Abstract
TonEBP is a Rel domain-containing transcription factor implicated in adaptive immunity, viral replication, and cancer. In the mammalian kidney, TonEBP is a central regulator of water homeostasis. Animals deficient in TonEBP suffer from life-threatening dehydration due to renal water loss. Ambient tonicity (effective osmolality) is the prominent signal for TonEBP in a bidirectional manner; TonEBP activity decreases in hypotonicity, whereas it increases in hypertonicity. Here we found that TonEBP displayed nuclear export in response to hypotonicity and nuclear import in response to hypertonicity. The nuclear export of TonEBP was not mediated by the nuclear export receptor CRM1 or discrete nuclear export signal. In contrast, a dominant nuclear localization signal (NLS) was found in a small region of 16 amino acid residues. When short peptides containing the NLS were fused to constitutively cytoplasmic proteins, the fusion proteins displayed tonicity-dependent nucleocytoplasmic trafficking like TonEBP. Thus, tonicity-dependent activation of the NLS is crucial in the nucleocytoplasmic trafficking of TonEBP. The novel NLS is present only in the vertebrates, indicating that it developed late in evolution.
TonEBP (tonicity-responsive enhancer-binding protein), also known as NFAT5 (nuclear factor of activated T cell 5), is a member of the Rel family of transcriptional activators (1, 2). Like other members of the family, NFκB and NFAT, TonEBP is involved in a variety of biological processes, including adaptive immunity (3, 4), viral replication (5), and cancer (6). TonEBP is most extensively characterized in the renal medulla, which is the only tissue in the body that is extremely hyperosmotic, routinely reaching over 1,000 mosmol/kg in humans and 3,000 mosomol/kg in rodents. The hyperosmolality is essential for the ability of the kidney to produce concentrated urine, a key function of the kidney for maintenance of body fluid volume and blood pressure. TonEBP is a major regulator in the renal concentration of urine via transcriptional stimulation of the aquaporine 2 water channel (7) and the UT-A urea transporters (8). TonEBP is also essential in the cellular protection from the deleterious effects of hyperosmolality in the renal medulla via promoting the cellular accumulation of organic osmolytes and the expression of HSP70 (9). Genetically modified mice with TonEBP deficiency in the renal medulla suffer from severe atrophy of the renal medulla because of failure to adapt to the hyperosmolality and life-threatening hydropenia due to excessive renal water loss (10, 11).
TonEBP is stimulated by hypertonicity (i.e. effective hyperosmolality that causes cell shrinkage due to net water loss across the plasma membrane) (12). This hypertonicity-induced stimulation is critical for the function of TonEBP in the immune (3, 4) and renal cells (9). The stimulation of TonEBP is associated with enhancement in the nuclear localization (13), transactivation (14), and protein abundance (12). The molecular mechanism underlying the stimulation of TonEBP has been elusive. For example, the role of phosphorylation is poorly understood, since sites of phosphorylation are not defined, and protein kinases involved are not definitively identified despite extensive studies over the years (9).
Changes in the nucleocytoplasmic localization represent a major mode of TonEBP regulation in the kidney and the brain. TonEBP in the renal medulla shifts to the cytoplasm in response to water diuresis, whereas it shifts to the nucleus in response to antidiuresis (15). Cytoplasmic shift of TonEBP is observed in the renal medullae of animals suffering from hypokalemia (16) and nephropathy caused by long term treatment with cyclosporine (17). These reports suggested that a reduction in the tonicity of the renal medullary interstitium signaled for the cytoplasmic shift of TonEBP. Similar mechanism appears to be present in the neurons. In the brain, TonEBP is exclusively localized in neurons, and the abundance of nuclear TonEBP increases in 45 min in response to systemic hypernatremia (18, 19). The rapid increase in the nuclear TonEBP suggests that nuclear trafficking of preexisting TonEBP takes place in the brain.
The nucleocytoplasmic localization of TonEBP has been studied in cultured cells. Earlier studies demonstrated the bidirectional nature: cytoplasmic shift in hypotonicity and nuclear shift in hypertonicity (12). A recent report described the role of a classical nuclear export signal (NES)3 that interacts with CRM1 in the nuclear export of TonEBP (20). In this study, however, we find no evidence for the CRM1-dependent NES in TonEBP. Instead, we find that the tonicity-dependent activation of the nuclear localization signal (NLS) is critical for the regulation of nucleocytoplasmic localization of TonEBP.
MATERIALS AND METHODS
DNA Constructs—All constructs were generated using standard cloning procedures and verified by restriction enzyme analysis and DNA sequencing. Hemagglutinin-tagged NFATc1 expression plasmid was a gift from Dr. Steffan Ho (University of California, San Diego). A cDNA encoding the c-form of human TonEBP mRNA (21) was cloned into a mammalian expression vector pCMV-Tag2 or pCMV-Tag3 (Stratagene, La Jolla, CA) or pEGFP-C2 (Clontech), which allows expression of TonEBP fused with FLAG, Myc, or green fluorescent protein (GFP) epitope, respectively. Site-directed mutants were made using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). TonEBP cDNA fragments Yc1 (amino acids 1–548) and Yc2 (amino acids 1–76 fused to amino acids 406–876) were generated by PCR and cloned into pCMV-Tag3 vector. DNA fragments containing the NLS of SV40 and the NLS of TonEBP (amino acids 181–234 and 194–209) were inserted into Yc2 as shown in Fig. 8A. A trimer of GFP (3GFP) was generated by cloning two tandem copies of GFP open reading frame into the pEGFP-C2 vector. A DNA fragment encoding Myc-SV40NLS or Myc-NLS was generated by PCR and cloned into the 3GFP vector to produce 3GFP-SV40NLS or 3GFP-NLS, respectively. The NES reporter construct pRev(1.4)-GFP and its positive control construct pRev(NES)-GFP (22) were kindly provided by Dr. Beric Henderson (University of Sydney, Australia). TonEBP-(194–209) and its mutant (R199A/K200A) were cloned in pRev(1.4)-GFP to test their NES activity.
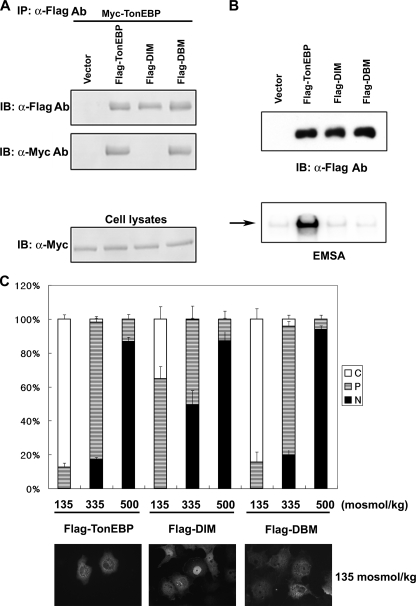
FIGURE 8.
Dimerization is required for nuclear export of TonEBP. A, FLAG-tagged TonEBP or its site-directed mutant, DIM (F464A/I466A) or DBM (T298A/E299A/R302A), was coexpressed with Myc-TonEBP in COS7 cells. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody. The immunoprecipitates, and cell lysates were immunoblotted (IB) for FLAG or Myc, as indicated. B, the FLAG-tagged constructs were transfected into COS7 cells. Top, cell lysates were immunoblotted for FLAG. Bottom, EMSA was performed using the cell lysates and 1 nm [32P]TonE. The arrow denotes the TonEBP·DNA complex. C, COS7 cells transfected as in B were seeded on coverslips and analyzed as in Fig. 3 using FLAG immunofluorescence. Fluorescence images of the cells in hypotonic conditions are shown at the bottom.
Cell Culture and Transfection—COS7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gemini Bio-Products, Woodland, CA), 100 μg/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). Cells were transfected using Lipofectamine 2000 (Invitrogen). 2 μg of plasmid DNA in 150 μl of Opti-MEM (Invitrogen) was mixed with 12 μl of Lipofectamine2000 dissolved in 150 μl of Opti-MEM and incubated at room temperature for 20–30 min. The mixture was added to a 1.5 ml of trypsinized cell suspension in antibiotic-free culture medium, mixed, seeded in a well of a 6-well cluster, and cultured for a day.
Immunoblot Analysis—To prepare cell extract, cells were washed once with ice-cold PBS and incubated for 30 min in lysis buffer containing 50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm dithiothreitol, protease inhibitor mixture (Roche Applied Science), and phosphatase inhibitor mixture (Sigma) at 4 °C. After being cleared by centrifugation for 5 min at 15,000 × g, the extracts were separated on a 6% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. Nonspecific binding was blocked with a 30-min incubation in 5% nonfat milk in TTBS (25 mm Tris, pH 7.4, 137 mm NaCl, 3 mm KCl, 0.05% Tween 20) at room temperature. The membrane was incubated for 60 min with a 1:2,000 dilution of anti-TonEBP antibody (1), anti-FLAG antibody (Sigma), anti-Myc antibody (Covance, Richmond, CA), or anti-GFP antibody (BD Biosciences) in the blocking solution at room temperature. The membrane was then incubated the same way with a 1:5,000 dilution of anti-rabbit IgG or anti-mouse IgG conjugated with horseradish peroxidase (Jackson Immunoresearch, West Grove, PA). An enhanced chemiluminescence assay was performed to visualize horseradish peroxidase using a commercial kit.
Northern Blot Analysis—Total RNA was isolated using Trizol reagent (Invitrogen). 5 μg of RNA was size-fractionated on a 1% agarose gel containing 2.2 m formaldehyde and was transferred to a nitrocellulose membrane. The membrane was hybridized overnight with 32P-labeled aldose reductase (AR) (GenBank™ accession number J05474), canine SMIT (M85068), or human HSP70 cDNA (M11717). After washing under stringent conditions (65 °C in 75 mm NaCl and 7.5 mm sodium citrate with 0.1% SDS), radioactivity was visualized and quantified using a PhosphorImager (Amersham Biosciences).
Immunocytochemistry—Cells grown on glass coverslips were treated with various conditions indicated. Cells were fixed for 15 min in 3% paraformaldehyde (Eastman Kodak Co.) in PBS. The cells were then permeabilized with 0.5% Triton X-100 in PBS for 5 min. Nonspecific binding was blocked with 3% fetal bovine serum in PBS for 20 min. The coverslips were incubated for 60 min with a 1:400 dilution of anti-TonEBP or anti-FLAG antibody or a 1:2,000 dilution of anti-Myc or anti-hemagglutinin antibody in blocking solution at room temperature. The coverslips were washed three times with PBS and incubated for 30 min with a 1:1,000 dilution of Alexafluor 568-conjugated goat anti-mouse IgG or anti-rabbit IgG (Molecular Probes, Inc., Eugene, OR). After three washes with PBS, the coverslips were mounted using the ProLong Antifade kit (Molecular Probes). A Nikon Eclipse E600 fluorescence microscope and Spot RT digital camera were used to observe and collect images.
Separation of Cytoplasmic and Nuclear Fractions—COS7 cells cultured in a 100-mm dish were cooled on ice and washed with ice-cold PBS. Nuclear and cytosolic fractions were separated using a commercial kit (NE-PER nuclear and cytoplasmic extraction reagents; Pierce), following the manufacturer's instructions. The fractions were immunoblotted using antibodies for Brg-1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and aldose reductase (16) to monitor nuclear and cytoplasmic proteins, respectively.
Electrophoretic Mobility Shift Assay—Double-stranded human TonE (1) was end-labeled using [γ-32P]ATP. The cell extract (5 μg of protein; see “Immunoblot Analysis”) was incubated for 10 min with 1 μg of poly(dA·dT) in 20 μl containing 20 mm HEPES (pH 7.9), 50 mm KCl, 5 mm MgCl2, 1 mm dithiothreitol, and 5% (v/v) glycerol. After the addition of 20 fmol of 32P-labeled human TonE, the reaction was incubated for 20 min at room temperature. The mixture was electrophoresed for 2.5 h on a 4% polyacrylamide gel in 45 mm Tris, 45 mm boric acid, and 1 mm EDTA at 150 V. Radioactivity of the TonEBP bands was visualized and quantified using a PhosphorImager.
RESULTS
Nuclear Import and Export of TonEBP in COS7 Cells—The goal of this study was to understand molecular basis for the nucleocytoplasmic trafficking of TonEBP in response to changes in ambient tonicity. We decided to use COS7 cells for this, because we found that COS7 cells, like many other cell lines, had functional TonEBP that was stimulated by ambient hypertonicity. When COS7 cells were switched to hypertonic medium, nuclear localization of TonEBP increased in 30 min, whereas the abundance of TonEBP increased gradually after 2 h (data not shown), as previously reported in Madin Darby canine kidney cells (12, 13). These changes stimulated TonEBP as demonstrated by increased expression of TonEBP target genes, such as the sodium/myoinositol cotransporter, aldose reductase, and HSP70 (data not shown).
In isotonic conditions, TonEBP was clearly localized in the cytoplasm as well as in the nucleus (Fig. 1A), as reported earlier (12, 13). It should be pointed out that TonEBP distribution was the same at 335 and 300 mosmol/kg (data not shown). Fig. 2A shows comparable distribution at 270 versus 335 mosmol/kg. TonEBP distribution shifted to predominantly cytoplasmic when the cells were switched to hypotonicity. The cytoplasmic shift was dose-dependent (Fig. 1A) and completed in 30 min at the ambient tonicity of 135 mosmol/kg (Fig. 1B). Since the abundance of TonEBP did not change in this time frame (Fig. 1C), the cytoplasmic shift was due to movement of TonEBP out of the nucleus (i.e. nuclear export).
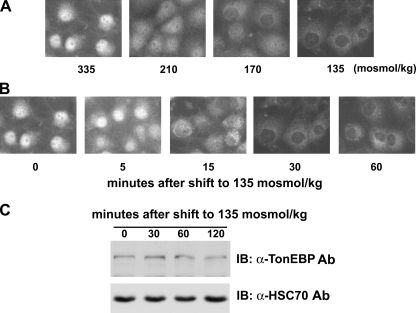
FIGURE 1.
Nuclear export of TonEBP in response to hypotonicity. A, dose dependence. COS7 cells grown on glass coverslips were switched for 30 min to hypotonic media of 210 to 135 mosmol/kg, made by deleting appropriate amounts of NaCl, or kept in isotonic medium (335 mosmol/kg), as indicated. TonEBP was visualized by immunofluorescence. B, time course. Cells were switched to hypotonic medium (135 mosmol/kg) for up to 60 min, as indicated, and TonEBP was visualized as above. C, cells grown on plastic dishes were switched to hypotonic medium for up to 120 min, as indicated, and immunoblotted (IB) for TonEBP and HSC70.
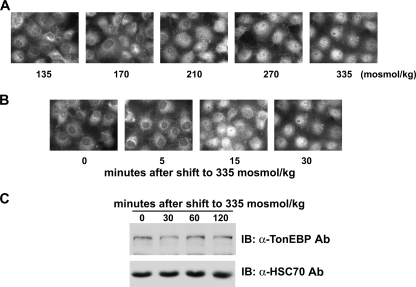
FIGURE 2.
Nuclear import of TonEBP in response to hypertonicity. A, dose response. COS7 cells had been incubated for 30 min in hypotonic medium (135 mosmol/kg) before the medium osmolality was increased up to 335 mosmol/kg, as indicated by the addition of NaCl. After 30 min, TonEBP was visualized by immunofluorescence. B, time course. Cells had been incubated for 30 min in hypotonic medium before they were switched to isotonic medium for 5, 15, or 30 min. C, cells grown on plastic dishes were treated as in B except that they were switched to isotonic medium for up to 120 min. TonEBP and HSC70 were detected by immunoblotting (IB).
The maximum cytoplasmic localization of TonEBP at 135 mosmol/kg of ambient osmolality provided an ideal start point to examine the nuclear shift of TonEBP. When the medium tonicity was raised to isotonicity, the TonEBP distribution shifted into the nucleus in a dose-dependent manner (Fig. 2A). The nuclear shift was completed in 30 min (Fig. 2B) without changes in the TonEBP abundance (Fig. 2C), indicating that it was due to movement into the nucleus (i.e. nuclear import). Taken together, the data in Figs. 1 and 2 define the bidirectional nucleocytoplasmic trafficking in terms of dose response and time course.
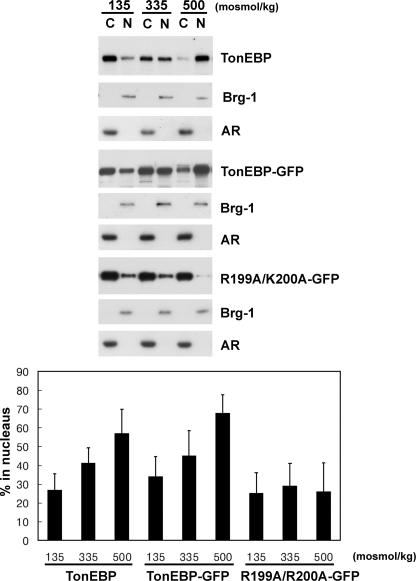
NLS in TonEBP Is Monopartite and Dominant—We found that overexpressed TonEBP in fusion with GFP showed mixed nuclear and cytoplasmic localization in isotonic conditions (Fig. 3D) much like the endogenous TonEBP (see above). The GFP-TonEBP became predominantly nuclear in response to hypertonicity, whereas it became predominantly cytoplasmic in response to hypotonicity (Fig. 3D). The same results were obtained when TonEBP was fused to FLAG (see Fig. 7) or Myc epitope (data not shown). Immunoblot analyses of cytoplasmic and nuclear fractions confirmed that the transfected and overexpressed TonEBP behaved like endogenous TonEBP; ∼30% of TonEBP was in the nucleus at 135 mosmol/kg compared with ∼60% at 500 mosmol/kg (Fig. 4). These data demonstrate that transfected TonEBP molecules exhibit the normal, tonicity-dependent nucleocytoplasmic trafficking. Doubling of TonEBP nuclear distribution from ∼30 to ∼60% is seen as predominantly cytoplasmic to predominantly nuclear in immunofluorescence analysis.
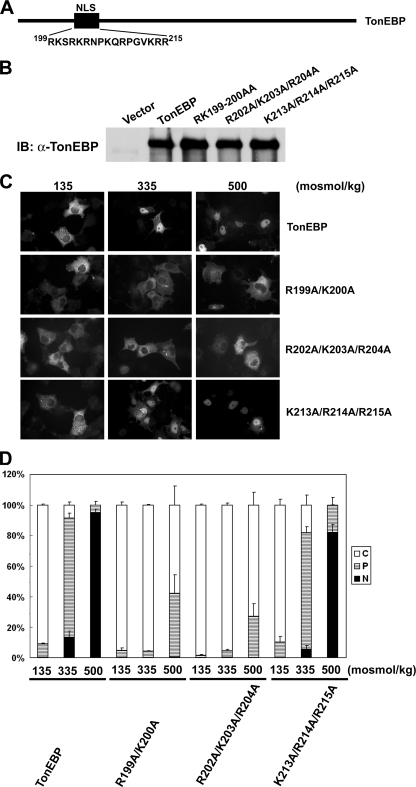
FIGURE 3.
NLS of TonEBP is monopartite. A, schematics of TonEBP and amino acid sequence of residues 199–215. B, immunoblot (IB) of COS7 cells transfected with TonEBP and its site-directed mutants (R199A/K200A, R202A/K203A/R204A, and K213A/R214A/R215A) in fusion with GFP. C, green fluorescence images of the transfected cells that had been incubated for 60 min in hypotonic (135 mosmol/kg H2O), isotonic (335 mosmol/kg H2O), or hypertonic medium (500 mosmol/kg H2O) before they were fixed for observation. D, the cells shown in C were scored as follows: C (open bars) for cells displaying predominantly cytoplasmic distribution of the fluorescence, P (hatched bars) for cells with both cytoplasmic and nuclear distribution, and N (filled bars) for cells with predominantly nuclear distribution. More than 200 cells were analyzed for each condition from a transfection. Values are mean ± S.D. from four independent experiments (n = 4).
FIGURE 7.
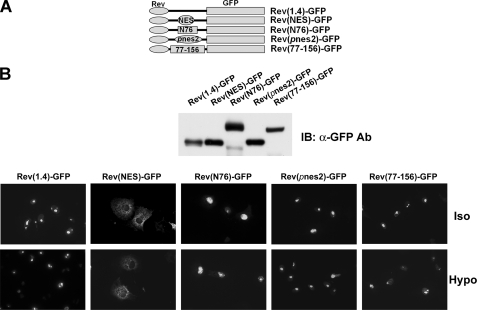
pnes1 and pnes2 do not promote nuclear export. A, schematics of constructs used. The nuclear export signal of human immunodeficiency virus Rev protein (NES), the N-terminal 76 amino acids of TonEBP, including pnes1 (N76; see Fig. 5A), pnes2, or TonEBP fragment containing amino acids 77–156 containing pnes2 (77–156) was inserted into the Rev(1.4)-GFP, as indicated. B, top, immunoblot of COS7 cells transfected with the constructs. The bottom panel shows green fluorescence images of the transfected cells seeded on glass coverslips. The cells were treated for 3 h with actinomycin D (5 μg/ml) and cycloheximide (15 μg/ml). Half of the cells were then switched to hypotonic medium (135 msomol/kg; Hypo) for 1 h or kept in isotonic medium (Iso) before fixation. In those cells transfected with Rev(NES)-GFP, the green fluorescence was localized diffusely in the cytoplasm. In other cells, the green fluorescence was concentrated in the nucleoli. A representative set of three independent experiments is shown. IB, immunoblot.
FIGURE 4.
Nuclear distribution of TonEBP, and GFP-fused TonEBP and K199A/R200A. COS7 cells were transfected without DNA (mock) or with expression vector for TonEBP-GFP or R199A/R200A-GFP and then incubated for 60 min in hypotonic, isotonic, and hypertonic medium as described in the legend to Fig. 3. Cytoplasmic (C) and nuclear fraction (N) were obtained from each condition and loaded at a 1:1 ratio for immunoblot analysis of TonEBP (mock-transfected samples) and GFP. In each condition, Brg-1 and aldose reductase (AR) were also immunoblotted to monitor separation of the nuclear versus cytoplasmic fractions. Intensity of TonEBP or GFP signal was quantified and expressed as percentage in the nucleus: 100 × (intensity in N)/(intensity in C + intensity in N) (mean ± S.D., n = 5 or 6). In TonEBP or TonEBP-GFP, all three values are different from each other (p < 0.01, Student's t test), whereas there are no statistical differences in R199A/K200A-GFP.
In order to delineate amino acid sequence elements involved in the nuclear import, we constructed and examined numerous mutant TonEBP molecules generated by serial deletions and truncations. From these studies, we found that the amino acids 199–215 (sequence shown in Fig. 3A) were required for the nuclear localization; mutant TonEBP molecules lacking these amino acids were constitutively cytoplasmic even in hypertonic conditions (data not shown). We noticed that there were two stretches of arginines and lysines in this region, as seen in many classical NLS sequences (23). We tested these residues by site-directed mutagenesis (Fig. 3A). Mutations of R199A/K200A or R202A/K203A/R204A led to constitutively cytoplasmic distribution, whereas K213A/R214A/R215A did not affect the trafficking (Fig. 3, C and D). Immunoblot analyses of cytoplasmic and nuclear fractions showed that less than 30% of R199A/K200A distributed in the nucleus regardless of ambient tonicity, confirming the analysis shown in Fig. 3. These data provide further characterization of previously recognized NLS (20) by showing that 199RKSRKR204 is the core of NLS in the context of the full TonEBP molecule. The aggregation of arginines and lysines in a short span conforms to the monopartite form of classical NLS (23). This NLS is dominant in that its mutations almost completely abolish the nuclear localization even in hypertonic conditions.
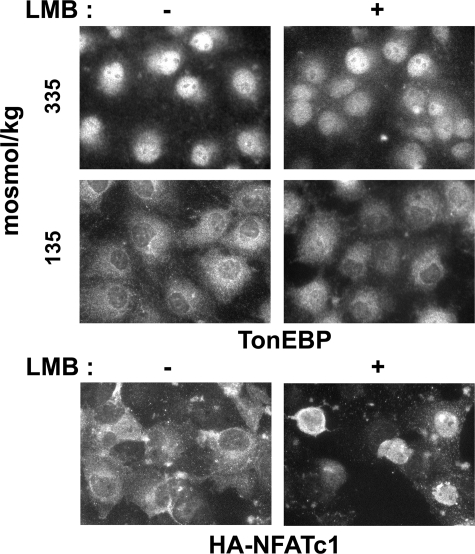
Nuclear Export of TonEBP Is Independent of CRM1 or Discrete NES—A recent report suggested that the nuclear export receptor CRM1 regulated the nuclear export of TonEBP via specific interaction with a leucine-rich NES-like motif at the N terminus (20). Since this was based on the behavior of an N-terminal truncated form of TonEBP, we reexamined this issue using two approaches. First, the role of CRM1 was examined using a specific inhibitor, leptomycin B (LMB). In COS7 cells, LMB inhibited nuclear export of transfected NFATc1 in response to decreased cell calcium concentrations (Fig. 5B), as reported earlier (24). However, the nuclear export of endogenous TonEBP was not affected by LMB (Fig. 5A), disputing the role of CRM1. Second, we examined the N-terminal leucinerich motif named pnes1 (pseudo-NES; 8LLSADLDL15) in the context of full-length TonEBP. For this, we made a couple of GFP-TonEBP fusion constructs where pnes1 was either deleted (TonEBP-a; this is the a-form of the TonEBP splice variant) (21) or mutated in the critical leucine residues (mpnes1), as depicted in Fig. 6A. These constructs did not display increased nuclear localization; instead, they displayed increased cytoplasmic distribution in isotonic conditions compared with wild type TonEBP. These data do not support the view that pnes1 is a functional NES or CRM1 is involved in the nuclear export of TonEBP.
FIGURE 5.
Nuclear export of TonEBP is insensitive to LMB. A, COS7 cells grown on glass coverslips were treated with vehicle or 10 ng/ml LMB for 30 min and then switched to hypotonic medium or kept in isotonic medium for another 30 min as indicated. TonEBP was visualized by immunofluorescence. B, COS7 cells transfected with hemagglutinin-tagged NFATc1 were seeded on glass coverslips. The cells were treated with 10 ng/ml ionomycin for 30 min in the absence or presence of LMB. They were then switched to medium without ionomycin for 30 min, followed by immunofluorescence visualization of the hemagglutinin tag.
FIGURE 6.
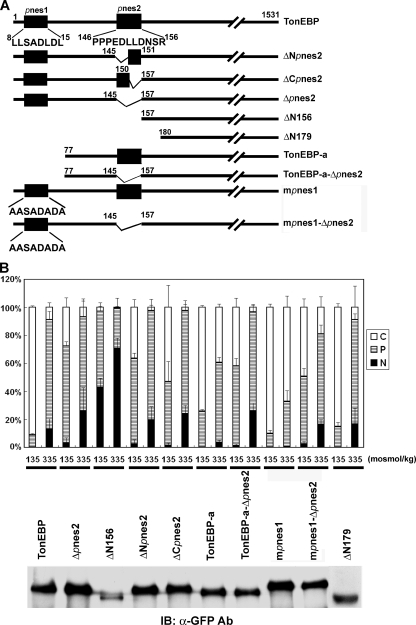
Putative nuclear export signals of TonEBP. A, schematics of TonEBP and its mutant constructs made by deletion and site-directed mutagenesis. Amino acid sequences are shown for pnes1 (residues 8–15) and pnes2 (residues 146–156). TonEBP-a is the a-form of the TonEBP splice variant. B, an immunoblot (IB) of COS7 cells transfected with the constructs in fusion with GFP is shown below. The transfected cells were analyzed as in Fig. 3, and results from cells in hypotonic and isotonic conditions are shown (mean ± S.D., n = 4). In hypertonic conditions, the distributions were identical in all constructs (not shown) (i.e. the same as TonEBP).
We searched for functional NES using deletion and truncation mutants of TonEBP. Initial results suggested that amino acids 146–156, named pnes2, were a functional NES (data not shown). To test pnes2 in the context of full-length TonEBP, we made several constructs shown in Fig. 5A: ΔNpnes2 (deletion of the N-terminal half of pnes2, residues 146–150), ΔCpnes2 (deletion of the C-terminal half of pnes2, residues 151–156), and Δpnes2. All of these deletion mutants displayed modestly but clearly increased nuclear localization in hypotonic and isotonic conditions (Fig. 6B). Essentially the same results were obtained in the context of TonEBP-a (TonEBP-a-Δpnes2) or mpnes1 (mpnes1-Δnes) (Fig. 6B). These results support the notion that pnes2 was a modest but functional NES. On the other hand, when the N-terminal 179 amino acids were deleted (ΔN179) to remove both pnes1 and pnes1 in a manner different from the TonEBP-a-Δpnes2 construct, the nucleocytoplasmic trafficking was normal. These data raised a question of whether pnes2 was an NES.
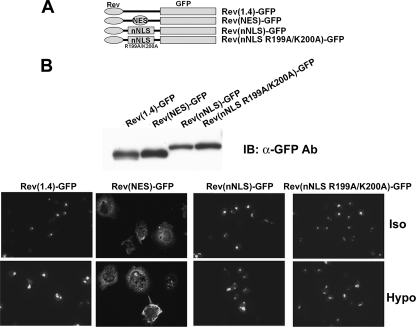
To clarify this issue, we sought to confirm the nuclear export activity of pnes2 using an established NES reporter vector pRev(1.4)-GFP (22). As shown in Fig. 7, the CRM1-interacting NES from human immunodeficiency virus Rev protein displayed a clear nuclear export activity, as shown by the cytoplasmic localization of green fluorescence in the transfected cells, as reported previously (22). On the other hand, a TonEBP fragment (N76) containing Δpnes1 did not display significant nuclear export activity, confirming that pnes1 is not an NES. In addition, two different TonEBP fragments containing Δpnes2 (TonEBP-(77–156) and Δpnes2 itself (see Fig. 7A)) did not show appreciable nuclear export activity either. These data do not support the notion that pnes2 was a functional NES.
We also examined the role of DNA binding and dimerization in the nucleocytoplasmic trafficking of TonEBP. Based on the crystal structure of the Rel-homology domain of TonEBP (25), we made a dimer interface mutant (DIM; F464A/I466A) and DNA binding mutant (DBM; T298A/E299A/R302A). DIM did not dimerize, whereas DBM did (Fig. 8A). As expected, neither of them bound DNA (Fig. 8B). Of interest, DBM trafficked normally, whereas DIM displayed increased nuclear localization in hypotonic and isotonic conditions (Fig. 8C). Thus, dimerization is required for full nuclear export of TonEBP, whereas DNA binding is dispensable. Taken together, our data presented in Figs. 5, 6, 7, 8 do not provide evidence that a discrete NES is involved in the nuclear export of TonEBP. This is in contrast with the NLS of TonEBP, which is discrete, dominant, and independent (see below).
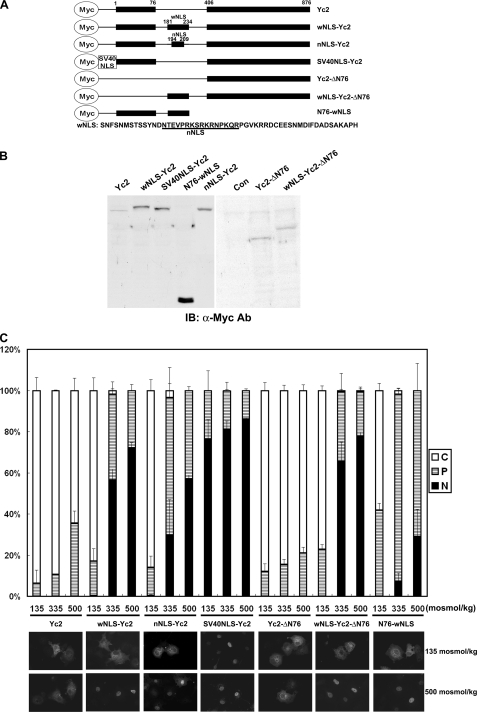
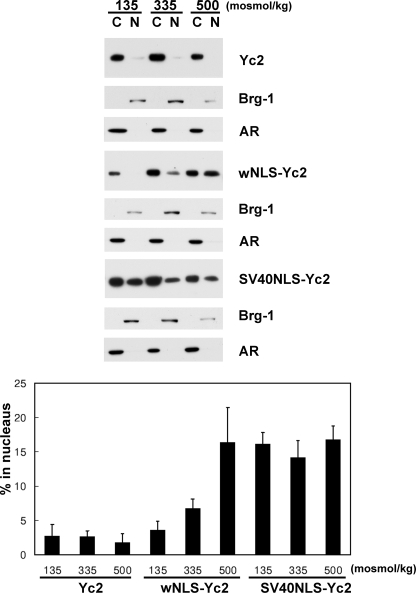
NLS Is Regulated by Tonicity—The primary goal of this study was to understand how the nuclear import and nuclear export of TonEBP was regulated by ambient tonicity. Since a functional and discrete NES was not found in TonEBP, we asked whether the NLS was regulated by tonicity. In order to measure the activity of NLS, we used the Yc2 construct, which was a fusion of the 1–76 and 405–786 fragments of TonEBP (see Fig. 9A). The 1–76 peptide was used to promote protein expression (21); expression of those constructs without this peptide was very low (Fig. 9B, right). Yc2 forms a dimer (26) with predicted molecular mass of ∼120 kDa, which is over the exclusion limit (∼60 kDa) of the nuclear pore (27). When expressed in COS7 cells, Yc2 was localized predominantly in the cytoplasm even in hypertonic conditions (Fig. 9C), presumably because it could not pass through the nuclear pore. When the TonEBP-(181–234) fragment containing NLS was inserted into Yc2, the resulting protein, wNLS-Yc2, displayed clear nuclear import in a tonicity-dependent manner. On the other hand, SV40NLS-Yc2 containing the NLS of SV40 T antigen (28) was constitutively nuclear regardless of the ambient tonicity (Fig. 9). These data were confirmed by immunoblot analyses of cytoplasmic and nuclear fractions (Fig. 10). Less than 5% of Yc2 was nuclear, whereas about 15% of SV40NLS-Yc2 was nuclear, regardless of ambient tonicity. On the other hand, wNLS-Yc2 showed a clear tonicity dependence: 3.5% at 135 mosmol/kg, 6.7% at 335 mosmol/kg, and 16.3% at 500 mosmol/kg (p < 0.05 in all pairwise comparisons). Thus, the activity of NLS in TonEBP is stimulated by ambient tonicity, whereas that of SV40 T antigen is not. Deletion of the 1–76 fragment from wNLS-Yc2 (wNLS-Yc2-ΔN76) did not affect its tonicity-responsive nuclear import (Fig. 9C); nor did deletion of the 406–876 fragment, although the resulting construct (N76-wNLS; smaller than 20 kDa) was small enough for free passage through the nuclear pore.
FIGURE 9.
The NLS of TonEBP is regulated by ambient tonicity. A, schematics of recombinant constructs. Yc2 is a fusion of amino acids 1–76 and 406–876 of TonEBP. TonEBP fragments containing NLS, corresponding to residues 181–234 and 194–209 (sequence shown at bottom), were inserted between the 1–76 and 406–876 fragments to produce wNLS-Yc2 and nNLS-Yc2, respectively. A peptide fragment (residues 119–135) from SV40 T antigen containing NLS was inserted at the N terminus of Yc2 to produced SV40NLS-Yc2. The 1–76 fragment of TonEBP was deleted from Yc2 and wNLS-Yc2 to produce Yc2-ΔN76 and wNLS-Yc2-ΔN76, respectively. The 406–876 fragment of TonEBP was deleted from wNLS-Yc2 to produce N76-wNLS. B, left, COS7 cells transfected with the constructs were immunoblotted (IB) for Myc. Right, transfected cells were immunoprecipitated with anti-Myc antibody and then immunoblotted for Myc. Con, vector-transfected cells. C, the transfected cells were analyzed as in Fig. 3 using Myc immunofluorescence. Fluorescence images in hypotonic and hypertonic conditions are shown at the bottom.
FIGURE 10.
Nuclear distribution of Yc2, wNLS-Yc2, and SV40NLS-Yc2. Cytoplasmic and nuclear fractions were prepared from COS7 cells transfected and treated with different osmolalities, as described in the legend to Fig. 9. The cytoplasmic and nuclear fractions were loaded at a 1:4 ratio and immunoblotted for Myc and loaded at 1:1 ratio and immunoblotted for Brg-1 and aldose reductase (AR) as described in the legend to Fig. 4. The intensity of the Myc signal was quantified and expressed as percentage in the nucleus: 100 × (intensity in N)/((4 × intensity in C) + intensity in N) (mean ± S.D., n = 3). In wNLS-Yc2, all three values are different from each other (p < 0.05; Student's t test), whereas there are no statistical differences in Yc2 or SV40NLS-Yc2.
We next examined a smaller fragment, TonEBP-(194–209), that contained 10 additional amino acid residues around the core of NLS by constructing nNLS-Yc2 (Fig. 9). Remarkably, nNLS-Yc2 displayed tonicity-dependent nuclear localization much like wNLS-Yc2. Both wNLS-Yc2 and wNLS-Yc2 displayed clear nuclear localization in isotonicity and, when switched to hypotonicity, decreased the nuclear localization acutely (i.e. within 30 min). This raised the possibility that there might be a cryptic NES in the nNLS peptide. To test this directly, we measure the NES activity of the nNLS peptide using the NES reporter construct, as described above. The results showed that neither nNLS nor its NLS-inactive mutant (R199A/K200A) displayed measurable NES activity (Fig. 11), demonstrating the lack of NES in the nNLS peptide. We conclude that the tonicity-dependent nucleocytoplasmic trafficking of wNLS-Yc2 and nNLS-Yc2 is due to the tonicity dependence of the NLS activity without the involvement of NES.
FIGURE 11.
The NLS of TonEBP does not promote nuclear export. A, schematics of constructs used. Rev(1.4)-GFP and Rev(NES)-GFP are described in the legend to Fig. 6. TonEBP fragment 194–209 (nNLS) or nNLS with R199A/K200A mutations (nNLS R199A/K200A) was inserted into Rev(1.4)-GFP as shown. B, the constructs were transfected into COS7 cells and analyzed as in Fig. 7.
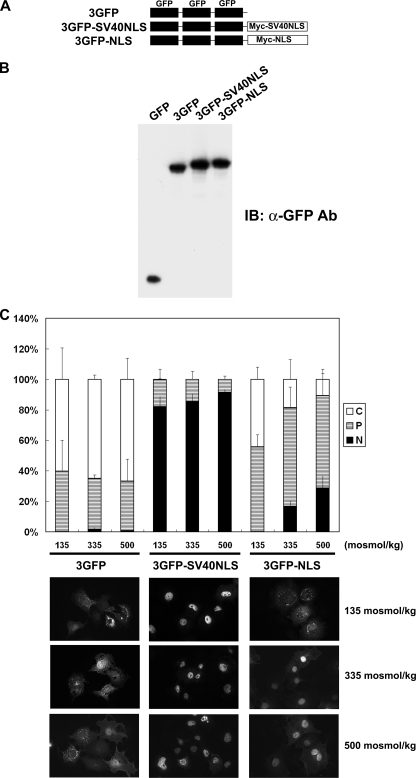
In order to further confirm the tonicity-dependent activity of NLS, we used another protein, an in-frame trimer of GFP, 3GFP, which is ∼80 kDa in size. As reported previously (29), monomeric GFP distributed evenly in the nucleus and cytoplasm (not shown), whereas 3GFP was predominantly cytoplasmic, regardless of tonicity (Fig. 12). In frame fusion of the SV40 NLS to 3GFP made the fusion protein (3GFP-SV40NLS) constitutively nuclear, whereas fusion of TonEBP NLS (amino acids 181–234; 3GFP-NLS) conferred tonicity-dependent nuclear localization (Fig. 12). These data demonstrate that the NLS of TonEBP is robust and independent in its response to ambient tonicity. The tonicity-dependent activity of NLS is the key in the nucleocytoplasmic trafficking of TonEBP in response to changes in ambient tonicity.
FIGURE 12.
Tonicity-responsive nuclear trafficking of GFP trimer (3GFP) by the NLS of TonEBP. A, schematics of recombinant constructs. 3GFP is an in-frame trimer of GFP. The Myc-SV40NLS fragment shown in Fig. 8A was used to produce 3GFP-SV40NLS. SV40NLS in 3GFP-SV40NLS was replaced by the TonEBP fragment 181–234 to produce 3GFP-NLS. B, COS7 cells transfected with pEGFP-C2 encoding a monomeric GFP or the 3GFP constructs were immunoblotted (IB) for GFP. C, the transfected cells were analyzed as in Fig. 3 using the green fluorescence. Representative fluorescence images in hypotonic, isotonic, and hypertonic conditions are shown at the bottom.
DISCUSSION
Here we have shown that TonEBP displays a highly predictable nucleocytoplasmic distribution as a function of ambient tonicity in the range 135–500 mosomol/kg: predominantly cytoplasmic localization (∼30% in the nucleus) at 135 mosmol/kg, predominantly nuclear localization (∼60% in the nucleus) at 500 mosmol/kg, and a gradient of mixed distribution in between. A change in the ambient tonicity leads to corresponding redistribution of TonEBP in 30 min. The redistribution is driven by intracellular trafficking mechanisms: nuclear import and nuclear export. The nuclear export of TonEBP in response to hypotonicity is not dependent on the exportin receptor CRM1 or a discrete NES. On the other hand, the nuclear import of TonEBP was mediated by a small motif containing monopartite NLS. A series of observations demonstrate that tonicity-dependent activity of the NLS is critical in the nucleocytoplasmic trafficking of TonEBP. The NLS functions in an autonomous manner (i.e. without the aid of other parts of TonEBP) in that it can drive the tonicity-dependent nucleocytoplasmic trafficking when fused to three different peptides: a large TonEBP fragment devoid of NLS or NES, an in frame triplet of GFP, and a small peptide containing the N-terminal 76 amino acids of TonEBP. Thus, we have discovered a novel NLS that is regulated by ambient tonicity.
The Rel family of transcription factors is found in only in higher animals, vertebrates and invertebrates (2). Although several NFκB and NFAT genes are present in the human genome, only one TonEBP gene is present in the human or other species down to Drosophila. The NLS of TonEBP is perfectly conserved in mammalian TonEBP genes and highly conserved in the zebrafish TonEBP (Fig. 13). However, it is absent in the Drosophila TonEBP homolog MSER1. When human TonEBP-GFP fusion protein was transfected into S2 cells, a Drosophila-derived cell line, it was predominantly localized in the nucleus regardless of ambient tonicity (data not shown). These observations support the view that the tonicity-dependent nucleocytoplasmic trafficking of TonEBP evolved only in the vertebrates. As such, the NLS is the first osmoregulatory mechanism shown to be unique to the vertebrates (see more below).
FIGURE 13.
NLS of TonEBP is conserved in vertebrates. Tonicity responsive nNLS (see Fig. 9) is conserved in zebrafish and mammals. Drosophila homolog MSER1 does not contain an NLS-like sequence.
As discussed under “Results,” our data do not agree with a recent study (20), which suggested that CRM1 interacted with the N terminus of TonEBP (pnes1 in Fig. 6) and inhibited the nucleus export of TonEBP. It is possible that the use of N-terminally truncated TonEBP (amino acids 1–581) in combination with overexpressed CRM1 led to experimental artifacts in the study. For example, mutations of pnes1 in the context of full-length TonEBP led to decreased nuclear localization (see Fig. 6 and “Results”), whereas the same mutation in the context of TonEBP-(1–581) increased the nuclear localization (20). In addition, LMB did not affect the nuclear export of the endogenous TonEBP (Fig. 5).
The lack of CRM1-interacting NES in TonEBP is somewhat surprising, because NFκB and NFAT are regulated by this type of NES in combination with NLS (24, 30). In fact, there appears be no discrete NES in TonEBP, as discussed under “Results.” The lack of clearly defined NES does not necessarily indicate that there is no functional NES in TonEBP. This is because the majority of known NESs are poorly defined in terms of critical amino acid residues involved and corresponding nuclear export receptors (23). In this regard, it is worth noting that dimerization is required for full nuclear export of TonEBP (Fig. 8).
Regulation of the nuclear localization in response to incoming signals is the hallmark of other Rel proteins NFAT and NFκB. It is well established that phosphorylation and dephosphorylation are central elements in the regulation of NFκB (31, 32) and NFAT (33, 34). A recent study (35) reported that protein kinase ATM (ataxia telangiectasia-mutated) stimulated the nuclear localization of TonEBP, raising the possibility that phosphorylation of TonEBP played a role in the nuclear trafficking of TonEBP. We tested whether phosphorylation of nNLS (see Figs. 9 and 13) affected its function. For this, we made site-directed mutants of nNLS-Yc2 (Fig. 9) as follows: T195A/S201A, singly and in combination, to prevent potential phosphorylation, and T195E/S201D, singly and in combination, to mimic phosphorylation. These mutants displayed normal tonicity-dependent trafficking (data not shown). At this point, there is no evidence that phosphorylation of TonEBP plays a role in the regulation of the tonicity-regulated NLS in TonEBP.
Cellular response to osmolality has been extensively studied in bacteria, yeast, and Caenorhabditis elegans. The power of genetic screening in these simple organisms has provided powerful tools to uncover signaling pathways of osmosensing. However, most of the mechanisms uncovered have had only limited relevance to higher animals. For example, the OmpR system in bacteria (36) or the two-component phosphorelay system in yeast (37) is not found in animals. Although the p38 mitogen-activated protein kinase cascade is critical in the cellular response to hypertonicity in yeast, its role in mammalian cells is controversial (9). At a minimum, inhibition of p38 using SB203580 did not affect the nuclear import or nuclear export (data not shown), excluding p38 in the tonicity regulation of the NLS of TonEBP. The tonicity-regulated NLS described here provides a paradigm for understanding the osmosensing mechanism unique to the vertebrates.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK61677. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NES, nuclear export signal; NLS, nuclear localization signal; GFP, green fluorescent protein; 3GFP, GFP trimer; PBS, phosphate-buffered saline; DIM, dimer interface mutant; DBM, DNA binding mutant; LMB, leptomycin B.
References
- 1.Miyakawa, H., Woo, S. K., Dahl, S. C., Handler, J. S., and Kwon, H. M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2538–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graef, I. S., Gastier, J. M., Francke, U., and Crabtree, G. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 5740–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Rodriguez, C., Aramburu, J., Jin, L., Rakeman, A. S., Michino, M., and Rao, A. (2001) Immunity 15 47–58 [DOI] [PubMed] [Google Scholar]
- 4.Go, W. Y., Liu, X., Roti, M. A., Liu, F., and Ho, S. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10673–10678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranjbar, S., Tsytsykova, A. V., Lee, S. K., Rajsbaum, R., Falvo, J. V., Lieberman, J., Shankar, P., and Goldfeld, A. E. (2006) PLoS Pathogens 2 e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jauliac, S., López-Rodriguez, C., Shaw, L. M., Brown, L. F., Rao, A., and Toker, A. (2002) Nat. Cell Biol. 4 540–544 [DOI] [PubMed] [Google Scholar]
- 7.Hasler, U., Jeon, U. S., Kim, J. A., Mordasini, D. Kwon, H. M., Feraille, E., and Martin, P. Y. (2006) J. Am. Soc. Nephrol 17 1521–1531 [DOI] [PubMed] [Google Scholar]
- 8.Nakayama, Y., Peng, T., Sands, J. M., and Bagnasco, S. M. (2000) J. Biol. Chem. 275 38275–38280 [DOI] [PubMed] [Google Scholar]
- 9.Jeon, U. S., Kim, J. A., Sheen, M. R., and Kwon, H. M. (2006) Acta Physiol. 187 241–247 [DOI] [PubMed] [Google Scholar]
- 10.Lam, A. K. M., Ko, B. C. B., Tam, S., Morris, R., Yang, J. Y., Chung, S. K., and Chung, S. S. M. (2004) J. Biol. Chem. 279 48048–48054 [DOI] [PubMed] [Google Scholar]
- 11.Lopez-Rodriguez, C., Antos, C. L., Shelton, J. M., Richardson, J. A., Lin, F., Novobrantseva, T. I., Bronson, R. T., Igarashi, P., Rao, A., and Olson, E. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2392–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo, S. K., Dahl, S. C., Handler, J. S., and Kwon, H. M. (2000) Am. J. Physiol. 278 F1006–F1012 [DOI] [PubMed] [Google Scholar]
- 13.Dahl, S. C., Handler, J. S., and Kwon, H. M. (2001) Am. J. Physiol. 280 C248–C253 [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. D., Colla, E., Sheen, M. R., Na, K. Y., and Kwon, H. M. (2003) J. Biol. Chem. 278 47571–47577 [DOI] [PubMed] [Google Scholar]
- 15.Cha, J. H., Woo, S. K., Han, K. H., Kim, Y. H., Handler, J. S., Kim, J., and Kwon, H. M. (2001) J. Am. Soc. Nephrol. 12 2221–2230 [DOI] [PubMed] [Google Scholar]
- 16.Jeon, U. S., Han, K.-H., Park, S. H., Lee, S. D., Sheen, M. R., Jung, J.-Y., Kim, W. Y., Sands, J. M., and Kwon, H. M. (2007) Am. J. Physiol. 293, F408–F415 [DOI] [PubMed] [Google Scholar]
- 17.Lim, S. W., Ahn, K. O., Sheen, M. R., Jeon, U. S., Kim, J., Yang, C. W., and Kwon, H. M. (2007) J. Am. Soc. Nephrol. 18 421–429 [DOI] [PubMed] [Google Scholar]
- 18.Loyher, M. L., Mutin, M., Woo, S. K., Kwon, H. M., and Tapaz, M. L. (2004) Neuroscience 124 89–104 [DOI] [PubMed] [Google Scholar]
- 19.Maallem, S., Mutin, M., Kwon, H. M., and Tappaz, M, (2006) Neuroscience 137 51–71 [DOI] [PubMed] [Google Scholar]
- 20.Tong, E. H. Y., Guo, J.-J., Huang, A.-L., Liu, H., Hu, C.-D., Chung, S. S. M., and Ko, B. C. B. (2006) J. Biol. Chem. 281 23870–23879 [DOI] [PubMed] [Google Scholar]
- 21.Maouyo, D., Kim, J. Y., Lee, S. D., Wu, Y., Woo, S. K., and Kwon, H. M. (2002) Am. J. Physiol. 282 F802–F809 [DOI] [PubMed] [Google Scholar]
- 22.Henderson, B. R., and Eleftheriou, A. (2000) Exp. Cell Res. 256 213–224 [DOI] [PubMed] [Google Scholar]
- 23.Nakielny, S., and Dreyfuss, G. (1999) Cell 99 677–690 [DOI] [PubMed] [Google Scholar]
- 24.Zhu, J., and McKeon, F. (1999) Nature 398 256–260 [DOI] [PubMed] [Google Scholar]
- 25.Stroud, J. C., López-Rodriguez, C., Rao, A., and Chen, L. (2002) Nat. Struct. Biol. 9 90–94 [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. D., Woo, S. K., and Kwon, H. M. (2002) Biochem. Biophys. Res. Commun. 294 968–975 [DOI] [PubMed] [Google Scholar]
- 27.Mattaj, I. W., and Englmeier, L. (1998) Annu. Rev. Biochem. 67 265–306 [DOI] [PubMed] [Google Scholar]
- 28.Kalderon, D., Roberts, B. L., Richardson, W. D., and Smith, A. E. (1984) Cell 39 499–509 [DOI] [PubMed] [Google Scholar]
- 29.Nakahara, S., Oka, N., Wang, Y., Hogan, V., Inohara, H., and Raz, A. (2006) Cancer Res. 66 9995–10006 [DOI] [PubMed] [Google Scholar]
- 30.Ossareh-Nazari, B., Bachelerie, F., and Dargemont, C. (1997) Science 278 141–144 [DOI] [PubMed] [Google Scholar]
- 31.Beg, A. A., and Baldwin, A. S., Jr. (1993) Genes Dev. 7 2064–2070 [DOI] [PubMed] [Google Scholar]
- 32.Huxford, T., Huang, D. B., Malek, S., and Ghosh, G. (1998) Cell 96 759–770 [DOI] [PubMed] [Google Scholar]
- 33.Beals, C. R., Clipstone, N. A., Ho, S. N., and Crabtree, G. R. (1997) Genes Dev. 11 824–834 [DOI] [PubMed] [Google Scholar]
- 34.Zhu, H., Shibasaki, F., Price, R., Guilemot, J., Yano, T., Dötsch, V., Wagner, G., Ferrara, P., and McKeon, F. (1998) Cell 93 851–861 [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Z., Ferraris, J. D., Irarrazabal, C. E., Dmitrieva, N. I., Park, J. H., and Burg, M. B. (2005) Am. J. Physiol. 289 F506–F511 [DOI] [PubMed] [Google Scholar]
- 36.Yoshida, T., Qin, L., Egger, L. A., and Inouye, M. (2006) J. Biol. Chem. 281 17114–17123 [DOI] [PubMed] [Google Scholar]
- 37.Posas, F., Wurgler-Murphy, S. M., Maeda, T., Witten, E. A., Thai, T. C., and Saito, H. (1996) Cell 86 865–875 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.