] [PubMed] [Google Scholar]
- 7.Xu, L., Kang, Y., Col, S., and Massagué, J. (2002) Mol Cell 10 271-282 [DOI] [PubMed] [Google Scholar]
- 8.Schmierer, B., and Hill, C. S. (2005) Mol. Cell. Biol. 25 9845-9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu, L., Yao, X., Chen, X., Lu, P., Zhang, B., and Ip, Y. T. (2007) J. Cell Biol. 178 981-994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu, L., Chen, Y. G., and Massagué, J. (2000) Nat. Cell Biol. 2 559-562 [DOI] [PubMed] [Google Scholar]
- 11.Xu, L., Alarcon, C., Col, S., and Massagué, J. (2003) J. Biol. Chem. 278 42569-42577 [DOI] [PubMed] [Google Scholar]
- 12.Xiao, Z., Liu, X., and Lodish, H. F. (2000) J. Biol. Chem. 275 23425-23428 [DOI] [PubMed] [Google Scholar]
- 13.Kurisaki, A., Kose, S., Yoneda, Y., Heldin, C. H., and Moustakas, A. (2001) Mol. Biol. Cell 12 1079-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe, M., Masuyama, N., Fukuda, M., and Nishida, E. (2000) EMBO Rep. 1 176-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierreux, C. E., Nicolas, F. J., and Hill, C. S. (2000) Mol. Cell. Biol. 20 9041-9054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaraman, L., and Massagué, J. (2000) J. Biol. Chem. 275 40710-40717 [DOI] [PubMed] [Google Scholar]
- 17.Das, P., Maduzia, L., Wang, H., Finelli, A., Cho, S. H., Smith, M., and Padgett, R. (1998) Development 125 1519-1528 [DOI] [PubMed] [Google Scholar]
- 18.Itoh, S., Landstrom, M., Hermansson, A., Itoh, F., Heldin, C. H., Heldin, N. E., and ten Dijke, P. (1998) J. Biol. Chem. 273 29195-29201 [DOI] [PubMed] [Google Scholar]
- 19.Zhang, S., Fei, T., Zhang, L., Zhang, R., Chen, F., Ning, Y., Han, Y., Feng, X. H., Meng, A., and Chen, Y. G. (2007) Mol. Cell. Biol. 27 4488-4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker, S. E., Lorenzen, J. A., Miller, S. W., Bunch, T. A., Jannuzi, A. L., Ginsberg, M. H., Perkins, L. A., and Brower, D. L. (2002) Genetics 162 285-296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorlich, D., Dabrowski, M., Bischoff, F. R., Kutay, U., Bork, P., Hartmann, E., Prehn, S., and Izaurralde, E. (1997) J. Cell Biol. 138 65-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman, N. D., and Yamamoto, K. R. (2004) Mol. Biol. Cell 15 2276-2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dean, K. A., von Ahsen, O., Gorlich, D., and Fried, H. M. (2001) J. Cell Sci. 114 3479-3485 [DOI] [PubMed] [Google Scholar]
- 24.Jakel, S., and Gorlich, D. (1998) EMBO J. 17 4491-4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakel, S., Albig, W., Kutay, U., Bischoff, F. R., Schwamborn, K., Doenecke, D., and Gorlich, D. (1999) EMBO J. 18 2411-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen, H. B., Rud, J. G., Lin, K., and Xu, L. (2005) J. Biol. Chem. 280 21329-21336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.