Abstract
ID1, inhibitor of differentiation/DNA binding, plays an important role in cell proliferation, differentiation, and tumor-igenesis. It has been shown that ID1 is de-regulated in multiple cancers and up-regulation of ID1 is correlated with high grades and poor prognosis of human cancers. In contrast, the p53 tumor suppressor was found to be mutated or inactivated in most human cancers and loss of p53 results in early onset of multiple cancers. Although the biological functions of the ID1 oncogene and the p53 tumor suppressor have been intensively investigated, little is known about the upstream regulators of ID1 and the cross-talk between ID1 and p53. Here, we showed that ID1 is down-regulated in cells treated with various DNA damage agents in a p53-dependent manner. Interestingly, we found that DEC1, which was recently identified as a p53 target and mediates p53-dependent cell cycle arrest and senescence, is capable of inhibiting ID1 expression. Conversely, we found that knockdown of DEC1 attenuates DNA damage-induced ID1 repression. In addition, we identified several potential DEC1 responsive elements in the proximal promoter region of the ID1 gene. Moreover, we showed that overexpression of ID1 or ID1′, an isoform of ID1, promotes cell proliferation potentially through inhibition of p21 expression. Finally, we found that the extent of DNA damage-induced premature senescence was substantially decreased by overexpression of ID1 or ID1′. Taken together, our study suggests that p53 trans-repressional activity can be mediated by its own target DEC1 and ID1 is an effector of the p53-dependent DNA damage response pathway.
ID1 is a member of the ID2 (inhibitor of differentiation or DNA binding) subfamily of the bHLH transcription factors (1). Like other subfamily members, ID1 contains a helix-loop-helix (HLH) motif, but not a DNA binding domain. Therefore, ID1 acts as a dominant negative regulator of other HLH transcription factors by forming inactive heterodimers (2–4). Multiple lines of evidence suggest that ID1 is an oncogene. ID1 was found to be up-regulated in various human cancers, including breast, prostate, cervical, and ovarian cancer (5–9). In addition, the increased levels of ID1 expression is correlated with cancer aggressiveness and high grades (6, 8, 9). Moreover, it has been shown that overexpression of ID1 confers cancer cells enhanced ability in cell proliferation and invasiveness (6). Conversely, knockdown of ID1 inhibits the metastatic potential of breast cancer cells (10). Furthermore, ID1 is down-regulated in arrested or senescent cells (11) and ectopic expression of ID1 extends the lifespan of human keratinocytes (12, 13). Although the biological functions of ID1 have been intensively studied, the upstream regulators of ID1 are not well defined (14).
The p53 tumor suppressor is a guardian of genomic integrity and is mutated or inactivated in over 50% of human cancers (15, 16). p53 functions as a sequence-specific transcription factor (17, 18). Mutations in the p53 gene or interactions with the products of viral and cellular oncogenes render p53 defective in its DNA binding and transcriptional activity. Genetic studies have shown that loss of p53 in both mutant mice and Li-Frau-mani syndrome patients leads to early onset of multiple tumors (19–22). In response to certain cytotoxic stresses such as DNA damage, p53 is activated and enhances the transcription of genes that are involved in the control of cell cycle progression and apoptosis (17, 23). For example, p21 cyclin-dependent kinase inhibitor is induced by p53, which inhibits the G1/S and G2/M transitions (24, 25). For p53-dependent apoptosis, both extrinsic and intrinsic apoptotic pathways were regulated through induction of membrane receptors (such as FAS and DR5) and BH3 group proteins (such as BAX, NOXA, and PUMA) (26–29).
Given the importance of ID1 and p53 in tumorigenesis, it is not surprising that both of them are attractive targets for cancer therapeutic strategies. However, little is known about the crosstalk between ID1 and p53. It has been reported that high levels of ID1 are associated with p53 immunoreactivity and mitotic index in clinical colorectal adenocarcinoma samples and ID1 expression is increased in normal p53-null mice (30). Consistently, ectopic expression of p53 mutants results in elevated ID1 expression (31). In addition, a cDNA microarray study showed that ID1 is up-regulated in cells treated with the chemotherapeutic agent 5-fluorouracil in a p53-dependent manner (32). But to date, direct or indirect regulation of ID1 by p53 has not been clarified.
In this study, we found that ID1 is down-regulated in HCT116 and U2OS cells treated with various chemotherapeutic drugs. Interestingly, inhibition of ID1 expression upon DNA damage is attenuated by p53-knockdown. In addition, we found that expression of ID1 is regulated by DEC1, a bHLH transcription factor and a p53 target. Conversely, we found that knockdown of DEC1 alleviates DNA damage-induced ID1 inhibition. Moreover, several potential DEC1 responsive elements were identified in the proximal promoter region of the ID1 gene. We also showed that the DEC1 protein binds to, and inhibits, the promoter of the ID1 gene. Finally, we showed that overexpression of ID1 or ID1′ promotes cell proliferation and inhibits p21 expression. Taken together, for the first time, our study suggests that ID1 is an effector of the p53-dependent DNA damage response pathway.
EXPERIMENTAL PROCEDURES
Plasmids—Untagged wild-type and mutant DEC1 cDNAs in pcDNA4 expression vectors were described previously (33, 34). To generate HA-tagged wild-type DEC1 cDNA in pcDNA4 for tetracycline-inducible expression (Invitrogen), a cDNA fragment was amplified from untagged wild-type DEC1 cDNA (34) with forward primer, 5′-AGGAATTCACCATGTACCCATACGATGTTCCAGATTACGCTGAGCGGATCCCCAGCGCGCAA-3′ (HA sequence in underline), and reverse primer, 5′-AGTCTAGAAGGAAGGAAAGCAAAGCAG-3′. To generate ID1 and ID1′ cDNAs in pcDNA4 for tetracycline-inducible expression, cDNA fragments were amplified from ID1 EST (MHS1011-59341) and ID1′ EST (H63146) (Openbiosystems), respectively. The primers for ID1 amplification were: forward primer, ID1-S, 5′-ACGGAATTCATCATGAAAGTCGCCAGTGGCAGCAC-3′, and reverse primer, ID1-AS, 5′-ATATAGCGGCCGCTTCAGCGACACAAGATGCGATCGTC-3′. The primers for ID1′ amplification were: the same forward primer as for ID1 amplification and reverse primer, ID1′-AS, 5′-AATAGCGGCCGCCTAGTGGTCGGATCTGGATCTCACC-3′. To generate three luciferase reporters under the control of the ID1 promoter (nucleotides –1389/+88, –1108/+88, and –146/+88), genomic DNA fragments were amplified from MCF7 cells with one of the three forward primers (ID1-P-1389, 5′-AGGTACCCTCGGCCTTCCAAATTGTTGGGATTACAG-3′; ID1-P-1108, 5′-AAGGTACCGAGGTAAGGTGACCCTTGCTCAGCGAC-3′; ID1-P-146, 5′-AAGGTACCAGCAGGCACTAGACGAGCAGGAGGC-3′) and a common reverse primer (ID1-P-AS, 5′-ATCTCGAGCGACTGGCTGAAACAGAATGGGCAAAG-3′), respectively. To transiently knockdown p53 and DEC1, 21-bp double-stranded RNA oligos (p53 siRNA, GACUCCAGUGGUAAUCUAdTdT; DEC1 siRNA, GCAAGGAGACCUACAAAUUdTdT) and a randomly selected scrambled siRNA were purchased from Dharmacon RNA Technologies.
Cell Culture—H1299, HCT116, U2OS, MCF7, and MCF7-p53KD were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. U2OS-DEC1 (clones 6 and 14), M7-DEC1 (clones 6 and 16), and M7-DEC1-R58P (clone 2) were used as previously described (34). MCF7-p53KD is a derivative of MCF7, in which p53 was stably knocked down by RNA interference. M7-TR-7, a derivative of MCF7 that expresses the tetracycline repressor, was used as described previously (35). To generate cell lines that inducibly express HA-tagged DEC1, ID1, or ID1′, M7-TR-7 cells were transfected with pcDNA4-HA-DEC1, pcDNA4-ID1, or pcDNA4-ID1′, and selected with medium containing 200 μg/ml Zeocin. Individual clones were screened for inducible expression of HA-DEC1, ID1, or ID1′ by Western blot analysis with anti-HA and anti-ID1 antibodies, respectively. The resulting cell lines were designated M7-HA-DEC1, M7-ID1, and M7-ID1′. One representative M7-HA-DEC1 (clone 2), two representative M7-ID1 (clones 10 and 21), and two representative M7-ID1′ (clones 1 and 13) were selected for this study.
Western Blot Analysis—Whole cell extracts were prepared with 2× SDS sample buffer and boiled for 5 min at 95 °C. The antibody against DEC1 was generated in rabbit (33). Antibodies against p53, p21, ID1, p130, and the HA epitope were purchased from Santa Cruz Biotechnology. Anti-actin and mouse IgG were purchased from Sigma. Anti-Rb (clone XZ-77) was used as described (36).
Northern Blot Analysis—Total RNAs were isolated by TRIzol reagent (Invitrogen). Northern blot analysis and preparation of GAPDH probe were described previously (37). Wild-type ID1 cDNA was used as probe and amplified as described above.
Colony Formation—Cells were seeded at 500 per well in 6-well plates maintained in 5% fetal bovine serum with or without doxycycline in triplicate for 14 days. Colonies were fixed with methanol:glacial acetic acid (7:1), washed in H2O, and stained with 0.02% crystal violet.
Luciferase Reporter Assay—The dual luciferase assay was performed in triplicate according to the manufacturer's instructions (Promega). Briefly, 0.25 μg of a luciferase reporter, 0.25 μg of empty pcDNA4 or pcDNA4 that expresses DEC1, DEC1-M, or DEC1-R58P, and 9 ng of an internal control Renilla luciferase assay vector pRL-CMV (Promega) were transfected into H1299 cells by ESCORT V transfection reagent according to the manufacturer's instruction (Sigma). Cells were seeded at 2 × 104 per well in 24-well plates 24 h before transfection. 18 h post-transfection, luciferase activity was measured with the dual luciferase kit and Turner Designs luminometer. The -fold change in relative luciferase activity is a product of the luciferase activity induced by DEC1, DEC1-M, or DEC1-R58P protein, divided by that induced by an empty pcDNA4 vector.
Chromatin Immunoprecipitation (ChIP) Assay—ChIP assay was performed as previously described (38). The binding of the DEC1 protein to the proximal ID1 promoter region was detected with forward primer, 5′-GACAAACTCTTCATACAGTGCCCGC-3′, and reverse primer, 5′-TCGCTGAGCAAGGGTCACCTTACCTC-3′. The binding of the DEC1 protein to the ID1 TATA box region was detected with forward primer, 5′-GACTGGCTGAAACAGAATGGGCAAAG-3′, and reverse primer, 5′-ACACTGCGAGCAGGCACTAGACGAG-3′. Primers for amplification of the DEC1 responsive elements within the Survivin promoter were described previously (39). Primers for amplification of the GAPDH promoter were used as previously described (40).
RESULTS
Inhibition of ID1 by DNA Damage Is Primarily p53-dependent—The p53 tumor suppressor is a key sensor of various stress signals, including those activated by chemotherapeutic drugs (28, 41). To uncover the potential cross-talk between ID1 and p53, we tested whether ID1 is regulated by DNA damage in cells treated with chemotherapeutic drugs, doxorubicin and camptothecin. Both doxorubicin (an inhibitor of topoisomerase II) and camptothecin (inhibitor of topoisomerase I) can induce DNA double strand breaks (42). We found that the expression levels of endogenous ID1 were decreased in HCT116 cells upon treatment with doxorubicin and camptothecin (Fig. 1A, ID1 panels). As expected, p53 was stabilized by doxorubicin and camptothecin in a time-dependent manner (Fig. 1A, p53 panels). p21, a well characterized p53 target, was up-regulated following p53 stabilization (Fig. 1A, p21 panels). The levels of actin were measured as a loading control (Fig. 1A, actin panels). Similarly, the levels of ID1 were decreased in U2OS cells treated with doxorubicin and camptothecin (Fig. 1B, ID1 and p53 panels). Again, p21 was up-regulated by p53 (Fig. 1B, p21 panels). Next, we wanted to examine whether p53 is required for inhibition of ID1 upon DNA damage. To address this, p53 was transiently knocked down by p53 siRNA along with scramble siRNA as a control. We found that p53 was efficiently knocked down in cells untreated or treated with doxorubicin or camptothecin and p21 was down-regulated in the presence of p53 siRNA (Fig. 1, C and D, p53 and p21 panels). In addition, DNA damage-induced inhibition of ID1 was significantly diminished by p53-knockdown (Fig. 1, C and D, ID1 panels). These data indicate that inhibition of ID1 by DNA damage is dependent on p53.
FIGURE 1.
ID1 expression is inhibited by DNA damage in a p53-dependent manner. A and B, ID1 expression is down-regulated upon DNA damage. Western blots were prepared with extracts from HCT116 and U2OS cells that were untreated (–) or treated (+) with doxorubicin (Dox) or camptothecin (CPT) for 12, 16, 20, and 24 h, respectively. ID1, p53, p21, and actin were detected by their respective antibodies. C and D, knockdown of endogenous p53 partially alleviates the inhibition of ID1 expression upon DNA damage. Western blots were prepared with extracts from HCT116 cells that were transiently transfected with control scramble siRNA and p53 siRNA for 3 days, and then untreated (–) or treated (+) with doxorubicin (0.3 μg/ml) for 24 h or with camptothecin (100 nm) for 6 h. The blots were analyzed as in A.
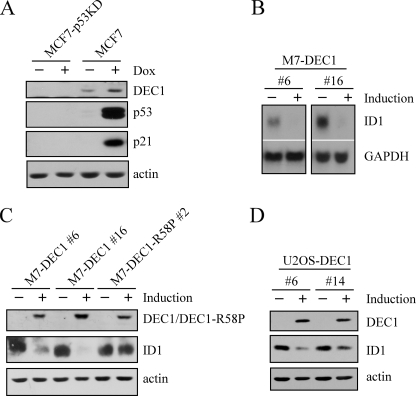
Expression of ID1 Is Down-regulated by DEC1—DEC1, a bHLH transcription factor, was recently identified as a p53 target, which mediates p53-dependent cell cycle arrest and senescence (34). Consistent with the previous study, DEC1 was up-regulated in p53-proficient but not p53-deficient MCF7 cells treated with doxorubicin (Fig. 2A, DEC1 panel). To identify potential DEC1 targets involved in inducing cell cycle arrest and senescence, an Affymetrix GeneChip assay was performed with U133 plus Chips with RNAs purified from MCF7 cells uninduced or induced to express DEC1. We found that ID1 was down-regulated upon overexpression of DEC1. To confirm this, Northern blot analysis was performed and showed that ID1 expression was repressed by DEC1 in MCF7 cells (Fig. 2B, ID1 panel). GAPDH was used as a loading control (Fig. 2B, GAPDH panel). Next, we examined whether a decrease in the ID1 transcript correlates with a decrease in ID1 protein. Western blot analysis was performed in MCF7 cells (M7-DEC1 numbers 6 and 16) in which wild-type DEC1 can be inducibly expressed and MCF7 cells (M7-DEC1-R58P number 2) in which DEC1-R58P can be inducibly expressed. DEC1-R58P has a point mutation at codon 58 (arginine to proline) within the DNA binding domain, which diminishes its DNA binding activity (33). We would like to note that the expression levels of mutant DEC1-R58P and wild-type DEC1 were comparable (Fig. 2C, DEC1/DEC1-R58P panel). We found that ID1 expression was down-regulated in MCF7 cells by DEC1, but not mutant DEC1-R58P (Fig. 2C, ID1 panel). Moreover, to rule out a potential cell type-specific effect, U2OS cells (U2OS-DEC1 numbers 6 and 14) in which wild-type DEC1 can be inducibly expressed were used. We showed that the level of ID1 was also decreased by DEC1 in U2OS cells (Fig. 2D, ID1 panel).
FIGURE 2.
ID1 expression is down-regulated by DEC1. A, DEC1 is induced by DNA damage in a p53-dependent manner. Western blots were prepared with extracts from MCF7 and MCF7-p53KD cells that were untreated (–) or treated (+) with 0.35 μg/ml doxorubicin (Dox) for 24 h. B, ID1 is inhibited by DEC1. Northern blots were prepared with RNAs purified from MCF7 cells that were uninduced (–) or induced (+) to express DEC1. The blots were probed with cDNAs derived from the ID1 and GAPDH genes, respectively. C and D, ID1 expression is down-regulated by DEC1. Western blots were prepared with extracts from MCF7 cells that were uninduced (–) or induced (+) to express DEC1 or mutant DEC1-R58P and from U2OS cells that were uninduced (–) or induced (+) to express DEC1. DEC1, ID1, and actin were detected by their respective antibodies.
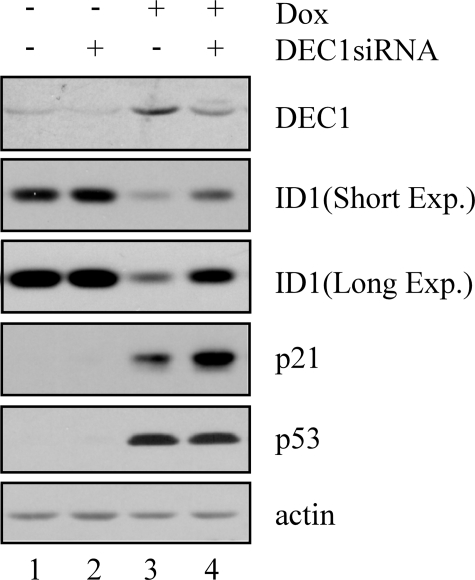
Because DEC1 can be induced by DNA damage in a p53-dependent manner (34), we hypothesized that inhibition of ID1 upon DNA damage might be dependent on DEC1. To address this, DEC1 was transiently knocked down in HCT116 cells by DEC1 siRNA along with scramble siRNA as a control (Fig. 3, DEC1 panel). The levels of p53 and p21 were measured as positive indicators of DNA damage (Fig. 3, p53 and p21 panels). Consistent with the above study, DEC1 was induced by DNA damage (Fig. 3, DEC1 panel, compare lane 1 with lane 3). In addition, we found that DEC1 expression was suppressed by DEC1 siRNA but not control siRNA in cells untreated or treated with doxorubicin (Fig. 3, DEC1 panel, compare lanes 1 and 3 with lanes 2 and 4, respectively). Interestingly, like p53-knockdown, DEC1-knockdown significantly inhibited DNA damage-induced repression of ID1 (Fig. 3, ID1 panel, compare lane 3 with lane 4). In summary, we concluded that inhibition of ID1 upon DNA damage is partially through the p53-DEC1 pathway.
FIGURE 3.
Knockdown of endogenous DEC1 partially attenuates the inhibition of ID1 expression upon DNA damage. Western blots were prepared with extracts from HCT116 cells that were transiently transfected with control scramble siRNA and DEC1 siRNA for 3 days, and then untreated (–) or treated (+) with doxorubicin (Dox) (0.3 μg/ml) for 24 h. The blots were analyzed as described in the legend to Fig. 1A.
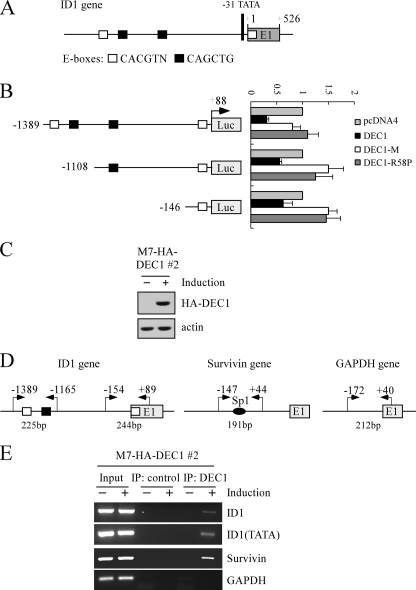
Identification of ID1 as a Direct Target of DEC1—DEC1 functions as a transcription repressor by binding to canonical E-boxes (CANNTG or CACGTN, where N is any nucleotide sequence) (43, 44). Thus, if ID1 is a direct target of DEC1, one or more DEC1-responsive elements (DEC1-REs) should exist in the ID1 gene. To test this, we analyzed the genomic locus of the ID1 gene and found several potential DEC1-REs within the proximal promoter region (Fig. 4A). To determine whether these potential DEC1-REs are responsive to DEC1, three DNA fragments from the ID1 promoter, that contain four E-boxes (–1389/+88), two E-boxes (–1108/+88), and one E-box (–146/+88), respectively, were cloned into pGL2-basic lucif-erase reporter. The resulting vectors were designated pGL2-ID1–1389, pGL2-ID1–1108, and pGL2-ID1–146 (Fig. 4B). Next, the luciferase reporter assay was performed and showed that the luciferase activity for each of the three luciferase reporters was inhibited by DEC1 (Fig. 4B). Interestingly, the ID1 promoter with four DEC1-REs was more sensitive to DEC1 than the promoter with one or two DEC1-REs (Fig. 4B). In contrast, mutants DEC1-R58P and DEC1-M were inert (Fig. 4B). DEC1-M lacks residues 53 to 65 in the DNA binding domain, and thus is transcriptionally inactive (33).
FIGURE 4.
ID1 is a direct target of DEC1. A, schematic presentation of the ID1 genomic structure with the location of the potential DEC1-REs (E-boxes). B, left panel, schematic presentation of three luciferase reporter constructs. See details in the text. Right panel, potential DEC1-REs in the ID1 gene are responsive to wild-type DEC1, but not mutant DEC1-M and DEC1-R58P. The lucifer-ase assay was performed as described under “Experimental Procedures.” C, generation of MCF7 cell lines that inducibly express HA-tagged DEC1. The levels of DEC1 were quantified with anti-HA. D, schematic presentation of the ID1, Survivin, and GAPDH promoters with the locations of potential DEC1-REs and PCR primers used for ChIP assays. E, DEC1 binds to the ID1 promoter in vivo. Upon induction or no induction of DEC1, MCF7 cells were cross-linked with formaldehyde followed by sonication. Chromatin was immunoprecipitated (IP) with anti-HA (HA-DEC1) or a control IgG. DEC1-responsive elements in the ID1 and Survivin genes were amplified by PCR. The binding of DEC1 to the GAPDH was quantified as a nonspecific binding control.
Next, we examined whether DEC1 can bind to these DEC1-REs in the ID1 gene in vivo. To test this, we generated multiple MCF7 cell lines that can inducibly express HA-tagged DEC1 under the control of a tetracycline-inducible promoter. One representative cell line, M7-HA-DEC1 number 2, was selected for this study (Fig. 4C). ChIP assay was performed with the primers shown in Fig. 4D (left panel). It has been shown that the Survivin gene is transcriptionally regulated by DEC1 primarily through multiple Sp1 sites (45). Therefore, the binding of DEC1 to the Survivin promoter was determined as a positive control (Fig. 4D, middle panel). Additionally, a region within the promoter of the GAPDH gene was amplified as a control for non-specific binding (Fig. 4D, right panel). To test the binding of DEC1 to the ID1 promoter, MCF7 cells were uninduced or induced to express HA-tagged DEC1 and the DEC1-DNA complexes were immunoprecipitated with anti-HA antibody or mouse IgG as a control. We found that the captured fragments containing the E-boxes in the proximal promoter region (nucleotides –1389 to –1165) and the region near the transcriptional start site (nucleotides –154 to +89) in the ID1 gene were significantly increased upon induction of DEC1 expression (Fig. 4E, ID1 panels). Similarly, DEC1 bound to the Survivin gene (Fig. 4E, Survivin panel). However, no fragments were enriched by control IgG (Fig. 4E, ID1 and Survivin panels). Furthermore, the GAPDH promoter was not recognized by DEC1 (Fig. 4E, GAPDH panel). Taken together, these data indicate that ID1 is likely to be a direct target gene of DEC1.
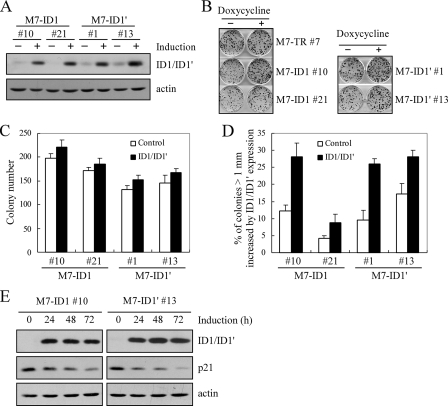
Overexpression of ID1 or ID1′ Promotes Cell Proliferation Potentially through Inhibition of p21 Expression—To analyze the biological activity of ID1 overexpression, we searched for a cell line in which a low level of endogenous ID1 is expressed. Thus, Western blot analysis was performed and showed that the expression level of ID1 was low in MCF7 cells but high in HCT116, U2OS, and MCF10A cells. Therefore, multiple MCF7 cell lines in which ID1 can be inducibly expressed were generated (Fig. 5A). In addition, the activity of ID1′, an alternative spliced isoform, was analyzed in MCF7 cells (Fig. 5A). ID1 and ID1′ have identical amino acid sequences except that ID1 and ID1′ contain 13 and 7 unique residues in their C termini, respectively (46). To analyze the effect of ID1 and ID1′ on the cell proliferation, colony formation assay was performed and showed that MCF7 cell proliferation was increased by overexpressed ID1 or ID1′ (Fig. 5B). Interestingly, we found that the size (>1 mm in diameter), but not the total number, of the colonies was markedly increased by overexpressed ID1 or ID1′ (Fig. 5, C and D). It has been shown that ID1 was able to promote cell proliferation partially through inhibiting p21 expression (47). Therefore, to further determine the molecular basis for their pro-growth function, Western blot analysis was performed and showed that both ID1 and ID1′ inhibited p21 expression (Fig. 5E).
FIGURE 5.
Overexpression of ID1 or ID1′ promotes cell proliferation potentially through inhibition of p21 expression. A, generation of MCF7 cell lines that inducibly express ID1 or ID1′. The levels of ID1 and ID1′ were quantified with anti-ID1. B, ID1 and ID1′ promote colony formation in MCF7 cells. Colony formation assay was performed with MCF7 cells uninduced or induced to express ID1 or ID1′ for 14 days, as described under “Experimental Procedures.” C, quantification of the total number of colonies shown in B. The number of colonies was calculated in triplicate for each cell line. D, quantification of the percentage of colonies with a diameter of >1 mm. The percentage of colonies with a diameter of >1 mm was calculated in triplicate for each cell line. The average was plotted as the percentage of colonies (>1 mm) increased by ID1 or ID1′ expression. E, over-expression of ID1 and ID1′ inhibits p21 expression. Western blots were prepared using extracts from MCF7 cells uninduced (–) or induced (+) to express ID1 or ID1′ for 0, 24, 48, or 72 h.
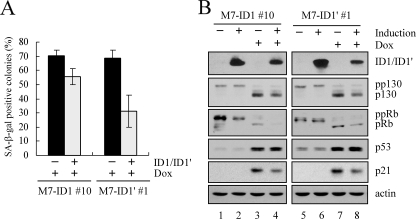
Overexpression of ID1 or ID1′ Attenuates DNA Damage-induced Premature Senescence—Our previous study showed that DEC1 promotes premature senescence and is required for DNA damage-induced senescence (34). In addition, ID1 was found to be down-regulated in senescent cells (11). To test whether ID1 plays a role in DNA damage-induced premature senescence, SA-β-galactosidase staining assay was performed and analyzed as previously described (34). We found that the extent of DNA damage-induced senescence was substantially reduced by overexpression of ID1 and ID1′ (Fig. 6A). To further analyze the effect of overexpression of ID1 or ID1′ on DNA damage-induced senescence, we examined the phosphorylation status of p130 and retinoblastoma protein (pRb). We found that the levels of hypophosphorylated p130 and pRb were significantly increased by treatment with doxorubicin in MCF7 cells (Fig. 6B, p130 and pRb panels, compare lanes 1 and 5 with 3 and 7, respectively). However, this increment was obviously reduced by overexpression of ID1 or ID1′ (Fig. 6B, p130 and pRb panels, compare lanes 3 and 7 with 4 and 8, respectively). The levels of p53 and p21 were measured as positive indicators of DNA damage (Fig. 6B, p53 and p21 panels). These data indicate that ID1 is a downstream effector of DEC1 and p53 to promote cell proliferation and inhibit DNA damage-induced premature senescence.
FIGURE 6.
Overexpression of ID1 or ID1′ attenuates DNA damage-induced premature senescence. A, overexpression of ID1 or ID1′ diminishes DNA damage-induced premature senescence. MCF7 cells, which were uninduced (–) or induced (+) to express ID1 or ID1′ for 2 days and then untreated (–) or treated (+) with 0.03 μg/ml of doxorubicin for 2 days, were analyzed by SA-β-galactosidase staining assay as described in a previous study (34). To quantify SA-β-galactosidase positive colonies, 150–200 colonies were counted and colonies containing ≥50% SA-β-galactosidase positive cells were defined as senescent colonies. B, overexpression of ID1 or ID1′ diminishes DNA damage-induced up-regulation of hypophosphorylated p130 and pRb. Western blots were prepared with extracts from MCF7 cells that were uninduced (–) or induced (+) to express ID1 or ID1′ for 2 days and then untreated (–) or treated (+) with 0.03 μg/ml doxorubicin for 1 day.
DISCUSSION
ID1 was found to be induced by serum and growth factors, including bone morphogenic protein and transforming growth factor β (11, 48–52). Moreover, a recent study showed that ID1 is also repressed by hypoxia (53). But to date, very little is known about a transcription factor(s) that directly regulates ID1 expression (14). Because we found that p53-knockdown attenuates DNA damage-induced repression of ID1, it is possible that ID1 is regulated by p53. As a transcriptional factor, p53 up-regulates gene expression by directly binding to a p53-responsive element (p53-RE) in the target gene promoter (17, 27). However, p53 down-regulates gene expression through diverse mechanisms, including binding directly to a p53-RE in the target gene promoter, interference with other transcriptional activators/coactivators, and modifications of the chromatin structure (54). In this study, we found that ID1 is repressed by DEC1, which is recently identified as a p53 target (34). In addition, we showed that like p53-knockdown, DEC1-knockdown alleviates inhibition of ID1 upon DNA damage. Furthermore, we found that DEC1 binds to, and inhibits, the promoter of the ID1 gene. These data indicate that ID1 is a direct target of DEC1. Therefore, for the first time, we found that p53 trans-repressional activity can be mediated by its own target DEC1 and ID1 is an effector of the p53-dependent DNA damage response pathway.
DEC1, a bHLH transcription factor, has been shown to play a role in the cell cycle regulation (33, 55), differentiation (56, 57), and apoptosis (33) in response to extracellular stimuli. In a previous study, we found that DEC1 is induced by DNA damage in a p53-dependent manner and mediates p53-dependent cell cycle arrest and senescence (34). Interestingly, ID1 is a well defined regulator of cell cycle progression and senescence. It has been shown that ID1 knock-out primary mouse embryonic fibroblasts undergo premature senescence (58). In addition, ID1 was found to be required for G1 phase progression and down-regulated in senescent cells (11). Moreover, overexpression of ID1 extends the lifespan of human keratinocytes (12, 13). Here, we showed that overexpression of ID1 and ID1′ promotes cell proliferation (Fig. 5). In addition, we found that over-expression of ID1 or ID1′ diminishes DNA damage-induced premature senescence (Fig. 6A). Furthermore, in response to DNA damage, overexpression of ID1 or ID1′ decreased the levels of hypophosphorylated p130 and pRb upon DNA damage (Fig. 6B), which is consistent with our previous study that p53-knockdown reduced the levels of hypophosphorylated p130 and pRb and DEC1-knockdown altered the level of hypophosphorylated p130 upon DNA damage (34). Taken together, inhibition of ID1 expression is a mechanism by which DEC1 and p53 inhibit cell proliferation and promotes premature senescence.
Human ID1 and its alternative splicing isoform ID1′ have an identical amino acid sequence except that ID1 and ID1′ have 13 and 7 unique residues in their C termini, respectively (11, 46). Although ID1′ has an expression pattern similar to ID1 during the cell cycle progression (11), the role of ID1′ in the control of the cell cycle has not been determined. A previous report showed that ID1 promotes cell proliferation partially through inhibition of p21 expression (47). Here, we found that like ID1, ID1′ also promotes cell proliferation by inhibiting p21 expression. Future study is needed to examine whether ID1 and ID1′ possess other common and/or distinct activities. Interestingly, DEC1-knockdown also leads to enhanced induction of p21 by DNA damage (Fig. 3), suggesting that p21 is inhibited by DEC1. Indeed, our preliminary study showed that as a repressor, DEC1 transcriptionally inhibits p21 expression.3 In summary, our study suggested that ID1 is an effector of the p53-dependent DNA damage response pathway. We believe that by revealing the interaction between p53 and ID1 in the DNA damage pathway, it will provide an insight to target ID1 for cancer therapeutic strategies.
Acknowledgments
We thank K. Harms for the M7-TR-7 cell line and W. Yan, A. Scoummane, and J. Zhang for suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA076069, CA081237, and CA102188. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ID, inhibitor of differentiation or DNA binding; bHLH, basis helix loop helix; HA, hemagglutinin; siRNA, small interfering RNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ChIP, chromatin immunoprecipitation; RE, responsive element; SA, senescence-associated; pRb, retinoblastoma protein.
Y. Qian and X. Chen, unpublished data.
References
- 1.Norton, J. D. (2000) J. Cell Sci. 113 3897–3905 [DOI] [PubMed] [Google Scholar]
- 2.Benezra, R., Davis, R. L., Lockshon, D., Turner, D. L., and Weintraub, H. (1990) Cell 61 49–59 [DOI] [PubMed] [Google Scholar]
- 3.Ellis, H. M., Spann, D. R., and Posakony, J. W. (1990) Cell 61 27–38 [DOI] [PubMed] [Google Scholar]
- 4.Garrell, J., and Modolell, J. (1990) Cell 61 39–48 [DOI] [PubMed] [Google Scholar]
- 5.Ouyang, X. S., Wang, X., Lee, D. T., Tsao, S. W., and Wong, Y. C. (2002) J. Urol. 167 2598–2602 [PubMed] [Google Scholar]
- 6.Lin, C. Q., Singh, J., Murata, K., Itahana, Y., Parrinello, S., Liang, S. H., Gillett, C. E., Campisi, J., and Desprez, P. Y. (2000) Cancer Res. 60 1332–1340 [PubMed] [Google Scholar]
- 7.Schindl, M., Oberhuber, G., Obermair, A., Schoppmann, S. F., Karner, B., and Birner, P. (2001) Cancer Res. 61 5703–5706 [PubMed] [Google Scholar]
- 8.Schoppmann, S. F., Schindl, M., Bayer, G., Aumayr, K., Dienes, J., Horvat, R., Rudas, M., Gnant, M., Jakesz, R., and Birner, P. (2003) Int. J. Cancer 104 677–682 [DOI] [PubMed] [Google Scholar]
- 9.Schindl, M., Schoppmann, S. F., Strobel, T., Heinzl, H., Leisser, C., Horvat, R., and Birner, P. (2003) Clin. Cancer Res. 9 779–785 [PubMed] [Google Scholar]
- 10.Fong, S., Itahana, Y., Sumida, T., Singh, J., Coppe, J. P., Liu, Y., Richards, P. C., Bennington, J. L., Lee, N. M., Debs, R. J., and Desprez, P. Y. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara, E., Yamaguchi, T., Nojima, H., Ide, T., Campisi, J., Okayama, H., and Oda, K. (1994) J. Biol. Chem. 269 2139–2145 [PubMed] [Google Scholar]
- 12.Alani, R. M., Hasskarl, J., Grace, M., Hernandez, M. C., Israel, M. A., and Munger, K. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9637–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickoloff, B. J., Chaturvedi, V., Bacon, P., Qin, J. Z., Denning, M. F., and Diaz, M. O. (2000) J. Biol. Chem. 275 27501–27504 [DOI] [PubMed] [Google Scholar]
- 14.Desprez, P. Y., Sumida, T., and Coppe, J. P. (2003) J. Mammary Gland Biol. Neoplasia 8 225–239 [DOI] [PubMed] [Google Scholar]
- 15.Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C., and Hainaut, P. (2002) Hum. Mutat. 19 607–614 [DOI] [PubMed] [Google Scholar]
- 16.Hollstein, M., Sidransky, D., Vogelstein, B., and Harris, C. C. (1991) Science 253 49–53 [DOI] [PubMed] [Google Scholar]
- 17.Ko, L. J., and Prives, C. (1996) Genes Dev. 10 1054–1072 [DOI] [PubMed] [Google Scholar]
- 18.Laptenko, O., and Prives, C. (2006) Cell Death Differ. 13 951–961 [DOI] [PubMed] [Google Scholar]
- 19.Li, F. P., and Fraumeni, J. F., Jr. (1969) Ann. Intern. Med. 71 747–752 [DOI] [PubMed] [Google Scholar]
- 20.Gasco, M., Yulug, I. G., and Crook, T. (2003) Hum. Mutat. 21 301–306 [DOI] [PubMed] [Google Scholar]
- 21.Birch, J. M., Alston, R. D., McNally, R. J., Evans, D. G., Kelsey, A. M., Harris, M., Eden, O. B., and Varley, J. M. (2001) Oncogene 20 4621–4628 [DOI] [PubMed] [Google Scholar]
- 22.Donehower, L. A., Harvey, M., Slagle, B. L., McArthur, M. J., Montgomery, C. A., Jr., Butel, J. S., and Bradley, A. (1992) Nature 356 215–221 [DOI] [PubMed] [Google Scholar]
- 23.Levine, A. J. (1997) Cell 88 323–331 [DOI] [PubMed] [Google Scholar]
- 24.el-Deiry, W. S., Tokino, T., Velculescu, V. E., Levy, D. B., Parsons, R., Trent, J. M., Lin, D., Mercer, W. E., Kinzler, K. W., and Vogelstein, B. (1993) Cell 75 817–825 [DOI] [PubMed] [Google Scholar]
- 25.Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K., and Elledge, S. J. (1993) Cell 75 805–816 [DOI] [PubMed] [Google Scholar]
- 26.Levine, A. J., Hu, W., and Feng, Z. (2006) Cell Death Differ. 13 1027–1036 [DOI] [PubMed] [Google Scholar]
- 27.Harms, K., Nozell, S., and Chen, X. (2004) Cell Mol. Life Sci. 61 822–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prives, C., and Hall, P. A. (1999) J. Pathol. 187 112–126 [DOI] [PubMed] [Google Scholar]
- 29.Harris, S. L., and Levine, A. J. (2005) Oncogene 24 2899–2908 [DOI] [PubMed] [Google Scholar]
- 30.Wilson, J. W., Deed, R. W., Inoue, T., Balzi, M., Becciolini, A., Faraoni, P., Potten, C. S., and Norton, J. D. (2001) Cancer Res. 61 8803–8810 [PubMed] [Google Scholar]
- 31.Tepper, C. G., Gregg, J. P., Shi, X. B., Vinall, R. L., Baron, C. A., Ryan, P. E., Desprez, P. Y., Kung, H. J., and deVere White, R. W. (2005) Prostate 65 375–389 [DOI] [PubMed] [Google Scholar]
- 32.Hernandez-Vargas, H., Ballestar, E., Carmona-Saez, P., von Kobbe, C., Banon-Rodriguez, I., Esteller, M., Moreno-Bueno, G., and Palacios, J. (2006) Int. J. Cancer 119 1164–1175 [DOI] [PubMed] [Google Scholar]
- 33.Li, Y., Zhang, H., Xie, M., Hu, M., Ge, S., Yang, D., Wan, Y., and Yan, B. (2002) Biochem. J. 367 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian, Y., Zhang, J., Yan, B., and Chen, X. (2008) J. Biol. Chem. 283 2896–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harms, K. L., and Chen, X. (2005) Mol. Cell. Biol. 25 2014–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu, Q. J., Bautista, C., Edwards, G. M., Defeo-Jones, D., Jones, R. E., and Harlow, E. (1991) Mol. Cell. Biol. 11 5792–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, X., Bargonetti, J., and Prives, C. (1995) Cancer Res. 55 4257–4263 [PubMed] [Google Scholar]
- 38.Liu, G., Xia, T., and Chen, X. (2003) J. Biol. Chem. 278 17557–17565 [DOI] [PubMed] [Google Scholar]
- 39.Li, Y., Xie, M., Yang, J., Yang, D., Deng, R., Wan, Y., and Yan, B. (2006) Oncogene 25 3296–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, G., and Chen, X. (2005) J. Biol. Chem. 280 20111–20119 [DOI] [PubMed] [Google Scholar]
- 41.Hofseth, L. J., Hussain, S. P., and Harris, C. C. (2004) Trends Pharmacol. Sci. 25 177–181 [DOI] [PubMed] [Google Scholar]
- 42.Nelson, W. G., and Kastan, M. B. (1994) Mol. Cell. Biol. 14 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zawel, L., Yu, J., Torrance, C. J., Markowitz, S., Kinzler, K. W., Vogelstein, B., and Zhou, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2848–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.St-Pierre, B., Flock, G., Zacksenhaus, E., and Egan, S. E. (2002) J. Biol. Chem. 277 46544–46551 [DOI] [PubMed] [Google Scholar]
- 45.Li, Y., Xie, M., Yang, J., Yang, D., Deng, R., Wan, Y., and Yan, B. (2006) Oncogene 25 3296–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura, Y., Sugimoto, M., Ohnishi, K., Sakai, T., and Hara, E. (1998) FEBS Lett. 436 169–173 [DOI] [PubMed] [Google Scholar]
- 47.Prabhu, S., Ignatova, A., Park, S. T., and Sun, X. H. (1997) Mol. Cell. Biol. 17 5888–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barone, M. V., Pepperkok, R., Peverali, F. A., and Philipson, L. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 4985–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korchynskyi, O., and ten Dijke, P. (2002) J. Biol. Chem. 277 4883–4891 [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Rovira, T., Chalaux, E., Massague, J., Rosa, J. L., and Ventura, F. (2002) J. Biol. Chem. 277 3176–3185 [DOI] [PubMed] [Google Scholar]
- 51.Ling, M. T., Wang, X., Tsao, S. W., and Wong, Y. C. (2002) Biochim. Biophys. Acta 1570 145–152 [DOI] [PubMed] [Google Scholar]
- 52.Damdinsuren, B., Nagano, H., Kondo, M., Natsag, J., Hanada, H., Nakamura, M., Wada, H., Kato, H., Marubashi, S., Miyamoto, A., Takeda, Y., Umeshita, K., Dono, K., and Monden, M. (2006) Oncol. Rep. 15 401–408 [PubMed] [Google Scholar]
- 53.Nemetski, S. M., and Gardner, L. B. (2007) J. Biol. Chem. 282 240–248 [DOI] [PubMed] [Google Scholar]
- 54.Ho, J., and Benchimol, S. (2003) Cell Death Differ. 10 404–408 [DOI] [PubMed] [Google Scholar]
- 55.Sun, H., and Taneja, R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 4058–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seimiya, M., Wada, A., Kawamura, K., Sakamoto, A., Ohkubo, Y., Okada, S., Hatano, M., Tokuhisa, T., Watanabe, T., Saisho, H., Tagawa, M., and O-Wang, J. (2004) Eur. J. Immunol. 34 1322–1332 [DOI] [PubMed] [Google Scholar]
- 57.Sun, H., Lu, B., Li, R. Q., Flavell, R. A., and Taneja, R. (2001) Nat. Immunol. 2 1040–1047 [DOI] [PubMed] [Google Scholar]
- 58.Alani, R. M., Young, A. Z., and Shifflett, C. B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7812–7816 [DOI] [PMC free article] [PubMed] [Google Scholar]