Abstract
The BCL6 oncogenic transcriptional repressor is required for development of germinal center centroblasts, which undergo simultaneous genetic recombination and massive clonal expansion. Although BCL6 is required for survival of centroblasts, its expression in earlier B-cells is toxic. Understanding these opposing effects could provide critical insight into normal B-cell biology and lymphomagenesis. We examined the transcriptional and biological effects of BCL6 in various primary cells. BCL6 repression of ATR was previously shown to play a critical role in the centroblast phenotype. Likewise, we found that BCL6 could impose an ATR-dependent phenotype of attenuated DNA damage sensing and repair in primary fibroblasts and B-cells. BCL6 induced true genomic instability because DNA repair was delayed and was qualitatively impaired, which could be critical for BCL6-induced lymphomagenesis. Although BCL6 can directly repress TP53 in centroblasts, BCL6 induced TP53 expression in primary fibroblasts and B-cells, and these cells underwent p53-dependent growth arrest and senescence in the presence of physiological levels of BCL6. This differential ability to trigger a functional p53 response explains at least in part the different biological response to BCL6 expression in centroblasts versus other cells. The data suggest that targeted re-activation of TP53 could be of therapeutic value in centroblast-derived lymphomas.
BCL65 (B-cell lymphoma 6) is a transcriptional repressor that belongs to the BTB/POZ (bric-á-brac, tramtrack, broad complex/Pox virus zinc finger) zinc finger family proteins. BCL6 is the most commonly involved proto-oncogene in diffuse large B-cell lymphomas (DLBCL) where it is often constitutively expressed through translocations, point mutations, and other mechanisms (1). In normal B-cell development, BCL6 expression is limited to germinal center B-cells (2). After T-cell-dependent antigenic stimulation, B-cells migrate to lymphoid follicles where they become centroblasts and form germinal centers. At this stage B-cells up-regulate BCL6 and undergo simultaneous clonal expansion and somatic hypermutation (2). Somatic hypermutation affects mainly the immunoglobulin loci but also mutates many other genomic loci including that of BCL6 (3).
Centroblasts have the unique ability to tolerate this state of proliferation and genomic instability due in part to BCL6-mediated repression of the ATR (ataxia telangiectasia and Rad3 related) (4), TP53 and p21 genes (5, 6). ATR is a master regulator of genomic damage sensing during replication, and its inactivation permits cells to transit through S-phase without undergoing arrest or triggering apoptosis (7, 8). Centroblasts and DLBCL cells exhibit a relative impairment in their ability to recognize and repair DNA damage, phosphorylate H2AX, and activate checkpoint pathways in response to DNA damage (4). Depletion of BCL6 in centroblasts and DLBCL cells re-activated ATR and reversed this phenotype (4). Conversely, ectopic expression of BCL6 in naïve B-cells repressed ATR and attenuated DNA damage sensing (4). The p53 transcription factor can induce cell cycle arrest, senescence, and cell death. Because DNA damage and genomic instability can induce TP53 expression and its downstream effects, BCL6 repression of TP53 almost certainly plays a critical role in facilitating the survival and proliferation of germinal center cells (5). Accordingly, BCL6 could protect lymphoma cells from p53 effects in response to DNA damage (5), and p53 blockade could partially alleviate the cell death that occurs with inhibition of BCL6.6
Collectively, these data suggest a model whereby BCL6 facilitates the germinal center B-cell phenotype by promoting cell survival and proliferation. However, contrasting data suggest an opposite role for BCL6 in inducing apoptosis and promoting cell cycle arrest. For example, expression of BCL6 in certain transformed or cancer cell lines such as HeLa, CV1, U20S, and others can induce cell death and growth arrest (10–12). Furthermore, BCL6 could immortalize Tp53-deficient but not wild type primary populations mixed murine splenic B-cells (13), raising questions regarding the physiological contribution of TP53 repression to lymphomagenesis and whether BCL6 regulation of target genes such as ATR and TP53 might be context specific in the germinal center but not other primary cells.
To determine whether BCL6-mediated repression of ATR and TP53 are intrinsic to BCL6 or to the centroblast milieu, we examined the phenotypic effects of BCL6 in non-centroblast cells: normal human fibroblasts and primary human tonsilar naïve B-cells. In these cells BCL6 could repress ATR and attenuate DNA damage sensing similar to its effect in centroblasts and DLBCL. However, in contrast to centroblasts these cells were unable to evade the p53 response to BCL6 including growth arrest and senescence. These effects were p53 dependent because they did not occur in Tp53-/- cells. Taken together these data indicate that when B-cells transit through the germinal center they acquire the ability to evade p53 checkpoint activity, which is not entirely dependent on the repressor activity BCL6 and mediated by some as of yet unknown factor. This allows the actions of BCL6 on DNA damage sensing and replication to proceed unopposed so that affinity maturation can proceed in these cells and presumably contributes to lymphomagenesis when BCL6 expression is constitutively sustained.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, Transfection, and Lentivirus Infection—WI-38 cells were grown in Dulbecco's modified Eagle's medium + 10% of fetal bovine serum + 1% sodium pyruvate. Primary mouse embryonic fibroblasts (MEFs) were maintained in Dulbecco's modified Eagle's medium + 15% of fetal bovine serum. BCL6-GFP and GFP were expressed by FUGW lentiviral transduction as in Ref. 4. A multiplicity of infection of 1.5 could infect human and murine fibroblasts with >95% efficiency. Infection was validated by fluorescence microscopy and FACS. For ATR rescue experiments WI-38 cells were transfected with pBJF-FLAG-ATR (ATRwt) or ATRkd (ATR kinase dead), a gift of Drs. Stuart Schreiber and Karlene Cimprich (14), using Amaxa electroporation. Primary MEFs were obtained after removing the uteri and the amnion, yolk sac, placenta, head, and liver from day 14.5 embryos, followed by mincing, incubation with trypsin-EDTA, and establishment of single cell suspensions.
Isolation of Primary B-cells—Human tonsils were obtained from routine tonsillectomies. Tissue was anonymous and obtained if not required for pathologic evaluation, with approval of the Institutional Review Board in accordance with the Helsinki protocols. After mincing, tonsilar mononuclear cells were isolated by HISTOPAQUE®-1077 (Sigma) density centrifugation. Naïve B-cells were isolated by magnetic cell separation using the MIdiMACS system (Miltenyi Biotec) following published protocols (15) and purity determined by FACScan (BD Biosciences) analysis. Naïve B cells were checked for IgD+, CD38low, and CD27-. Antibodies used for FACS analysis were: anti-IgD-FITC, CD27-FITC, and CD38-PE (Pharmingen). A multiplicity of infection of 3.4 for the GFP and GFP-BCL6 lentivirus could infect human naïve B-cells with 80–90% efficiency as reported in Ref. 4.
Reporter Assay—293T were transfected with 100 ng of a (GAL4)5-TK-LUC reporter construct, 50 ng of GFP expressing or BCL6-GFP expressing lentiviral backbone plasmids, and a Renilla reporter as internal control using Superfect (Qiagen). After 48 h the cells were lysed and submitted to dual luciferase assays (Promega) using a POLARstar Optima (BMG Labtechnologies) luminometer.
Chromatin Immunoprecipitation—Chromatin immunoprecipitation assays were performed in Toledo cells transduced with GFP or GFP-BCL6 lentivirus using BCL6 (N-3, Santa Cruz) or actin (Santa Cruz) rabbit polyclonal antibodies and our standard BCL6 chromatin immunoprecipitation protocol (4). The percent enrichment with respect to the input of the BCL6 binding site from the CCL3 promoter was detected by performing quantitative PCR using an Opticon 2 thermal cycler (MJ Research) and the Qiagen SYBR Green Kit (Qiagen). The CCL3 primers were previously reported (16).
DNA Damage Induction—5 Gy of γ-radiation were administered using a Shepherd Mark I, irradiator. Culture medium was changed after radiation and cells were maintained at 37 °C until they were harvested for the corresponding experiments.
H2A.X Phosphorylation Assay—2 h after DNA damage, cells were resuspended in permeabilization solution (0.5% saponin, 10 mm HEPES, pH 7.4, 0.14 m NaCl, 2.5 mm CaCl2) stained for P-Ser139-H2A.X (referred to as γH2A.X) at a density of 2 × 106/ml cells using a rabbit polyclonal αP-Ser139-H2A.X primary antibody (Upstate) and Cy3-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch) followed by FACscan analysis (BD Bioscience).
Comet Assay—Cells were diluted to a density of 2.5 × 104 cells/ml. Microscope slides were pre-coated with 1% type IA agarose and 0.5 ml of cells mixed with 1 ml of 1% type-VII agarose and spread over a pre-coated slide in quadruplicate. A coverslip was added and agarose was allowed to solidify. Coverslips were removed and slides were placed in lysis solution (100 mm disodium EDTA, 2.5 m NaCl, 10 mm Tris-HCl, pH 10.0 containing 1% Triton X-100 at 4 °C and incubated for 25 min; washed three times in ice-cold water for 15 min, transferred to an electrophoresis tank containing ice-cold alkaline solution (50 mm NaOH, 1 mm disodium EDTA, pH 12.5), and incubated for 45 min. Electrophoresis was carried out for 25 min at 18 V (0.5 V/cm) and 250 mA. Slides were removed and 1 ml of neutralizing solution (0.5 m Tris-HCl, pH 7.5) was added and incubated for 10 min. Slides were then rinsed twice in phosphate-buffered saline, stained with ethidium bromide (2.5 μg/ml), covered with a coverslip and placed at 4 °C overnight. Comets were analyzed using a NIKON DIAPHOT TMD epifluorescent microscope at ×20 magnification, 50–100 cells were analyzed per slide using KOMET-5 Assay software (Kinetic Imaging). Olive tail moment was recorded and the standard deviation calculated using Excel.
Cell Cycle Profile Analysis—1 × 106 cells were fixed in cold 70% ethanol, treated with ribonuclease A (Sigma) for 30 min at 37 °C (100 units), and stained with propidium iodide (20 μg/ml, Invitrogen) and analyzed by FACScan (BD Biosciences) using CellQuest and ModFit software.
Real-time PCR—RNA was prepared using TRIzol (Invitrogen). Superscript III First strand cDNA synthesis kit (Invitrogen) was used to prepare cDNA. Amplification was performed using QuantiTect SYBR® Green PCR kit (Qiagen). Gene expression was normalized to hypoxanthine-guanine phosphoribosyltransferase and data indicated as -fold change relative to corresponding controls using the 2-ΔΔCT method (for primers, see supplemental Table 1).
Western Blot—10 × 106 infected and control cells were lysed in TNG buffer (50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 50 mm β-glycerophosphate, 1% Tween 20, 0.2% Nonidet P-40 containing protease inhibitors). Primary antibodies used were: αBCL6 rabbit polyclonal (N-3) (Santa Cruz) (1:100 dilution), αATR rabbit polyclonal (SEROTEC, Oxford, UK) (1:100 dilution), αp21 rabbit polyclonal (Santa Cruz) (1:100 dilution), αCyclin D1 rabbit polyclonal (1:500 dilution) (Cell Signaling), and α-horseradish peroxidase-actin conjugated as a loading control (Santa Cruz) (1:1000 dilution). Secondary antibodies used were: peroxidase-conjugated goat anti-mouse (115-035-003) and anti-rabbit (111-035-003) 1:7000 (Jackson ImmunoResearch).
Viability and Apoptosis Assays—1 × 106 infected cells and controls were resuspended in 25 μl of culture medium and 2 μl of dye mixture (100 μgml-1 acridine orange plus 100 μgml-1 ethidium bromide in phosphate-buffered saline) and visualized by fluorescence microscopy. At least 200 cells counted were examined for apoptotic and/or necrotic cells.
Growth Cell Curves—Human and murine fibroblasts were plated in triplicates at a density of 200,000 cells. Counting was performed at the indicated time points using trypan blue. The cell growth curves presented are representative of three independent experiments.
Senescence Assay—β-Galactosidase activity as a biomarker for cellular senescence (17) was assayed using the senescence β-galactosidase staining kit (Cell Signaling) following the manufacturer's instructions.
Plasmid Misrepair and Deletion Assays—Nuclear cell extracts were prepared from 106 wild type naïve B-cells, FUGW-GFP, and BCL6 infected using the CelLytic NuCLEAR Extraction kit (Sigma). The pUC18 plasmid (Invitrogen) was linearized by introducing DNA double strand breaks (DSB) within the LacZ gene using the EcoRI restriction enzyme (MBI Fermentes) and dephosphorylated with calf intestine alkaline phosphatase (Promega). For the assay, 2 μg of EcoRI linearized pUC18 were incubated with 3 mg of nuclear extract. Reactions (10 μl) were carried out in 50 mm triethanolamine-HCl (pH 7.5), 60 mm KOAc, 50 μm deoxynucleotide triphosphates, 2 mm ATP, 1 mm dithiothreitol, and 100 μg/ml bovine serum albumin. The mixture was incubated for 24 h at 18 °C. Plasmid DNA was purified from extract by passing it down a filter column (Qiagen-Minelute) and then diluted 3-fold in 10 mm Tris (pH 8), -1 mm EDTA buffer and 10 ng were used to transform Escherichia coli strain DH5. Transformed bacteria were plated out on Luria-Bertani (LB) agar plates, including 100 g/ml ampicillin, 20 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal), and 200 mg/ml isopropyl 1-thio-d-galactopyranoside. To allow for spontaneous rejoining/incomplete EcoRI cutting, assay controls were conducted without nuclear extract. The number of colonies generated was subtracted from extract-treated plasmid colonies. To correct for bacterial plating numbers and determine whether nuclease activity was affecting plasmid efficacy, cells were plated on LB agar without ampicillin. Primers around the EcoRI site were designed to give a PCR product of 628 bp corresponding to nucleotides 150–777. Colony PCR was performed on white colonies (misrepaired) to determine the size of the deletion, compared with the blue (correctly repaired) colony controls.
RESULTS
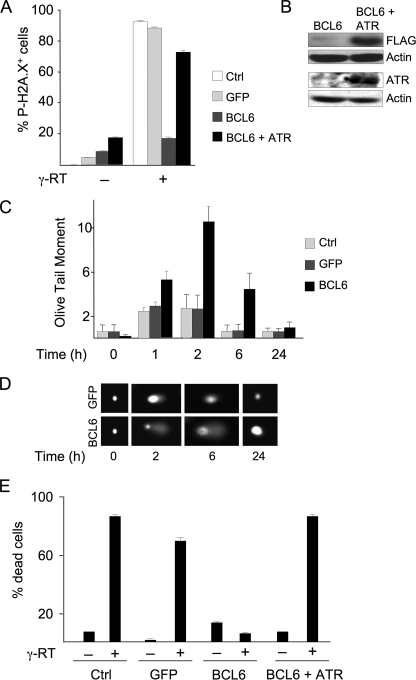
BCL6 Attenuates DNA Damage Sensing and Repair in Normal Diploid Fibroblasts—To determine the phenotype induced by BCL6 in non-centroblast cells we delivered a GFP-tagged form of BCL6 to WI-38 human diploid fibroblasts by lentiviral transduction (versus GFP control). GFP-BCL6 can repress a BCL6 reporter construct and bind to a BCL6 target gene (supplemental Fig. S1, A and B). The GFP-BCL6 fusion protein expressed by this construct was also previously shown to be fully biologically functional comparable with wild type BCL6 (4, 18). Transduced WI-38 cells expressed BCL6-GFP at physiological levels as evidenced by its microspeckled nuclear staining similar to that of endogenous BCL6 (Fig. 1, A and B). The level of BCL6 protein expression is fairly similar (although slightly lower) than endogenous BCL6 expression detected in primary human centroblasts (supplemental Fig. S1C). BCL6-GFP repressed ATR at both the mRNA and protein levels (Fig. 1, C and D). As a consequence, BCL6-transduced WI-38 cells displayed an attenuated ability to sense DNA damage. Damage sensing in response to 5 Gy γ-radiation was detected by measuring phosphorylation of H2A.X on serine 139, which is mediated in part by ATR and plays an important role in recognizing and repairing DNA DSBs (19). Control cells exhibited 90–95% H2A.X phosphorylation after γ-radiation, whereas BCL6-transduced cells only displayed 20% phosphorylation (Fig. 2A). The effect was at least partly ATR-dependent because H2AX phosphorylation was rescued by expressing ATR together with BCL6. Expression of ATR expression in the transfected cells was confirmed by immunoblotting (Fig. 2B). To determine whether defective damage sensing led to a delay in DNA repair we performed Comet assays, a method that can quantify DNA DSBs at the level of a single cell. The extent of DNA damage was examined over a 24-h time course after γ-radiation. BCL6-transduced cells displayed delayed repair at 1, 2, and 6 h and returned to normal after 24 h (Fig. 2, C and D). Extensive DNA damage triggers cell death through ATR and other pathways. However, BCL6 transduction allowed damaged WI-38 cells to survive, even while exhibiting considerable DNA breakage as evidence by the Comet assay (Fig. 2E). Expression of ATR restored the ability of BCL6-transduced cells to undergo cell death. Taken together, expression of BCL6 in normal fibroblasts represses ATR and emulates the DNA damage attenuation phenotype of centroblasts.
FIGURE 1.
BCL6 represses ATR in normal human fibroblasts. WI-38 normal diploid fibroblasts were transduced with BCL6-GFP or GFP expressing lentivirus. BCL6 was detected by quantitative PCR (A), represented as -fold increase in BCL6 compared with untransduced cells, and microscopy (B), showing phase contrast for untransduced cells and fluorescence for GFP and BCL6 GFP-transduced cells. All BCL6-GFP transductions are referred to as “BCL6.” C and D show quantitative PCR and Western blots performed to detect ATR mRNA and protein abundance, respectively. D includes a BCL6 Western blot to demonstrate the amount of BCL6 protein and actin as loading control. Ctrl refers to un-transduced WI-38 cells.
FIGURE 2.
BCL6 attenuates DNA damage response in fibroblasts in an ATR-dependent manner. A, WI-38 fibroblasts were transduced as indicated, harvested before and after 5 Gy γ-radiation, and submitted to FACS to detect the percentage of cells staining for Ser139 phosphorylation of H2A.X. B, immunoblots were performed in BCL6-transduced WI-38 cells transfected with either pCDNA or pCDNA-FLAG-ATR. The top immunoblot was performed with FLAG antibody (because ATR is FLAG tagged) and the bottom, a Western blot with ATR antibody. The data confirm that ATR was expressed in ATR-transfected cells. C, repair of DSBs was detected in transduced WI-38 cells at the indicated time points before and after 5 Gy γ-radiation. Quantitative Comet assays were performed in quadruplicate and the mean ± S.D. calculated. The y axis represents the abundance of DNA double strand breaks in arbitrary (Olive tail moment) units. D, representative photomicrographs of single cells, stained with ethidium bromide from the Comet assay experiment of panel B. E, WI-38 cells were transduced as indicated and ATR transfected 24 h later. The percentage of dead cells were evaluated by acridine orange/ethidium bromide staining 24 h after 5 Gyγ-radiation (γ-RT; +) or after no radiation (-). Ctrl refers to untransduced WI-38 cells.
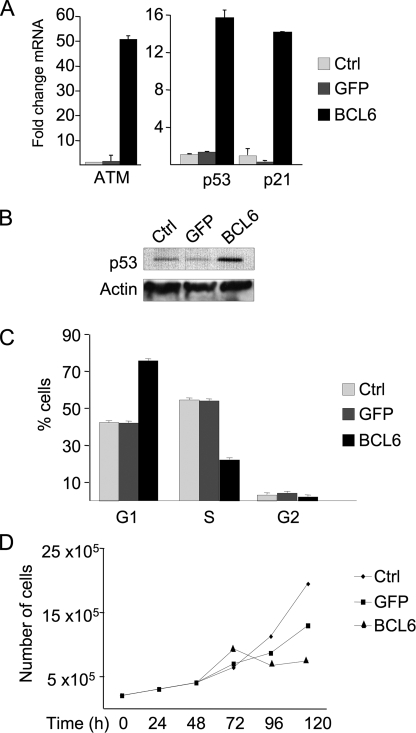
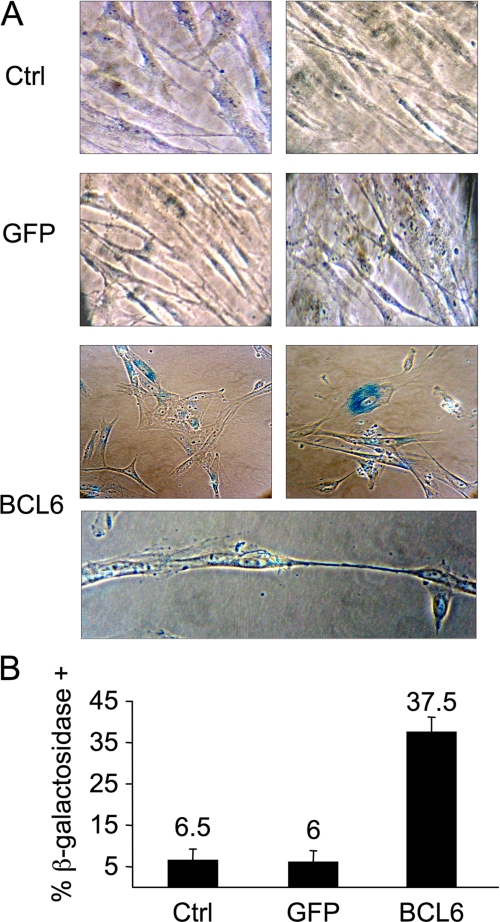
BCL6 Triggers Growth Suppression and Senescence in Normal Diploid Fibroblasts—In centroblasts expression of BCL6 facilitates sustained proliferation despite mutagenesis occurring as a byproduct of affinity maturation. To determine whether BCL6 mediate this effect outside of the centroblast context we examined its impact on cell checkpoints and proliferation. mRNA extracted from BCL6-transduced or control cells was examined for transcript abundance of ATM, TP53, and its downstream target P21. Although BCL6 can repress TP53 and P21 in germinal center and lymphoma cells, expression of BCL6 in WI-38 cells induced ATM 50-fold and TP53 and p21 about 15-fold (Fig. 3A), suggesting that these cells mount an oncogenic stress response (20) able to overcome BCL6 repressor effects. p53 protein levels were correspondingly increased in the presence of BCL6 (Fig. 3B). ATM protein levels were also increased after BCL6 transduction (supplemental Fig. S1D). Consistent with the ability of p53 to induce a G1 arrest, BCL6-transduced WI-38 cells exhibited an increase in the G1 phase fraction from 74 versus 42% in control cells (Fig. 3C); and a progressive reduction in their proliferative rate (Fig. 3D) that could not be overcome by increasing serum concentration or passage in culture (data not shown). Also, certain morphological features, such as cytoplasmic vacuoles and enlarged cell size and flattening were observed. These changes were suggestive of cellular senescence, which was further confirmed by detection of pH-dependent β-galactosidase levels (Fig. 4A). Altogether, within 8 days, 37.5% of BCL6-transduced WI-38 cells displayed evidence of senescence as compared with 6% of GFP transduced or untransduced cells (Fig. 4B). Therefore, the effects of BCL6 can be physiologically countered by a response that includes induction of TP53 and cellular senescence in human diploid fibroblasts.
FIGURE 3.
BCL6 triggers a p53 response and growth arrest in WI-38 cells. A, quantitative PCR was performed in WI-38 fibroblasts 48 h after transduction as indicated to determine the -fold change in transcript abundance for ATM, TP53, and P21, expressed relative to untransduced cells. B, Western blot was performed to detect the amount of p53 protein, as compared with actin loading control. C, the percent of cells in G1, S, and G2/M phase was determined by propidium iodide FACS. D, the proliferation of WI-38 cells was assessed over a 5-day time course at the indicated time points. Ctrl refers to untransduced WI-38 cells.
FIGURE 4.
BCL6 induces senescence in WI-38 cells. A, light microscopy of WI-38 cells stained for acid β-galactosidase activity 8 days after transduction with BCL6-GFP or GFP. B, quantification of percent of cells positive for acid β-galactosidase activity (i.e. senescent cells). Ctrl refers to untransduced WI-38 cells.
Induction of Senescence by BCL6 Is Dependent on p53—We wondered whether BCL6-induced senescence was dependent on p53. Therefore, we compared and contrasted the effects of BCL6 in freshly prepared Tp53+/+ MEFs versus Tp53-/- MEFs. As in WI-38 cells, transduction of BCL6 in normal MEFs resulted in proliferative arrest within 72–96 h (Fig. 5A). In contrast, transduction of BCL6 in Tp53-/- MEFs failed to provoke growth arrest (Fig. 5B). As before, growth arrest was associated with cellular senescence, because 52% of MEFs were β-galactosidase positive after 8 days as compared with 1% of controls (Fig. 5, C and E). Senescence was largely p53-dependent because no more than 10% of Tp53-/- MEFs became positive for β-galactosidase (Fig. 5, D and E). Like WI-38 cells, Tp53+/+ MEFs displayed up-regulation of Tp53 upon transduction of BCL6 (Fig. 5F). Not only did BCL6-transduced Tp53-/- MEFs maintain their proliferative potential, but also acquired the ability to form colonies on tissue culture plates in contrast to control transduced cells (Fig. 5G). These data indicate that primary non-germinal center cells can trigger a p53-dependent cellular senescence response capable of overcoming the oncogenic effects of BCL6.
FIGURE 5.
Induction of senescence in fibroblasts is dependent on p53. A and B, the proliferation of Tp53+/+ and Tp53-/- MEFs, respectively, transduced as indicated was measured over an 8-day period. C and D, quantification of percent Tp53+/+ and Tp53-/- MEFs cells, respectively, positive for acid β-galactosidase activity (i.e. senescent cells) after 10 days. E, representative photomicrographs of MEFs from panel B stained for acid β-galactosidase activity. F, Western blots were performed to determine protein abundance of BCL6 and p53 in wild type MEFs transduced as indicated. G, microscopic images of Tp53-/- MEFs transduced as indicated after 10 days. Upper panels are phase-contrast and lower panels are fluorescence microscopy to determine growth patterns of transduced cells as indicated after 10 days. Ctrl refers to untransduced MEFs.
BCL6 Induces a p53 Response in Primary Human Naïve B-cells—We next examined whether this biological phenomenon was relevant to the context of primary B-cells. We isolated human primary tonsilar naïve B-cells, which include B-cells that have not yet entered into the germinal center reaction and have not yet up-regulated BCL6. Transduction of BCL6 silenced expression of ATR in these cells (Fig. 6A), and we previously showed that BCL6 could attenuate DNA damage sensing as detected by H2AX phosphorylation and Comet assays (4). However, just as in fibroblasts, BCL6 transduction powerfully induced TP53 mRNA and protein up-regulation as well as P21 (Fig. 6, A and B). To determine whether TP53 induction was associated with a similar growth suppression response, we examined the growth curves of naïve B-cells in culture after BCL6 transduction. Once removed from tonsils and cultured under standard conditions, naïve B-cells initially proliferate but within 48 h subsequently gradually decline in number (Fig. 6C). Transduction of BCL6 suppressed proliferation of naïve B-cells and led to a sharp decline in cell numbers compared with control within 4 days. Part of the reason for this reduction in numbers was loss of viability induced by BCL6 transduction (Fig. 6D). These results suggest that normal mature B-cells as well as fibroblasts react to BCL6-mediated attenuation of DNA damage sensing by triggering a p53-associated growth suppressive reaction.
FIGURE 6.
BCL6 induces p53 response and growth arrest in naïve B-cells. Purified human tonsilar naïve B-cells were transduced with BCL6-GFP or GFP lentivirus. A, Western blots were performed to detect abundance of BCL6, ATR, and p53 in naïve B-cells 48 h after transduction as indicated. B, quantitative PCR was performed to detect ATM, TP53, and P21 mRNA transcript -fold change after BCL6-GFP versus GFP lentiviral transduction. C, naïve B-cell proliferation was determined at various time points over a 96-h time course after transduction with BCL6-GFP or GFP lentivirus versus control cells. D, the percent of dead cells as determined by ethidium bromide acridine orange staining is shown 48 h after the indicated transductions. Ctrl refers to untransduced naïve B-cells.
Altered regulation of TP53 and ATR could potentially affect the ability of cells to properly repair DNA damage. To determine in fact whether BCL6 causes misrepair, we directly measured the quality of DNA repair of DNA DSB in the nuclear extracts of BCL6-transduced naïve B-cells. We used an in vitro assay for end-joining, one of the main pathways for the repair of DSB in mammalian cells. We exposed a linearized LacZ containing plasmid to cell extracts of naïve B-cells with or without transduced BCL6 or GFP and compared and contrasted the ability of these extracts to repair the plasmid. BCL6 expressing naïve B-cell extracts exhibited a 2-fold increase in misrepair, as evidenced by loss of LacZ activity upon plasmid re-ligation and transformation to E. coli (Fig. 7A). There was also a 2-fold increase in large deletions versus control cells (Fig. 7B). These data indicate that BCL6 not only attenuates DNA damage sensing and delays repair, but that the presence of BCL6 also affects repair qualitatively. These effects of BCL6 are not tolerated by primary B-cells, except upon initiation of the germinal center reaction, whereupon they must acquire the ability to at least transiently facilitate BCL6-mediated repression of p53 responses.
FIGURE 7.
BCL6 causes misrepair and deletion of double-strand breaks. Cellular extracts were obtained from naïve B-cells transduced with BCL6, GFP, or mock transduced. A linearized LacZ containing plasmid was incubated in these extracts, and then recovered and analyzed by blue/white colony forming assays and PCR. The left panel shows the percent of colonies that have lost the ability to express functional LacZ, which has doubled in the BCL6 expressing cells. The right panel indicates the percentage of recovered plasmids that have undergone extensive deletions, which again has doubled in the presence of BCL6. Ctrl refers to untransduced naïve B-cells.
DISCUSSION
This work explores the effect of cellular context on the activities of the BCL6 transcriptional repressor. BCL6 was reported to play a critical role in survival and proliferation of centroblasts at least in part by repressing TP53 and P21 to evade cellular checkpoints triggered in germinal center B-cells by rapid proliferation and affinity maturation (5, 6). BCL6 also contributes to this effect by directly repressing ATR, resulting in attenuation of DNA damage sensing and repair (4). BCL6 can bind to several other DNA damage response genes including CHEK1, although their contribution to BCL6 biological actions remains unknown (16). Peptide inhibitors of BCL6 induce TP53 and ATR in lymphoma cells, resulting in cell death and showing that maintenance of BCL6 is required for survival of lymphoma cells (21). In contrast, other groups have reported that BCL6 mediates induction of apoptosis and growth arrest in other cell types (as reviewed in Ref. 22). To understand why BCL6 yields opposing effects in these different cellular contexts we examined the effect of BCL6 in primary non-GC cells using similar assays and conditions as recently reported in centroblasts and lymphoma cells (4).
Herein we show that BCL6 can repress ATR in both normal diploid fibroblasts and naïve B-cells. Similar to centroblasts, this leads to attenuation of DNA damage sensing (H2AX phosphorylation), as well as quantitative (Comet assay) and qualitative (plasmid ligation assay) defects in DNA repair after exposure to genotoxic damage. One way that cells respond to genotoxic stress or to oncogenes like constitutively active RAS and MYC is by triggering checkpoint responses, in which p53 plays a critical role (23). Such responses can direct cells to exit the cell cycle and undergo senescence or cell death. An ATM-p53 pathway was recently shown to play a critical role in the early response of cells to oncogenes that could induce genomic instability (20). BCL6 can evade these checkpoints in centroblasts by repressing TP53. However, our results suggest a more complex scenario, because expression of BCL6 in normal fibroblasts resulted in the opposite effect, with up-regulation of ATM, TP53, and P21 and consequent cell cycle arrest and senescence. This effect was dependent on Tp53 because BCL6 failed to trigger senescence in p53 null fibroblasts or primary human naïve B-cells. The fact that both TP53 mRNA and protein abundance were increased in the presence of BCL6 is consistent with previous reports showing that several types of cellular stress can cause a similar effect (24–26). ATM and ATR are not bio-equivalent proteins, and consistent with previous reports (e.g. Ref. 4 and references therein), the presence of ATM does not rescue cells from the effects of BCL6-induced ATR depletion.
These results are consistent with those of Kusam et al. (27) demonstrating that BCL6 could only immortalize primary splenic B-cells in a p53 null background. In contrast, Shvarts et al. (28) reported that BCL6 might be able to bypass p53-induced senescence in primary MEFs and in a mixed population of human B-cells, and that BCL6-mediated induction of cyclin D1 could explain this effect (presumably as a secondary effect because BCL6 is not a transcriptional activator). In our experiments BCL6 could not bypass p53 and did not induce cyclin D1 in primary human fibroblasts, primary MEFs, or purified naïve B-cells (supplemental Fig. S2). One possible explanation for this difference could be BCL6 expression levels, which in this case was similar to physiological levels. Our current results are also consistent with a genetic model where BCL6 expression prior to the germinal center resulted in toxicity and loss of B-cells, similar to the effects we report herein.7 If BCL6 could truly bypass p53 than such mice would instead be expected to develop lymphomas, which is not the case.
Our data demonstrate that the effects of BCL6 on TP53 expression is dependent on the cellular context. In contrast to primary fibroblasts and naïve B-cells, centroblasts fail to mount a functional p53 response. The reason for this is likely multifactorial. First, BCL6 is uniquely able to inhibit or attenuate TP53 induction in the centroblast milieu, because blockade of BCL6 could induce p53 to various degrees (5, 9). This is likely due to direct repression of the TP53 gene by BCL6. BCL6 might also attenuate p53 biological effects by directly repressing p53 targets. For example, the P21 and GADD45A genes are both direct transcriptional targets of BCL6 and p53 (6, 9). Also likely to contribute are defects in p53 activation through post-translational modifications. For example, loss of ATR might impair p53 phosphorylation through the CHEK1 pathway. It seems very likely that additional as of yet unidentified factors or biological pathways active in centroblasts but not in naïve B-cells or fibroblasts contribute to attenuate p53 and facilitate physiological genomic instability mediated by BCL6 (supplemental Fig. S3). Identification of factors that enhance the activity of BCL6 in centroblasts will be critical for understanding how these cells survive affinity maturation. Such factors might include BCL6 interacting corepressor proteins, such as HDAC9 and MTA3, which are expressed preferentially in the germinal center compartment with BCL6 (21, 29, 30). Alternatively, other signaling or transcription factors that do not interact with BCL6 could independently affect factors that might otherwise induce the oncogene response. For example, the BAFF (B cell-activating factor of the tumor necrosis factor family) protein can inhibit p53 in B-cell lymphoma cells (31). Pro-survival effects may not be limited to centroblasts, because BCL6 was reported to maintain viability of differentiating myocytes, although the mechanism in this case is not known (32).
In summary, our data suggest that when BCL6 represses ATR with the consequent defect in DNA damage sensing and repair, normal cells respond by generating a p53 response. It seems that the failure of centroblasts to respond in a similar way explains at least in part the results obtained by different groups in BCL6 ectopic expression systems. The ability of BCL6 to repress both ATR and TP53 probably plays a central role in its tendency to cause DLBCL (33, 34). The likely engine for DNA breaks in centroblasts is their rapid replication in the absence of ATR and the mutagenic effects of AID. The importance of these factors in BCL6-mediated lymphomagenesis is highlighted by a report that loss of AID can attenuate lymphomagenesis in BCL6 constitutively expressing mice (35). These observations could have therapeutic implications. We have developed a specific targeted therapy agent called BPI (BCL6 peptide inhibitor) that blocks the transcriptional repressor activity of BCL6 (9). Agents that rescue the p53 pathway might be expected to enhance the anti-lymphoma activity of BPI. Along these lines we have shown that a p53 activating peptide synergistically kills DLBCL cells when administered in combination with BPI (9). Dissection of B-cell checkpoint pathways could thus lead to improved therapeutic regimens for B-cell lymphomas.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA100885 (to M. C.) and R01 CA104348 (to A. M.) from NCI. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
Footnotes
The abbreviations used are: BCL6, B-cell lymphoma 6; DLBCL, diffuse large B-cell lymphomas; DSB, double strand break; ATR, ataxia telangiectasia- and Rad3-related; GFP, green fluorescent protein; FACS, fluorescence-activated cell sorter; MEF, mouse embryonic fibroblast; Gy, gray.
L. Cerchietti, J. Polo, G. DaSilva, P. Farinha, P. Juszczynski, R. Shaknovich, R. Gascoyne, S. Dowdy, and A. Melnick, unpublished data.
L. Bergsagel, personal communication.
References
- 1.Ye, B. H. (2000) Cancer Investig. 18 356-365 [DOI] [PubMed] [Google Scholar]
- 2.Allman, D., Jain, A., Dent, A., Maile, R. R., Selvaggi, T., Kehry, M. R., and Staudt, L. M. (1996) Blood 87 5257-5268 [PubMed] [Google Scholar]
- 3.Pasqualucci, L., Neumeister, P., Goossens, T., Nanjangud, G., Chaganti, R. S., Kuppers, R., and Dalla-Favera, R. (2001) Nature 412 341-346 [DOI] [PubMed] [Google Scholar]
- 4.Ranuncolo, S. M., Polo, J. M., Dierov, J., Singer, M., Kuo, T., Greally, J., Green, R., Carroll, M., and Melnick, A. (2007) Nat. Immunol. 8 705-714 [DOI] [PubMed] [Google Scholar]
- 5.Phan, R. T., and Dalla-Favera, R. (2004) Nature 432 635-639 [DOI] [PubMed] [Google Scholar]
- 6.Phan, R. T., Saito, M., Basso, K., Niu, H., and Dalla-Favera, R. (2005) Nat. Immunol. 6 1054-1060 [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Muse, T., and Boulton, S. J. (2005) EMBO J. 24 4345-4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shechter, D., and Gautier, J. (2005) Cell Cycle 4 235-238 [PubMed] [Google Scholar]
- 9.Polo, J. M., Dell'Oso, T., Ranuncolo, S. M., Cerchietti, L., Beck, D., Da Silva, G. F., Prive, G. G., Licht, J. D., and Melnick, A. (2004) Nat. Med. 10 1329-1335 [DOI] [PubMed] [Google Scholar]
- 10.Yamochi, T., Kaneita, Y., Akiyama, T., Mori, S., and Moriyama, M. (1999) Oncogene 18 487-494 [DOI] [PubMed] [Google Scholar]
- 11.Albagli, O., Lantoine, D., Quief, S., Quignon, F., Englert, C., Kerckaert, J. P., Montarras, D., Pinset, C., and Lindon, C. (1999) Oncogene 18 5063-5075 [DOI] [PubMed] [Google Scholar]
- 12.Zhang, H., Okada, S., Hatano, M., Okabe, S., and Tokuhisa, T. (2001) Biochim. Biophys. Acta 1540 188-200 [DOI] [PubMed] [Google Scholar]
- 13.Kusam, S., Toney, L. M., Sato, H., and Dent, A. L. (2003) J. Immunol. 170 2435-2441 [DOI] [PubMed] [Google Scholar]
- 14.Cliby, W. A., Roberts, C. J., Cimprich, K. A., Stringer, C. M., Lamb, J. R., Schreiber, S. L., and Friend, S. H. (1998) EMBO J. 17 159-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein, U., Tu, Y., Stolovitzky, G. A., Keller, J. L., Haddad, J., Jr., Miljkovic, V., Cattoretti, G., Califano, A., and Dalla-Favera, R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2639-2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polo, J. M., Juszczynski, P., Monti, S., Cerchietti, L., Ye, K., Greally, J. M., Shipp, M., and Melnick, A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3207-3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dimri, G. P., Lee, X., Basile, G., Acosta, M., Scott, G., Roskelley, C., Medrano, E. E., Linskens, M., Rubelj, I., Pereira-Smith, O., Peacock, M., and Campisi, A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9363-9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo, T. C., Shaffer, A. L., Haddad, J., Jr., Choi, Y. S., Staudt, L. M., and Calame, K. (2007) J. Exp. Med. 204 819-830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Capetillo, O., Lee, A., Nussenzweig, M., and Nussenzweig, A. (2004) DNA Repair 3 959-967 [DOI] [PubMed] [Google Scholar]
- 20.Bartkova, J., Horejsi, Z., Koed, K., Kramer, A., Tort, F., Zieger, K., Guldberg, P., Sehested, M., Nesland, J. M., Lukas, C., Orntoft, T., Lukas, J., and Bartek, J. (2005) Nature 434 864-870 [DOI] [PubMed] [Google Scholar]
- 21.Parekh, S., Polo, J. M., Shaknovich, R., Juszczynski, P., Lev, P., Ranuncolo, S. M., Yin, Y., Klein, U., Cattoretti, G., Dalla Favera, R., Shipp, M. A., and Melnick, A. (2007) Blood 110 2067-2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albagli-Curiel, O. (2003) Oncogene 22 507-516 [DOI] [PubMed] [Google Scholar]
- 23.Schmitt, C. A. (2003) Nat. Rev. Cancer 3 286-295 [DOI] [PubMed] [Google Scholar]
- 24.Takaoka, A., Hayakawa, S., Yanai, H., Stoiber, D., Negishi, H., Kikuchi, H., Sasaki, S., Imai, K., Shibue, T., Honda, K., and Taniguchi, T. (2003) Nature 424 516-523 [DOI] [PubMed] [Google Scholar]
- 25.Boggs, K., and Reisman, D. (2006) Oncogene 25 555-565 [DOI] [PubMed] [Google Scholar]
- 26.Oh, S. M., Pyo, C. W., Kim, Y., and Choi, S. Y. (2004) Oncogene 23 8282-8291 [DOI] [PubMed] [Google Scholar]
- 27.Kusam, S., Vasanwala, F. H., and Dent, A. L. (2004) Oncogene 23 839-844 [DOI] [PubMed] [Google Scholar]
- 28.Shvarts, A., Brummelkamp, T. R., Scheeren, F., Koh, E., Daley, G. Q., Spits, H., and Bernards, R. (2002) Genes Dev. 16 681-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gil, V., Howell, L., Yeung, J., Petrie, K., Smith, A., Sebire, N., Matutes, E., Wotherspoon, A., Enver, T., So, E., and Zelent, A. (2007) Blood 110 118a [Google Scholar]
- 30.Fujita, N., Jaye, D. L., Geigerman, C., Akyildiz, A., Mooney, M. R., Boss, J. M., and Wade, P. A. (2004) Cell 119 75-86 [DOI] [PubMed] [Google Scholar]
- 31.He, B., Chadburn, A., Jou, E., Schattner, E. J., Knowles, D. M., and Cerutti, A. (2004) J. Immunol. 172 3268-3279 [DOI] [PubMed] [Google Scholar]
- 32.Kumagai, T., Miki, T., Kikuchi, M., Fukuda, T., Miyasaka, N., Kamiyama, R., and Hirosawa, S. (1999) Oncogene 18 467-475 [DOI] [PubMed] [Google Scholar]
- 33.Cattoretti, G., Pasqualucci, L., Ballon, G., Tam, W., Nandula, S. V., Shen, Q., Mo, T., Murty, V. V., and Dalla-Favera, R. (2005) Cancer Cell 7 445-455 [DOI] [PubMed] [Google Scholar]
- 34.Baron, B. W., Anastasi, J., Montag, A., Huo, D., Baron, R. M., Karrison, T., Thirman, M. J., Subudhi, S. K., Chin, R. K., Felsher, D. W., Fu, Y. X., McKeithan, T. W., and Baron, J. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14198-14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasqualucci, L., Compagno, M., Mo, T., Smith, P., Morse, H., III, Murty, V., and Dalla-Favera, R. (2008) Nat. Genet. 40 108-112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.