Abstract
In T4 phage, coordinated leading and lagging strand DNA synthesis is carried out by an eight-protein complex termed the replisome. The control of lagging strand DNA synthesis depends on a highly dynamic replisome with several proteins entering and leaving during DNA replication. Here we examine the role of single-stranded binding protein (gp32) in the repetitive cycles of lagging strand synthesis. Removal of the protein-interacting domain of gp32 results in a reduction in the number of primers synthesized and in the efficiency of primer transfer to the polymerase. We find that the primase protein is moderately processive, and this processivity depends on the presence of full-length gp32 at the replication fork. Surprisingly, we find that an increase in the efficiency of primer transfer to the clamp protein correlates with a decrease in the dissociation rate of the primase from the replisome. These findings result in a revised model of lagging strand DNA synthesis where the primase remains as part of the replisome after each successful cycle of Okazaki fragment synthesis. A delay in primer transfer results in an increased probability of the primase dissociating from the replication fork. The interplay between gp32, primase, clamp, and clamp loader dictates the rate and efficiency of primer synthesis, polymerase recycling, and primer transfer to the polymerase.
The T44 replisome has served as a highly useful model system for studying coupled DNA replication (1). The T4 replisome is made up of eight proteins, all of which have counterparts in more complex organisms such as Escherichia coli, Saccharomyces cerevisiae, and humans (2). DNA synthesis is carried out in a 5′ to 3′ direction by T4 DNA polymerase (gp43), which together with the clamp protein (gp45) makes up the holoenzyme complex (3). The holoenzyme can form through several different routes, all dependent on the activity of the clamp loader protein (gp44/62) (4, 5). The clamp loader is an AAA+ protein that uses energy derived from ATP hydrolysis to chaperone the holoenzyme assembly process (6). The T4 primosome moves along the lagging strand DNA template in the 5′ to 3′ direction and is composed of a hexameric helicase (gp41) that unwinds the duplex DNA and an oligomeric primase (gp61) that synthesizes pentaribonucleotide primers at 5′-GTT and 5′-GCT sequences to initiate repetitive Okazaki fragment synthesis (7, 8). The helicase is loaded onto the lagging strand template by the helicase loader protein (gp59) (9, 10). gp59 plays an additional role as a “gatekeeper” of the replisome by coordinating the assembly of the primosome with the initiation of leading strand DNA synthesis through a direct interaction with the leading strand polymerase (11-13). Finally, gp32 plays a central role in most aspects of DNA metabolism, including DNA replication (14). The gp32 protein coats the ssDNA produced by the primosome and is thought to be involved in the coordination of lagging strand synthesis (15). gp32 is made up of N-terminal, C-terminal, and core domains (16). The N-terminal domain (domain B for “basic”) is involved in cooperative ssDNA binding. Removal of residues 1-21 completely eliminates ssDNA binding cooperativity (17, 18). The major function of the highly acidic C-terminal domain (residues 254-301, domain A for “acidic”) is to interact with other T4 proteins (19). Affinity chromatography using gp32-agarose has detected interactions between gp32 and itself, gp43, gp45, and gp59 (14, 20). gp32 also has been shown to co-purify with gp61 (21). Removal of domain A abolishes interaction with all of these proteins (19). The core domain is responsible for the recognition and binding of ssDNA (22).
The T4 replisome appears to be the most dynamic of the well characterized replisomes, with the primase, clamp loader, clamp, and ssDNA-binding protein all exchanging with solution proteins during coupled leading and lagging strand synthesis (23, 24). Additionally, the DNA polymerase is “dynamically processive,” meaning solution polymerase is capable of displacing the replicating polymerase during active replication without prior disassembly of the holoenzyme complex (25). Under normal conditions, only the hexameric helicase remains at the replication fork for the lifetime of the replisome (26). It is thought that many of these dynamic processes are related to the mechanism of repeated lagging strand synthesis. However, very little is known about the timing and rates of protein dissociation or how they may control lagging strand synthesis.
Because of the opposite polarities of the leading and lagging strand template, the two polymerase holoenzymes must copy their templates in opposite directions (27). This fact, coupled with the observation that the lagging strand polymerase is resistant to dilution, led to the proposal of the “trombone model” for DNA replication (14). This model links the two holoenzyme complexes and loops the lagging strand template into a structure resembling a trombone slide so that replication on both strands can occur in the same apparent direction. This model adequately explains why the lagging strand is synthesized in short 1-2-kb Okazaki fragments, whereas the leading strand is continuous. The lagging strand polymerase must repeatedly release from its position on the lagging strand template and recycle to the newly synthesized primer to begin a new round of Okazaki fragment synthesis. Presumably, interactions between the lagging strand polymerase and other components at the replication fork allow the polymerase to remain as part of the replisome during the recycling process.
The signal for lagging strand polymerase release and recycling has been a subject of intense investigation (28-31). Several models have been proposed to act as the trigger for lagging strand release and recycling. The two with the most support are the collision and the signaling models. The collision model states that the collision of the lagging strand polymerase with the 5′ end of the previous Okazaki fragment causes the primase to synthesize an RNA primer and the polymerase to release from the lagging strand template and recycle to the newly made primer. In the signaling model, the lagging strand polymerase releases from the DNA as the result of events that are associated with the synthesis or capture of the RNA primer.
There is substantial support for both of these models, and it is likely that both mechanisms are operable during lagging strand synthesis (28). Support for the collision model comes from data indicating that the dissociation rate of the holoenzyme is drastically increased upon collision with the 5′ end of both DNA and RNA primers (29, 31). Recent work from our laboratory has demonstrated that new Okazaki fragments can be initiated without completion of the previous fragment, indicating that the polymerase recycling via collision is not necessary (28). The responsiveness of primer utilization efficiency and Okazaki fragment size to the concentration of clamp and clamp loader led us to suggest that clamp loading onto the RNA primer could serve as the signal for lagging strand polymerase release and recycling (28). Computer simulations of lagging strand synthesis using a simple stochastic model incorporating the rate of primase association, primer synthesis, and clamp loading onto the newly synthesized primer accurately described the observed distribution of Okazaki fragments produced in vitro (28). If clamp loading does serve as the signal for lagging strand polymerase dissociation, then primer handoff must follow an indirect pathway where the primase transfers the primer to the clamp loader and clamp proteins before the release and recycling of the lagging strand polymerase. This pathway is similar to that as described in the E. coli system and contrasts that of the T7 replisome (32, 33).
Here we report on experiments that highlight the interplay between the ssDNA-binding protein, primase, clamp, and clamp loader during the initiation of lagging strand synthesis. We examined the effect of removing the protein interaction domain of gp32 (gp32-A) on primer synthesis, primer utilization, and primase processivity. We found that total primer synthesis is drastically reduced in the presence of gp32-A, and a reduction in the efficiency of primer transfer from the primase to the polymerase is also observed. These effects combine to produce abnormally long Okazaki fragments with a broad distribution ranging from 0.2 to 10 kb. Using an inactive primase trap protein, we find that the rate of dissociation of the primase from the replisome is dependent on the concentration of clamp and clamp loader proteins, as well as the presence of intact gp32. The fast dissociation rate of primase in the presence of gp32-A can be compensated for by high concentrations of clamp and clamp loader. Based on these results, we present a more elaborate model for lagging strand DNA synthesis where the primase remains as part of the replisome through successful cycles Okazaki fragment initiation. If primer handoff is delayed, the primase has a higher probability of dissociating from the replication fork, and if this occurs, a new primase must enter the replisome from solution to begin a new round of primer synthesis.
MATERIALS AND METHODS
[α-32P]dCTP, [α-32P]dGTP, and [α-32P]CTP were purchased from PerkinElmer Life Sciences. Unlabeled ribonucleotides were purchased from Roche Applied Science. Bacteriophage T4 replication proteins gp41, gp61, gp43, gp44/62, gp45, gp32, and primase trap protein were purified as described previously (24, 34). The gp32-A-petIMPACT and gp32-B-petIMPACT plasmids were provided by Jingsong Yang. The minicircle substrate was prepared as described previously (15). Single-stranded M13 phage DNA (ssM13) was purified from infected XL1-Blue cells by polyethylene glycol precipitation and phenol extraction as described (35). The sequence of the oligonucleotide used in the priming assays was 5′-AGAGGGAGATTTAGATGAGATGATTGAGGATGGAGATGTTGATGGAGAGATGATGAATGATGAGATGAGGG-3′.
Expression and Purification of gp32-A and gp32-B Mutant Proteins—The gp32-A-petIMPACT or gp32-B-petIMPACT plasmids were transformed into BL21(DE3) cells and grown (separately) in 20 ml of Luria broth (LB) overnight at 37 °C. The expression and purification of gp32-A and gp32-B were identical. The overnight cultures were diluted 100-fold into two 1-liter flasks of LB and grown at 37 °C to an A600 of 0.8. The cultures were then allowed to cool to 18 °C, and protein expression was induced with 0.2 mm of isopropyl 1-thio-β-d-galactopyranoside. After 16 h of shaking, cells were collected by centrifugation at 6000 × g and resuspended in 80 ml of chitin column binding buffer containing one protease inhibitor pellet (Roche Applied Science). Cells were lysed using sonication, and cell debris was pelleted at 25,000 × g. Cell-free extract was loaded onto a 5-ml chitin column and washed with 200 column volumes of chitin binding buffer. The chitin resin was then resuspended in binding buffer plus 75 mm β-mercaptoethanol and incubated for 48 h at 4 °C to facilitate intein-mediated cleavage. Following cleavage, gp32-A or gp32-B proteins were eluted, dialyzed into storage buffer, and analyzed for purity using SDS-PAGE. Protein concentrations were determined by measuring the absorbance at 280 nm using an extinction coefficient of 38690 m-1 cm-1 for both gp32-A and gp32-B.
Standard Minicircle Replication Reactions and Measurement of Primer Utilization—Replication reactions were carried out in a standard replication buffer (25 mm Tris acetate (pH 7.8), 125 mm KOAc, and 10 mm Mg(OAc)2) at 37 °C. The standard conditions used to assemble and initiate the replication reaction consisted of the following: 100 nm minicircle substrate; 200 nm each of gp43, gp45 (as trimer), and gp44/62; 600 nm each of monomer gp41, gp61, and gp59; 0.5 μm of gp32; 100 μm each of CTP, GTP, and UTP; 2 mm ATP; 50 μm each of dATP, dGTP, dCTP, and dTTP, in a typical reaction volume of 12 μl. The reaction was allowed to proceed for 2.5 min followed by a 10-fold dilution into replication buffer containing gp43, gp44/62, gp45, gp61, gp59, ATP, CTP, UTP, and GTP at the standard concentrations. Wild-type or gp32 mutant was included in the dilution buffer at a concentration of 2 μm along with 25 μCi of [α-32P]dGTP or [α-32P]dCTP for visualization of leading or lagging strands, respectively. 20-μl aliquots were removed at the indicated time points and quenched with a half-volume of 0.5 m EDTA. The quenched reactions were analyzed with either DE81 filter binding assays or alkaline agarose gel electrophoresis (34). For the filter binding assay, an 8-μl sample of the quenched reaction aliquot was spotted onto DE81 filters and allowed to air dry for ∼5 min. The filters were then washed 10 times with 100 ml of 300 mm ammonium formate or until the wash had an insignificant level of radioactivity as determined by a hand-held Geiger counter. Following the ammonium formate washes, the filters were washed once with 100% EtOH and allowed to air-dry for 30 min. Once dried, the filters were placed in LSC vials with 5 ml of Ecoscint LSC mixture in each vial and counted with single channel liquid scintillation counting. For the analysis of replication products using alkaline-agarose gel electrophoresis, 10 μl of the quenched reaction aliquot was mixed with 10 μl of loading dye (50% glycerol and 0.1% Orange G) and loaded onto a 15 × 25-cm 1% agarose gel in 30 mm NaOH and 1 mm EDTA. The samples were run at 40 V for 24 h in 30 mm NaOH and 1 mm EDTA buffer. After running, the gels were removed from their trays and neutralized by soaking in 1× TBE buffer for 1 h at room temperature. The neutralized gels were then dried onto a sheet of DE81 paper using a stack of paper towels to facilitate gel drying. After 4 h of drying with the paper towels, the gel and filter sheet were vacuum-dried. The dried gel and DE81 filter paper were then exposed overnight to a PhosphorImager plate and analyzed using a Storm 800 PhosphorImager system (Amersham Biosciences).
To measure primer utilization, the standard replication reaction was performed by replacing [α-32P]dCTP with [α-32P]CTP in the dilution buffer. The reactions were quenched with an equal volume of 0.5 m EDTA at 2, 4, and 8 min after dilution. Reaction products were then mixed with loading dye (formamide with 0.2% xylene cyanol FF and bromphenol blue) and run on a 20% urea-acrylamide gel at 25 mA for 3 h at room temperature. The gel was then removed from the glass plates, wrapped in plastic film, and directly exposed to a PhosphorImager plate overnight. The PhosphorImager plate was analyzed using the Storm PhosphorImager. The total number of primers synthesized was calculated by summing the signal from the free and utilized primers. The percent primer utilization was determined by dividing the signal from utilized primers by the total number of primers.
Single-stranded DNA Priming Assays—Priming reactions were carried out in the standard replication buffer containing 0.4 μm gp41, 0.4 μm gp61, 100 nm gp59 (ssM13 reactions only), 2 μm gp32 (wild-type or mutant where indicated), 2 mm ATP, 100 μm each of CTP, GTP, and UTP, and either 10 nm M13 ssDNA or 3 μm ssDNA oligonucleotide (strands) in a typical reaction volume of 25 μl. Approximately 20 μCi of [α-32P]CTP per reaction was used for labeling the primer. For dilution experiments using the primase trap protein, the reactions were initiated using the same protein, nucleotide, and ssM13 concentrations as above except that wt-gp32 was used exclusively in the pre-dilution reaction. The reactions were allowed to proceed for 2.5 min before a 10-fold dilution into buffer containing only nucleotides, trap protein (3 μm), and either wt-gp32 or gp32-A. The reactions were carried out at 37 °C, and aliquots were withdrawn at the indicated times and quenched with an equal volume of 0.5 m EDTA, pH 8.0. Priming products were analyzed by 20% urea-acrylamide sequencing gel electrophoresis. Autoradiography was accomplished as described above.
RESULTS
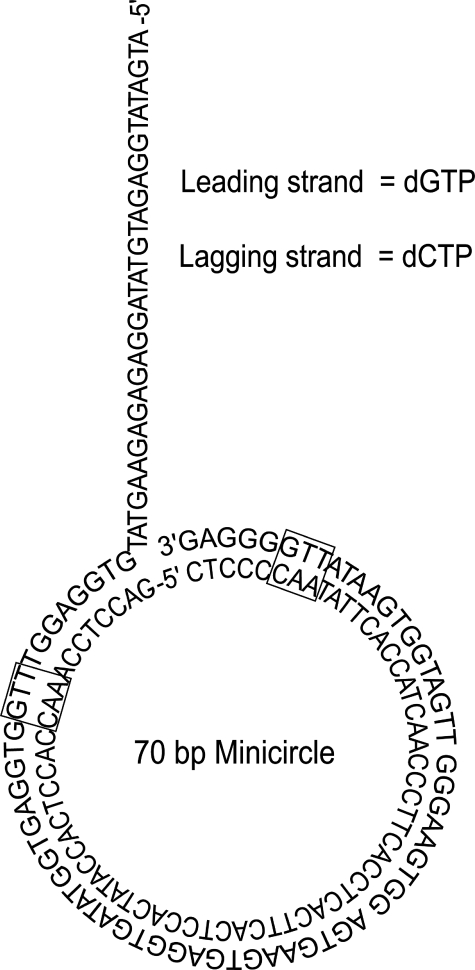
Leading and Lagging Strand Replication with Wild-type and Mutant gp32 Proteins—The replisome was assembled using a low concentration wild-type gp32 (0.5 μm), followed by 10-fold dilution into replication buffer containing 2 μm wild-type or mutant gp32. This allows for the assembly and initiation of leading and lagging strand synthesis under normal conditions, followed by a rapid exchange of ssDNA-bound protein with the excess of protein in the dilution buffer (23). The minicircle DNA substrate was used so that the leading and lagging strand could be independently monitored using alkaline agarose gel electrophoresis or DE81 filter binding assays (Fig. 1).
FIGURE 1.
The minicircle DNA substrate. The minicircle substrate is 70 bases long and contains two priming sites (GTT, black boxes) located equidistant from each other. The leading strand template contains only cytosine, adenine, and thymine bases so only dGTP, dTTP, and dATP are incorporated into the leading strand. The lagging strand contains only guanine, adenine, and thymine bases so only dCTP, dTTP, and dATP are incorporated into the lagging strand.
We examined the leading and lagging strand replication products with gp32-A, gp32-B, wt-gp32, and no gp32 in the dilution buffer (Fig. 2). Omission of gp32 from the dilution buffer leads to the inhibition of both leading and lagging strand DNA synthesis. This is a consequence of the rapid production of uncoated ssDNA that chelates the distributive replisomal proteins (e.g. gp45, gp44/62) and has been previously observed by us and others (15, 36). Inclusion of gp32-A in the dilution buffer results in helicase-dependent leading strand DNA synthesis that is very similar to the reaction containing wt-gp32 in the dilution buffer. However, when compared with the wt-gp32 reaction, there is a significant increase in the amount of helicase-independent strand displacement synthesis (indicated by the black arrows in Fig. 2). This is likely because of a reduction in the ability of gp59 to “lock” the polymerase in an inhibited state in the absence of a gp59-gp32 interaction (37). Lagging strand DNA synthesis in the presence of gp32-A is reduced compared with the amount of leading strand synthesis produced in the same reaction, although the fragments tend to be longer. This indicates that the leading and lagging strand polymerases have become uncoupled (28). Leading and lagging strand replication in the presence of gp32-B protein is reduced in a manner very similar to the reaction performed with gp32 omission. Presumably, the loss of cooperativity in ssDNA binding reduces the overall affinity to a level where gp32-B does not bind to ssDNA. For this reason, we focused our efforts at understanding the effects of gp32-A on lagging strand synthesis.
FIGURE 2.
Leading and lagging strand DNA replication with wild-type and mutant gp32 proteins. The replication products were produced in reactions carried out as described under “Materials and Methods.” The three time points for each reaction are 3, 6, and 9 min after dilution. The black arrows indicate the helicaseindependent DNA synthesis that is increased in reactions containing gp32-A.
We next examined the lagging strand products of reactions carried out with wt-gp32 or gp32-A in the dilution buffer in more detail. To equalize the relative amounts of signal in the reactions, we increased the amount of α-32P by 5-fold in the gp32-A reaction (Fig. 3). This enables an accurate determination of both the average Okazaki fragment length and the size distribution. As shown, the average length of Okazaki fragments in the gp32-A reaction is 2.3 times longer than the wt-gp32 reaction, and the size distribution is much broader in the presence of gp32-A (0.1-8 kb) as compared with the wild-type enzyme (0.1-2 kb).
FIGURE 3.
Length of Okazaki fragments produced in wt-gp32 and gp32-A replication reactions. On the left is a comparison of the length distribution of Okazaki fragments from the reactions containing gp32-A (lane 2) and wt-gp32 (lane 3). On the right is a plot of the product distribution from the replication reactions containing gp32-A (blue) and wt-gp32 (red). The most abundant fragment size is given above the distribution curve. rfu, relative fluorescence units.
The production of longer Okazaki fragments can be caused by a decrease in either the primer synthesis rate or in the efficiency of primer handoff, or both (28). To determine what mechanism is responsible for the increase in Okazaki fragment length in the presence of gp32-A, we carried out full replisome replication assays in the presence of [α-32P]CTP to label RNA primers produced by the primase. The products of these reactions were analyzed using urea-PAGE and quantitated as shown in Fig. 4. This analysis reveals both a decrease in the total primers synthesized (Fig. 4A) and in the percentage of primers being used for Okazaki fragment initiation by gp32-A (Fig. 4B). The decrease in primer number is not the result of fewer replication forks or a decrease in the amount of lagging strand template produced because the assembly of the replisome is carried out under identical conditions for both gp32-A and wt-gp32 reactions, and leading strand DNA synthesis is not affected by the gp32-A mutant (Fig. 2). The decrease in total primer synthesis could be the result of a reduction in catalytic efficiency of the primase protein itself (presumably through the loss of interaction with gp32) or a reduction in the number of replisomes containing primase (i.e. a decrease in binding rate or increase in dissociation rate). To determine whether gp32 directly affects the catalytic activity of the primase protein, we examined the activity of the primase using an ssDNA oligonucleotide containing a single priming recognition site. Under the conditions of this assay, the formation of a helicase-primase-DNA complex is efficient, and thus the overall rate of primer synthesis depends on the activity of the primase protein itself (i.e. the kcat for the priming reaction). Additionally, the loading of primase and helicase onto the ssDNA oligonucleotide does not require gp59 and therefore does not require a specific gp32-gp59 interaction. Compared with a reaction performed without gp32, the rate of primer synthesis is unaffected by the presence of either wt-gp32 or gp32-A (Fig. 4C). This indicates that the decrease in the priming rate of the gp32-A-containing replisome is not because of a reduction in the intrinsic activity of primase, suggesting that a change in the equilibrium between the replisome-bound and -unbound forms of the primase is responsible.
FIGURE 4.
Priming activity with wt-gp32 and gp32-A proteins. A, rate of total priming activity using the minicircle substrate as described under “Materials and Methods.” The error bars represent the standard deviation of a fit to the linear portion of the reaction. B, quantification of the percentage of primers utilized from the reactions shown in A. The error bars represent the standard error of three determinations. C, rate of total priming using an ssDNA oligonucleotide substrate as described under “Materials and Methods.” The reactions containing wt-gp32, gp32-A, and gp32 omission are represented by closed triangles, squares, and diamonds, respectively. The solid lines are fits to a linear equation with calculated slopes of 55,461, 55,828, and 49,865 cpm/min for wt-gp32, gp32-A, and gp32 omission reactions, respectively.
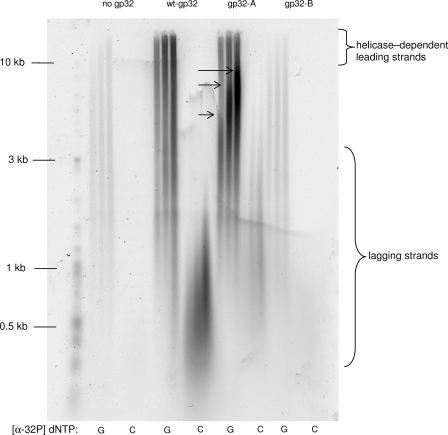
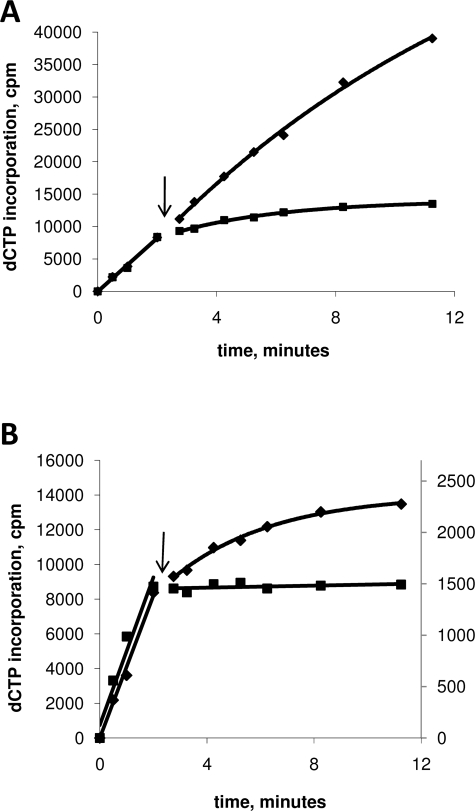
The equilibrium between the replisome-bound and unbound forms of primase is controlled by the binding and dissociation rates of the protein. Although it is difficult to determine the rate of binding of the primase to the replisome because of the non-uniform nature of the ensemble, the dissociation rate of the primase can be determined using a previously engineered trap protein (24). The trap protein is an active site mutant of the primase (E234Q) that is normal in every way except for a total absence of priming activity (24). Primase processivity in the presence of wild-type or gp32-A protein was determined by allowing the replisome to assemble and perform leading and lagging strand synthesis for 2.5 min under normal conditions, followed by a 10-fold dilution into buffer containing the primase trap and either wt-gp32 or gp32-A. To prevent the assembly of new replication forks or reassembly of collapsed forks, helicase was omitted from the dilution buffer. In the absence of trap protein, the rate of leading and lagging strand synthesis is relatively unchanged over the time period of the assay (Table 1 and Fig. 5A). Thus, in the presence of trap protein, a time-dependent decrease in lagging strand DNA synthesis can be attributed to the dissociation of wild-type primase from the replisome and incorporation of the primase trap (24). The presence of a single trap subunit within the primase oligomer has been shown to inhibit the activity of the entire primase oligomer (38). The results of this experiment indicate that the primase protein is much less processive in the presence of gp32-A than wt-gp32 (Table 1 and Fig. 5B), suggesting that the decrease in total primer synthesis in the presence of gp32-A is caused by a reduction in the amount of time the primase spends bound to the helicase within the replisome.
TABLE 1.
Rate constants for inactivation of lagging strand synthesis
| Conditiona | kinactb |
|---|---|
| min−1 | |
| No trapc | 0.08 ± 0.02 |
| Trap,c wt32, 200 nm clamp/clamp loader | 0.27 ± 0.04 |
| Trap, wt32, 800 nm clamp/clamp loader | 0.13 ± 0.03 |
| Trap, gp32-A, 200 nm clamp/clamp loader | d |
| Trap, gp32-A, 800 nm clamp/clamp loader | 0.20 ± 0.06 |
Reaction conditions are described under “Materials and Methods.” Unless noted, standard concentrations for all proteins and nucleotides were used.
kinact was determined by fitting the data found in Figs. 5 and 6 to a standard single exponential equation.
Trap refers to the E234Q primase mutant at a concentration of 3 μm.
The rate of inactivation is too rapid to be accurately determined (>6 min−1).
FIGURE 5.
Sensitivity to primase trapping in replication reactions containing wt-gp32 and gp32-A. A plot of [α-dCTP] incorporation (lagging strand synthesis) versus time after dilution. The replication reactions were carried out as described under “Materials and Methods.” The time points before and after trap or buffer addition (indicated by arrow) are fitted to linear and single exponential equations, respectively. The rate constants from the single exponential fits can be found in Table 1. A, replication reactions containing only wt-gp32 in the dilution buffer. Either buffer (diamonds) or primase trap (squares) was added at 2.5 min. B, reactions containing wt-gp32 (diamonds) and gp32-A (squares) in the dilution buffer. To ease comparison between the two reactions, the plot is shown with two y axes. On the left y axis are the values from the reaction containing wt-gp32, and on the right are the values from the reaction containing gp32-A.
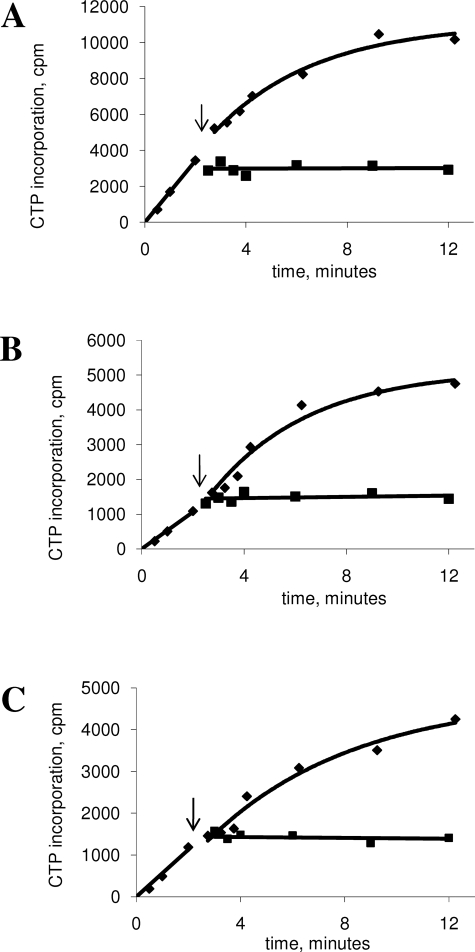
Because we observed a decrease in primer handoff efficiency in reactions performed in the presence of gp32-A, we next examined the processivity of the primase protein in the presence of either gp32-A or wt-gp32 under conditions where primer handoff efficiency is increased. We have previously shown that the efficiency of primer handoff is increased at high concentrations of the clamp loader and clamp proteins (28). Interestingly, we found that both the wt-gp32- and the gp32-A-containing replisomes are less sensitive to the primase trap when the concentration of clamp and clamp loader is increased from 200 to 800 nm (Table 1 and Fig. 6, A and B). The increase in clamp and clamp loader concentrations nearly compensates for the difference in primase processivity in reactions performed with wt-gp32 and gp32-A (kinact of 0.13 and 0.2 min-1, respectively).
FIGURE 6.
Sensitivity to primase trapping in replication reactions with 200 and 800 nm clamp/clamp loader proteins. A plot of [α-dCTP] incorporation (lagging strand synthesis) versus time after dilution. The replication reactions were carried out as described under “Materials and Methods.” The time points before and after trap addition (indicated by arrow) are fitted to linear and single exponential equations, respectively. The rate constants from the single exponential fits can be found in Table 1. Replication reactions carried out in the presence of wt-gp32 (A) or gp32-A (B). Lagging strand replication products were monitored at 200 (squares) and 800 nm (diamonds) clamp and clamp loader proteins. To ease comparison between the two reactions plotted on each graph, the plots are shown with two y axes. On the left y axis are the values from the 800 nm clamp/clamp loader reaction, and on the right are the values from the 200 nm clamp/clamp loader reaction.
To investigate the processivity of the primase-helicase complex protein outside of the context of the replisome, we examined the activity of the primase using ssM13 as a DNA substrate. These experiments were carried out in a similar fashion as the processivity experiments that used the complete replisome and the minicircle DNA substrate. The primase and helicase were allowed to assemble and initiate primer synthesis for a period of 2.5 min, followed by a 10-fold dilution into buffer without gp32, with gp32-A, or with wt-gp32. Helicase, helicase loader, and primase were omitted from the dilution buffer to reduce the likelihood of primosome reassembly. The results from this experiment demonstrate that the primase dissociates very rapidly from the primosome outside of the context of the replisome, regardless of the presence or absence of gp32 (Fig. 7, A-C). Primer synthesis ceases immediately upon addition of the primase trap, indicating a half-life of less than 10 s (kinact > 6 min-1). The half-life of the helicase traveling along ssDNA has been estimated to be around 30-60 s, suggesting that the primase dissociates from the helicase, which then continues to travel along the ssDNA template (39, 40). This suggests that gp32-induced increase in primase processivity requires a unique feature(s) of the replisome or the replication fork that only occurs during coupled leading and lagging strand DNA synthesis.
FIGURE 7.
Sensitivity to primase trapping on ssM13 DNA substrates using wt-gp32, gp32-A, and gp32 omission. The priming reactions were carried out as described under “Materials and Methods.” The time points before and after trap addition (indicated by arrow) are fitted to linear and single exponential equations, respectively. Reactions were performed in the presence of wt-gp32 (A), no gp32 (B), or gp32-A (C). In each case, the diamond and square symbols represent the reactions where either buffer or primase trap was added to the reaction, respectively. The first three time points are taken from a single reaction. The reaction was then divided, and either buffer or primase trap was added. The single exponential rate constants for the reactions without primase trap are 0.25 ± 0.08, 0.26 ± 0.04, and 0.18 ± 0.09 min-1 for (A-C), respectively. The rate of inactivation of reactions with the primase trap is too rapid to determine.
DISCUSSION
The initiation of lagging strand synthesis is a multistep process involving priming, clamp loading, polymerase recycling, and holoenzyme assembly. As a central player of the replisome, ssDNA-binding protein (gp32) is likely involved in any or all of these steps (1). The usual method for determining the effect of a particular protein on a complex system is to simply omit the protein from the process and observe the results. With regard to gp32, there are two problems with this approach. First, a specific interaction between gp59 and gp32 is required for the proper assembly of the replisome (41, 42). Second, removal of gp32 from replisome-dependent leading and lagging strand DNA synthesis results in the production of a large amount of uncoated ssDNA, which acts as a trap for the distributive replisomal proteins (23, 34). For the latter reason, we examined the effect of a truncated gp32 protein lacking its protein-interacting domain (gp32-A) on lagging strand synthesis. The gp32-A mutant retains the ability to cooperatively bind ssDNA, thus coating the ssDNA produced by the helicase and leaving the distributive replisomal proteins unperturbed. gp32 has been shown to bind several replisomal proteins, including the polymerase, helicase loader, clamp protein, and the primase (43, 44). All of these interactions are abolished by removal of domain A in gp32. To achieve proper assembly of the replisome, we took advantage of the distributive nature of gp32 by first assembling the replisome in the presence of wt-gp32 and then exchanging it with gp32-A by a 10-fold dilution into buffer containing chosen replisome components and gp32-A.
The role of gp32 on replisomal DNA synthesis has been studied previously (44, 45). However, in that study the gp32-A was used at both the stage of replisomal assembly and DNA synthesis, which for the reasons just stated, prevents the assembly of the helicase and results in very low levels of helicase-dependent DNA synthesis (44). Here, our dilution protocol isolates the effect of the gp32-A mutant to that of lagging strand synthesis. We find that the replisomal DNA synthesis in the presence of gp32-A results in a 2.5-fold increase in the average length of Okazaki fragments and more broadly a distributed range of lengths as compared with reactions performed with wt-gp32. Examination of the primers produced during coupled replisomal DNA synthesis indicates that the source of the Okazaki fragment lengthening is a combination of a reduction in total priming and lower primer utilization.
The primase protein has been shown to be somewhat distributive, being recruited from solution during Okazaki fragment synthesis (24). Here we find that the dissociation rate of primase is dramatically increased in the presence of gp32-A, indicating that an interaction between gp32 and a replisomal protein (presumably the primase) is responsible for the moderate processivity of the primase. The dissociation rate of the primase in the presence of wt-gp32 is 0.27 min-1, whereas Okazaki fragments are synthesized every 5-10 s. This indicates that many rounds of Okazaki fragment synthesis will occur before the dissociation of a single primase subunit. When wt-gp32 is replaced with gp32-A, the dissociation rate becomes too fast to measure with our assay, suggesting that a primase subunit dissociates after each cycle of Okazaki fragment synthesis. We have previously observed that Okazaki fragment length is dependent on the concentration of primase, suggesting that in the event of primase dissociation, its rebinding rate is a partially rate-limiting step in the primer synthesis cycle (24, 33). Therefore, an increase in the dissociation rate of the primase will result in a decrease in the overall priming rate, which is what we have observed here. It is therefore likely that the decrease in primase processivity is directly responsible for the reduced amount of total priming activity in reactions containing gp32-A.
Surprisingly, we found that high levels of the polymerase accessory proteins, clamp and clamp loader, decreased the dissociation rate of the primase 2-fold (0.27 to 0.13 min-1). The most likely mechanism for a clamp/clamp loader-induced increase in primase processivity involves the handoff of the RNA pentamer from the primase to the polymerase. We have previously demonstrated that high concentrations of clamp and clamp loader increase the efficiency of primer handoff (28). That result, combined with the signaling mechanism for the release and recycling of the lagging strand polymerase, favors a primer handoff pathway where the RNA primer is first transferred to the clamp loader and clamp proteins before the association of the polymerase. The loading of clamp/clamp loader signals the lagging strand polymerase to release its DNA template and recycle to the new primer. The increase in primase processivity is likely linked to the increase in primer handoff efficiency. The primase protein must release the RNA primer so that the clamp loader can recognize the primer/template and load/chaperone the clamp protein into position on the RNA/DNA duplex. At the point of primer release, the primase could remain as part of the replisome or dissociate into solution. The data here indicate that the primase is more likely to remain in the replisome when handoff efficiency is increased. This requires a mechanism that allows the primase to respond to the presence of the clamp loader. This could be through a direct interaction between the primase and the clamp loader or indirectly through the competition for a mutual binding partner (e.g. gp32). We favor a direct, yet transient, interaction between the clamp loader and the primase because the clamp/clamp loader-induced increase in primase processivity remains when gp32 is replaced with the gp32-A mutant. The increase in trapping rate at low clamp and clamp loader concentrations indicates that when primer handoff is less efficient, the probability of primase dissociation is increased.
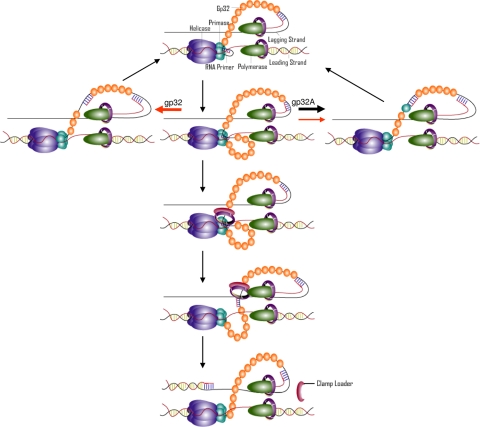
The simplest mechanism to account for these data is a “timing” mechanism where the primase is bound to the RNA primer within the replisome for a finite time (Fig. 8). This model is speculative but is consistent with all of the data presented here and with the lagging strand polymerase recycling mechanism previously demonstrated. The average Okazaki fragment cycle is completed in less than 10 s with the majority of this time devoted to the synthesis of the Okazaki fragment. Therefore, the time required to proceed through the steps shown in Fig. 8 is likely only a few seconds. The start of an Okazaki fragment cycle occurs when the primase recognizes a priming site and synthesizes a RNA pentamer (step 1). The primase remains bound to both the helicase and the RNA primer, although the helicase continues to unwind the duplex DNA. The production of ssDNA by the helicase produces a second loop between the helicase and primase (step 2). Presumably, this loop becomes coated with gp32 in the same manner as the lagging strand loop. Next, the clamp and clamp loader enter the replisome from solution and displace the primase from the RNA primer, causing the priming loop to collapse (steps 5 and 6). The primase remains bound to the helicase ready for the next round of primer synthesis. Once the clamp loader has chaperoned the clamp onto the primer/template, the lagging strand polymerase releases the lagging strand template and recycles to the newly synthesized RNA primer (step 7). However, if the clamp and clamp loader proteins are slow to enter the replisome and fail to displace the primase from the RNA primer, then the primase will either release the primer and remain with the replisome (step 4) or will dissociate from the replisome with the primer (step 3). In the presence of a functional interaction between gp32 and the primase, the pathway where the primase remains with the replisome is strongly favored. In addition to explaining primase processivity, this mechanism accounts for the decrease in primer utilization when wt-gp32 is replaced with gp32-A. In the absence of a primase-gp32 interaction, the dissociation rate of the primase (and therefore the primer) increases, which narrows the window of opportunity for primer capture by the clamp loader and therefore primer utilization efficiency decreases.
FIGURE 8.
Model for the initiation of lagging strand DNA synthesis in the T4 replisome. Initiation of lagging strand synthesis begins with the synthesis of an RNA pentamer by primase (step 1). The lagging strand polymerase is synthesizing an Okazaki fragment, causing the lagging strand loop to increase in size. The helicase continues to unwind the DNA duplex, causing a second loop to form (step 2). If the clamp loader protein does not load the clamp in time, the primase either releases from the helicase and dissociates from the replisome (step 3) or releases the primer and remains with the replisome (step 4). Route (step 4) is favored in the presence of wt-gp32, whereas route (step 3) is favored in the presence of gp32-A. Arrow size for steps 3 and 4 represent their relative rates, and arrow color represents replication with wt-gp32 (red) or gp32-A (black). If the primase dissociates, a new primase subunit must be recruited from solution to reset the replisome and allow for another round of RNA primer synthesis. If the clamp loader loads the clamp (step 5), the primase hands off the RNA primer to the clamp and remains bound to the helicase (step 6). The successful loading of the clamp onto the RNA primer causes the lagging strand polymerase to release the lagging strand template and recycle to the new clamp-loaded RNA primer (step 7).
This work was supported, in whole or in part, by National Institutes of Health Grant GM013306 (to S. J. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: T4, bacteriophage T4; gp32, T4 single-stranded DNA-binding protein; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; wt-gp32, wild-type gp32; gp32-A, residues 1-253 of gp32; gp32-B, residues 22-301.
References
- 1.Nossal, N. G. (1992) FASEB J. 6 871-878 [DOI] [PubMed] [Google Scholar]
- 2.Benkovic, S. J., Valentine, A. M., and Salinas, F. (2001) Annu. Rev. Biochem. 70 181-208 [DOI] [PubMed] [Google Scholar]
- 3.Kaboord, B. F., and Benkovic, S. J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 10881-10885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smiley, R. D., Zhuang, Z., Benkovic, S. J., and Hammes, G. G. (2006) Biochemistry 45 7990-7997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang, Z., Berdis, A. J., and Benkovic, S. J. (2006) Biochemistry 45 7976-7989 [DOI] [PubMed] [Google Scholar]
- 6.Kaboord, B. F., and Benkovic, S. J. (1996) Biochemistry 35 1084-1092 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, Z., Spiering, M. M., Trakselis, M. A., Ishmael, F. T., Xi, J., Benkovic, S. J., and Hammes, G. G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 3254-3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinton, D. M., and Nossal, N. G. (1987) J. Biol. Chem. 262 10873-10878 [PubMed] [Google Scholar]
- 9.Barry, J., and Alberts, B. (1994) J. Biol. Chem. 269 33049-33062 [PubMed] [Google Scholar]
- 10.Nossal, N. G., Hinton, D. M., Hobbs, L. J., and Spacciapoli, P. (1995) Methods Enzymol. 262 560-584 [DOI] [PubMed] [Google Scholar]
- 11.Dudas, K. C., and Kreuzer, K. N. (2005) J. Biol. Chem. 280 21561-21569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi, J., Zhang, Z., Zhuang, Z., Yang, J., Spiering, M. M., Hammes, G. G., and Benkovic, S. J. (2005) Biochemistry 44 7747-7756 [DOI] [PubMed] [Google Scholar]
- 13.Nelson, S. W., Yang, J., and Benkovic, S. J. (2006) J. Biol. Chem. 281 8697-8706 [DOI] [PubMed] [Google Scholar]
- 14.Alberts, B. M., Barry, J., Bedinger, P., Formosa, T., Jongeneel, C. V., and Kreuzer, K. N. (1983) Cold Spring Harbor Symp. Quant. Biol. 47 655-668 [DOI] [PubMed] [Google Scholar]
- 15.Salinas, F., and Benkovic, S. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7196-7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waidner, L. A., Flynn, E. K., Wu, M., Li, X., and Karpel, R. L. (2001) J. Biol. Chem. 276 2509-2516 [DOI] [PubMed] [Google Scholar]
- 17.Giedroc, D. P., Khan, R., and Barnhart, K. (1990) J. Biol. Chem. 265 11444-11455 [PubMed] [Google Scholar]
- 18.Villemain, J. L., Ma, Y., Giedroc, D. P., and Morrical, S. W. (2000) J. Biol. Chem. 275 31496-31504 [DOI] [PubMed] [Google Scholar]
- 19.Krassa, K. B., Green, L. S., and Gold, L. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 4010-4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrical, S. W., Beernink, H. T., Dash, A., and Hempstead, K. (1996) J. Biol. Chem. 271 20198-20207 [DOI] [PubMed] [Google Scholar]
- 21.Burke, R. L., Munn, M., Barry, J., and Alberts, B. M. (1985) J. Biol. Chem. 260 1711-1722 [PubMed] [Google Scholar]
- 22.Shamoo, Y., Friedman, A. M., Parsons, M. R., Konigsberg, W. H., and Steitz, T. A. (1995) Nature 376 362-366 [DOI] [PubMed] [Google Scholar]
- 23.Kadyrov, F. A., and Drake, J. W. (2001) J. Biol. Chem. 276 29559-29566 [DOI] [PubMed] [Google Scholar]
- 24.Trakselis, M. A., Roccasecca, R. M., Yang, J., Valentine, A. M., and Benkovic, S. J. (2003) J. Biol. Chem. 278 49839-49849 [DOI] [PubMed] [Google Scholar]
- 25.Yang, J., Zhuang, Z., Roccasecca, R. M., Trakselis, M. A., and Benkovic, S. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8289-8294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrock, R. D., and Alberts, B. (1996) J. Biol. Chem. 271 16678-16682 [DOI] [PubMed] [Google Scholar]
- 27.Alberts, B. M. (1987) Philos. Trans. R Soc. Lond. B Biol. Sci. 317 395-420 [DOI] [PubMed] [Google Scholar]
- 28.Yang, J., Nelson, S. W., and Benkovic, S. J. (2006) Mol. Cell 21 153-164 [DOI] [PubMed] [Google Scholar]
- 29.Carver, T. E., Sexton, D. J., and Benkovic, S. J. (1997) Biochemistry 36 14409-14417 [DOI] [PubMed] [Google Scholar]
- 30.Chastain, P. D., Makhov, A. M., Nossal, N. G., and Griffith, J. D. (2000) Mol. Cell 6 803-814 [DOI] [PubMed] [Google Scholar]
- 31.Hacker, K. J., and Alberts, B. M. (1994) J. Biol. Chem. 269 24221-24228 [PubMed] [Google Scholar]
- 32.Yuzhakov, A., Kelman, Z., and O'Donnell, M. (1999) Cell 96 153-163 [DOI] [PubMed] [Google Scholar]
- 33.Kato, M., Ito, T., Wagner, G., Richardson, C. C., and Ellenberger, T. (2003) Mol. Cell 11 1349-1360 [DOI] [PubMed] [Google Scholar]
- 34.Yang, J., Trakselis, M. A., Roccasecca, R. M., and Benkovic, S. J. (2003) J. Biol. Chem. 278 49828-49838 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J. F., Maniatis, T., and Sambrook, J. F. (1989) Molecular Cloning: A Laboratory Manual, pp. 431-432, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 36.Kadyrov, F. A., and Drake, J. W. (2002) Nucleic Acids Res. 30 4387-4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre, S. D., Wong, M. L., and Morrical, S. W. (1999) J. Biol. Chem. 274 22830-22838 [DOI] [PubMed] [Google Scholar]
- 38.Yang, J., Xi, J., Zhuang, Z., and Benkovic, S. J. (2005) J. Biol. Chem. 280 25416-25423 [DOI] [PubMed] [Google Scholar]
- 39.Lionnet, T., Spiering, M. M., Benkovic, S. J., Bensimon, D., and Croquette, V. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19790-19795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young, M. C., Schultz, D. E., Ring, D., and von Hippel, P. H. (1994) J. Mol. Biol. 235 1447-1458 [DOI] [PubMed] [Google Scholar]
- 41.Ma, Y., Wang, T., Villemain, J. L., Giedroc, D. P., and Morrical, S. W. (2004) J. Biol. Chem. 279 19035-19045 [DOI] [PubMed] [Google Scholar]
- 42.Jones, C. E., Mueser, T. C., and Nossal, N. G. (2004) J. Biol. Chem. 279 12067-12075 [DOI] [PubMed] [Google Scholar]
- 43.Richardson, R. W., and Nossal, N. G. (1989) J. Biol. Chem. 264 4732-4739 [PubMed] [Google Scholar]
- 44.Burke, R. L., Alberts, B. M., and Hosoda, J. (1980) J. Biol. Chem. 255 11484-11493 [PubMed] [Google Scholar]
- 45.Cha, T. A., and Alberts, B. M. (1990) Biochemistry 29 1791-1798 [DOI] [PubMed] [Google Scholar]