Abstract
High lactate generation and low glucose oxidation, despite normal oxygen conditions, are commonly seen in cancer cells and tumors. Historically known as the Warburg effect, this altered metabolic phenotype has long been correlated with malignant progression and poor clinical outcome. However, the mechanistic relationship between altered glucose metabolism and malignancy remains poorly understood. Here we show that inhibition of pyruvate dehydrogenase complex (PDC) activity contributes to the Warburg metabolic and malignant phenotype in human head and neck squamous cell carcinoma. PDC inhibition occurs via enhanced expression of pyruvate dehydrogenase kinase-1 (PDK-1), which results in inhibitory phosphorylation of the pyruvate dehydrogenase α (PDHα) subunit. We also demonstrate that PDC inhibition in cancer cells is associated with normoxic stabilization of the malignancy-promoting transcription factor hypoxia-inducible factor-1α (HIF-1α) by glycolytic metabolites. Knockdown of PDK-1 via short hairpin RNA lowers PDHα phosphorylation, restores PDC activity, reverts the Warburg metabolic phenotype, decreases normoxic HIF-1α expression, lowers hypoxic cell survival, decreases invasiveness, and inhibits tumor growth. PDK-1 is an HIF-1-regulated gene, and these data suggest that the buildup of glycolytic metabolites, resulting from high PDK-1 expression, may in turn promote HIF-1 activation, thus sustaining a feed-forward loop for malignant progression. In addition to providing anabolic support for cancer cells, altered fuel metabolism thus supports a malignant phenotype. Correction of metabolic abnormalities offers unique opportunities for cancer treatment and may potentially synergize with other cancer therapies.
Cancer is a disease whereby genetic mutation results in uncontrolled cell growth combined with malignancy. High lactate accumulation, despite adequate oxygen availability, is a metabolic pattern commonly associated with malignant transformation of the uncontrolled dividing cell. This metabolic phenotype, termed aerobic glycolysis and historically known as the Warburg effect, is characterized by high glycolytic rates and reduced mitochondrial oxidation (1, 2), features that may favor cell survival in the hypoxic microenvironments found in tumors. This phenotype also favors the routing of key metabolic intermediates away from oxidative destruction and toward anabolic processes required by rapidly dividing cells (2). Hypoxia and growth factors may select for this phenotype by activating hypoxia-inducible transcription factor-1 (HIF-1),3 which induces transcription of glucose transporters, glycolytic enzymes, and many other genes associated with hypoxic survival, angiogenesis, and tissue invasion (3). Hypoxia, HIF-1 activation, and high lactate levels in tumors are all independently correlated with poor clinical outcome for many human cancers (3–5). A causative role for hypoxia and HIF-1 stabilization in tumor formation and progression has been demonstrated (reviewed in Ref. 3). However, the buildup of glycolytic metabolites, such as lactate or pyruvate, in tumors has historically been viewed as a secondary phenomenon, rather than an active contributor to the malignant process (5). Recently, several intermediary glucose metabolites have been shown to promote HIF-1 activation independently of hypoxia by interacting with the HIF prolyl hydroxylases (PHD 1–3). The PHD enzymes regulate the continuous oxygen-dependent degradation of the HIF-1α subunit (6). PHDs require oxygen, iron, ascorbate, and 2-oxoglutarate for their activity. Fumarate and succinate have been shown to inhibit PHDs by competing with 2-oxoglutarate (7, 8), whereas lactate, pyruvate, and oxaloacetate appear to inactivate PHDs in an ascorbate-reversible manner (9). The discovery of these novel signaling actions of metabolites suggests that, like hypoxia, alterations of glucose routing in cancer cells may directly impact malignant progression. This has been demonstrated for the rare familial syndromes of succinate dehydrogenase deficiency and fumarate hydratase deficiency, where the buildup of succinate and fumarate, respectively, may contribute to formation of paragangliomas through sustained HIF-1α stabilization (7, 8).
The pyruvate dehydrogenase complex (PDC) controls pyruvate entry into the tricarboxylic acid cycle and thus sits at the interface between glycolysis and glucose oxidation. The PDC is composed of several enzymatic complexes and their regulatory proteins. Several differentially expressed pyruvate dehydrogenase kinases (PDK-1–4) and pyruvate dehydrogenase phosphatases (PDP1,2) tightly regulate PDC activity via reversible inhibitory phosphorylation of the pyruvate dehydrogenase E1α (PDHα) subunit (10). This regulation of PDC activity provides a convenient way for cells to divert pyruvate into anabolic pathways, while favoring oxidation of other fuels. Pyruvate dehydrogenase kinase-1 (PDK-1) was recently identified as an HIF-1-regulated gene, which directly ties HIF-1 activation to decreased glucose oxidation through PDHα phosphorylation and PDC inhibition (11, 12). We hypothesize that in cancer, by allowing accumulation of glycolytic end products that can stabilize HIF-1α, persistent PDHα phosphorylation may also potentially establish a feed-forward stimulatory loop between glucose metabolites and HIF-1, thereby accelerating malignant progression. We tested this hypothesis in human head and neck cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments—Human UM-22A, UM-22B, and JHU-O22 head and neck squamous cell carcinoma cell (HNSCC) lines were cultured in Dulbecco's modified Eagle's medium (DMEM) 1× glutamine, high glucose, no pyruvate supplemented with 10% FBS. and 1% (v/v) penicillin/streptomycin (Invitrogen). The UM-22A and UM-22B were kindly provided by Thomas Carey, University of Michigan. Cell lines were maintained in 21% O2, 5% CO2, and 74% N2 in a humidified cell incubator at 37 °C. For hypoxia treatments, culture dishes were sealed in a humidified chamber, incubated at 37 °C and continuously flushed with a gas mixture of 1% O2, 5% CO2, and 94% N2 for the indicated times. Dichloroacetic acid (DCA), l-ascorbic acid, d(+)-glucose, and pyruvate were purchased from Sigma. MG-132 was purchase from Calbiochem.
Cell Number and Viability Determination—Cells were plated in 6-well plates at 0.1–0.25 × 106 cells per well. At various times points, cells were trypsinized and counted. The cell number and viability were performed using the Vi-CELL Series cell viability analyzer (BeckmanCoulter), which determines viability based on the trypan blue dye exclusion method.
Colony Formation—Cells (104) from each cell line were placed in 1 ml of DMEM with 0.3% low melting agarose (soft agar) and 10% FBS and overlaid onto 1 ml/well DMEM with 0.5% agarose and 10% FBS. After 18 days, colonies were counted using the Nikon SMZ1500 microscope and photographed with the Nikon DXM camera.
Lactate Measurements—At various time points, 50 μl of medium was taken from cells grown in 2 ml of medium in 6-well cell culture plates and frozen until the time of measurement. Measurements were performed using the CMA 600 Analyzer (CMA-Microdialysis). This instrument performs an automated enzymatic conversion of lactate to H2O2. Peroxidase catalyzes a reaction between H2O2 and other substrates to form the red violet-colored quinonediimine. The rate of formation of quinonediimine per min is measured at 546 nm.
Preparation of Whole Cell, Nuclear, and Cytoplasmic Extracts—Nuclear and cytoplasmic extracts were prepared according to manufacturer's instructions using the NE-PER kit (Pierce). Whole cell extracts were prepared by lysing cell pellets in RIPA buffer (0.1% SDS, 1% Nonidet P-40, 5 mm EDTA, 0.5% sodium deoxycholate, 150 mm NaCl, 50 mm Tris-HCl, freshly supplemented with 2 mm dithiothreitol, protease inhibitor mixture (Roche Applied Science), 1 mm NaVO4, and 10 mm NaF) for 30–60 min on ice. Lysates were centrifuged (4 °C) at 16,000 × g for 10 min, and supernatant was collected for Western blot analysis. Protein concentration was determined using the Bradford assay (Bio-Rad).
Western Blot Analysis—Western blots were performed by running 50–75 μg of nuclear or 35–50 μg of whole cell or cytoplasmic protein extracts on 4–12% SDS-polyacrylamide gels and transferred to nitrocellulose membranes by standard procedures. Membranes were blocked with TBS/Tween with 5% nonfat milk for 1 h and then either incubated overnight at 4°C or 1 h at room temperature with the respective primary antibody: PDH E1α phosphoserine 293, 1:5000 (Novus Biologicals)4; PDK-1, 1:2500 (Assay Designs); PDK-2, 1:1000, Robert Harris Lab (Indiana University School of Medicine); PDK-3, 1:1000 (Abgent); PDH E1α, 1:5000 (Invitrogen); HIF-1α, 1:400 (BD Transduction Laboratories); HIF-1β, 1:1000 (Novus Biologicals); and β-actin, 1:10,000 (Abcam). Immunoreactive bands were visualized using horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody, 1:10,000 and enhanced chemiluminescence reagent (Pierce). Densitometry was performed using Image J software (National Institutes of Health).
Cell Transfections—Validated plasmids encoding short hairpin RNA (shRNA) for PDK-1, PDK-2, and negative control (SuperArray) were purified for transfection into 22B cells using the QIAfilter maxi plasmid kit (Qiagen Sciences) and introduced into the cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were placed under selective pressure 24 h post-transfection, using 1.5 mg/ml Geneticin® (Invitrogen).
PDC Activity Assay—PDC activity was assayed according to previously published methods with minor modifications for use with culture cells (13). Cells were grown to confluency in 100-mm dishes and rinsed with phosphate-buffered saline (PBS) (without Ca2+/Mg2+). Extraction buffer (400 μl) was added directly to the plates, and the cells were scraped and pelleted. Samples were homogenized with a motor-driven homogenizer and then centrifuged at 13,000 rpm for 10 min with a tabletop microcentrifuge. Phenylmethylsulfonyl fluoride (1 μm final concentration) was added to the PDC extraction and resuspension buffer just before use.
Glucose Oxidation—Glucose oxidation was assayed via 14CO2 capture using uniformly labeled glucose according to previously published methods (14) with the exception that Krebs buffer was used as the assay buffer, and protein contents were determined using the Bradford assay.
VEGF Enzyme-linked Immunosorbent Assay—Culture medium VEGF was measured via an enzyme-linked immunosorbent assay according manufacturer's instructions (R & D Systems). Cells were plated in 6-well dishes, and medium was replaced at the start of the 24-h experiment. At the end point, media samples were taken and frozen to assay VEGF, and cells were trypsinized and counted for normalization.
Cell Invasion Assay—Cell invasion experiments were performed using 24-well Biocoat Matrigel™ invasion chambers with an 8-μm pore polycarbonate filter according to the manufacturer's instructions (BD Biosciences). Regular growth factor Matrigel invasion chambers were used (BD Biosciences). Prior to experimentation all invasion chamber inserts were hydrated according to the manufacturer's protocol. Briefly, cells in the growing phase were trypsinized and suspended at a concentration of 4 × 105 cells/ml. The lower compartment of the plates received 750 μl of serum-free medium and/or the treatment condition designated in each experiment. 2 × 105 cells (for serum-free medium experiments) and 2.5 × 104 cells (for 10% serum medium experiments) were plated in each insert and allowed to invade for 48 h at 37 °C in a humidified incubator with 21% O2.
Xenograft of Human Tumors—BALB/c male nude mice were purchased from NCI-Frederick (Frederick, MD) and housed in a pathogen-free room controlled for temperature and humidity. All animal protocols and handling were performed under approved Uniformed Services University of the Health Sciences IACUC guidelines. Twenty (22B control shRNA, n = 10, 22B PDK-1 shRNA, n = 10) 6–8-week-old mice were injected subcutaneously in the scruff with 1 × 107 cells in 100 μl of PBS. Tumor bearing mice (9 from each group) were used for the experiment. One week after injection, tumor volume was assessed bi-weekly for 2 months using a digital caliper and calculated using the following formula: volume = (a × b2)/2, where a is the widest diameter of the tumor, and b is the diameter perpendicular to a. The results from individual mice were graphed as average tumor volume over 60 days.
1H NMR Spectroscopy—Six replicate and three replicate flasks were cultured for the 22B control shRNA and 22B PDK-1 shRNA cell lines, respectively. Cells were plated at 70% confluency, and media and cells were collected when cells reached confluency. Cells were rinsed with PBS and then harvested by scraping into 5 ml of PBS and centrifuging for 5 min at 600 × g. PBS was removed, and cells were stored at -80 °C until extraction procedure.
CHCl3/Methanol Cell Extraction—Cells were suspended in 200 μl of CHCl3 and 100 μl of methanol on ice and mixed vigorously. The cells were thoroughly mixed with 300 μl of H2O before the resulting suspension was centrifuged at 9500 × g for 10 min. The upper aqueous layer was separated from the lower organic layer and allowed to stand overnight to allow methanol to evaporate before freeze-drying. The resultant aqueous extract was reconstituted for NMR in 500 μl of deuterated 0.1% sodium phosphate buffer containing 0.002% 3,3,3-trimethylsilyl propionate, as an internal reference peak, and 0.02% azide.
NMR Data Acquisition—NMR spectra were acquired on a Bruker DRX600 spectrometer operating at 600.13-MHz 1H NMR frequency and 300 K. 1H NMR spectra of the samples were acquired using a one-dimensional nuclear Overhauser effect spectroscopy pulse sequence (RD-90°-t1-90°-tm-90°-acquire), which generates an unedited spectrum with improved solvent peak suppression. For all spectra 256 free induction decays were collected into 32 K complex data points, using a spectral width of 12,019 Hz (20 ppm), with a 2-s relaxation delay between pulses. A water presaturation pulse was applied throughout the relaxation delay.
NMR Spectral Data Processing—NMR spectra were acquired on a Bruker DRX600 spectrometer using the standard one-dimensional pulse sequence and processed as described previously (15). Spectra were referenced internally to the anomeric proton signal of α-glucose at 5.23 ppm. Spectra were normalized to an internal reference peak. Significant differences in resonance intensities were determined using the probabilistic quotient normalization method (16).
All averaged results are presented as mean ± S.D. Differences in means were statistically analyzed using a two-tailed Student's t test. A probability level of 0.05 was considered significant.
RESULTS
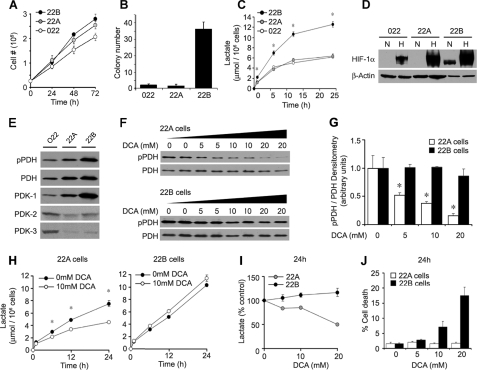
PDC and HIF-1 in Human HNSCC Cells—To elucidate a possible role for PDHα phosphorylation in the malignant phenotype of cancer cells, we utilized the human head and neck squamous cell carcinoma (HNSCC) cells lines UM-22A, UM-22B, and JHU-O22. Two of these were generated from a primary (UM-22A) and metastatic lesion (UM-22B) in the same patient (17). Although these cancer cell lines showed comparable anchorage-dependent growth properties (Fig. 1A), the 22B cells displayed much greater anchorage-independent growth in soft agar colony formation assays (Fig. 1B) and twice the lactate accumulation seen with the other cell lines (Fig. 1C). Also, the 22B cells alone accumulated nuclear HIF-1α protein after 12 h in culture under normal oxygen conditions (Fig. 1D). Normoxic or basal HIF-1α expression can be mediated by products of glucose metabolism and is additive with hypoxia-induced HIF-1α accumulation (9). In keeping with this, 22B cells also displayed the highest induction of HIF-1α following 4-h exposures to hypoxia (1% O2) (Fig. 1D). To determine the respective PDHα phosphorylation status of the HNSCC cell lines, we used a PDHα phosphoserine 293 (site 1)-specific antisera since phosphorylation of this site produces nearly complete inhibition of PDC activity (18). A monoclonal antibody was used to measure total PDHα expression, and densitometry was used to determine the ratio of phospho-PDHα to total PDHα. The highest ratio of phospho-PDHα to total PDHα was observed in the 22B cells (Fig. 1E). Although differential expression of PDK isoforms 1–3 was seen among the head and neck cancer cell lines, we observed the highest basal expression of PDK-1 in 22B cells (Fig. 1E).
FIGURE 1.
Growth, metabolic phenotype, and PDHα phosphorylation in HNSCC cell lines. A, 22A, 22B, and O22 cells demonstrate similar anchorage-dependent growth, but only the 22B cells (B) develop significant anchorage-independent colonies (n = 3). C, 22B cells demonstrate higher lactate production than O22 and 22A cells (n = 3). D, Western blot analysis of nuclear extracts shows 22B cells have greater normoxic (N) and hypoxic (H) HIF-1α protein expression than O22 and 22A cells. E, Western blot analysis of whole cell lysate for phosphorylated PDHα (pPDHα), PDHα, and PDK-1–3 protein expression shows 22B cells to have the highest pPDHα and PDK-1 expression. F, Western blot analysis of pPDHα and PDHα protein expression in whole cell lysates from 22A and 22B cells treated with 0, 5, 10, and 20 mm DCA for 24 h shows dose-dependent decrease of PDHα phosphorylation in 22A but not 22B cells. G, densitometric ratio of immunoreactive pPDHα/PDHα from F. H and I, DCA lowers lactate production in 22A but not 22B cells. J, DCA shows dose-dependent toxicity in 22B cells but not 22A cells.
DCA Treatment in HNSCC Cells—Recent studies with the weak PDK inhibitor DCA have suggested that inhibition of PDHα phosphorylation may activate PDC, shift cancer cell metabolism toward aerobic respiration, promote cancer cell apoptosis, and reduce tumor growth (11, 12, 19, 20). However, none of these studies documented direct changes in PDHα phosphorylation or PDC enzymatic activity. Using the phospho-specific antibody, we found that DCA decreased PDHα phosphorylation dose-dependently in 22A cells but was surprisingly without significant effect in 22B cells, even at concentrations up to 20 mm (Fig. 1, F and G). Also, the time- and dose-dependent inhibition of lactate release by DCA observed in 22A cells was absent in 22B cells (Fig. 1, H and I). Moreover, at the high doses required for its effects, DCA produced toxicity in the 22B cells but not the 22A cells (Fig. 1J). In addition to being a weak inhibitor of PDK, DCA also blocks glutathione S-transferase ζ (21), inhibits cholesterol biosynthesis (22), promotes oxidative stress (23), and activates peroxisome proliferator-activated receptors (24). Both these published observations and our data persuaded us to use more specific strategies for implicating a role for PDHα phosphorylation in cancer biology.
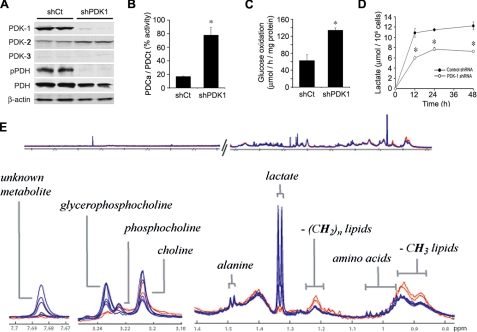
PDK Inhibition with shRNA—We therefore utilized shRNA to knock down PDK-1 expression in 22B cells. Cells were transfected with a vector under control of the U1 promoter containing a neomycin-resistant gene, and stably expressing cell lines were selected by antibiotic treatment. Western blot analysis showed the effective reduction of PDK-1 expression in the stably transfected PDK-1 shRNA (shPDK-1) cells when compared with cells transfected with control shRNA (shCt) (Fig. 2A). PDK-2 expression was slightly increased in the shPDK-1 cells, whereas PDK-3 expression remained low in both cell lines (Fig. 2A). Western blotting confirmed reduction of PDHα phosphorylation but not PDHα expression following PDK-1 knockdown (Fig. 2A). Measurement of PDC enzymatic activity in cell lysates showed that shCt cell extracts had 60% lower PDC activity than the shPDK-1 cells.5 However, after addition of recombinant PDP-1 to maximally dephosphorylate PDHα, and thus fully activate PDC, the shCt cell extracts actually showed greater total activity than the shPDK-1 cell extracts,5 confirming strong phosphorylation-dependent inhibition. The ratio of active to total PDC was four times higher in the 22B shPDK-1 cells compared with the 22B shCt cells (Fig. 2B). Consistent with this, the shPDK-1 cells were found to oxidize uniformly labeled [14C]glucose to 14CO2 at twice the rate of shCt cells (Fig. 2C). Furthermore, whereas 22B shCt cells generated similar amounts of medium lactate as the nontransfected 22B cells, lactate secretion by the shPDK-1 cells was decreased to levels similar to the 22A and O22 cell lines (Fig. 2D). To rule out the influence of differential lactate transport rates between the cells, we also analyzed the aqueous component of the cell extracts by 1H NMR spectroscopy. This approach confirmed a pronounced reduction in cellular lactate following PDK-1 knockdown but few other changes in metabolite accumulation patterns (Fig. 2E). Notable other changes were a decrease in glycerophosphocholine and branched-chain amino acids (isoleucine, leucine, and valine) and a rise in free CH2 and CH3 lipids following PDK-1 knockdown. An unidentified metabolite at corresponding to a singlet at 7.69 ppm was also markedly reduced in the shPDK-1-transfected cells.
FIGURE 2.
Dependence of Warburg metabolic phenotype on PDK expression. A, Western blot of 22B cells stably transfected with shPDK-1 show markedly reduced PDK-1 and pPDHα expression when compared with shCt cells, whereas PDK-2 and -3 expression changes little. B, ratio of active to total PDC activity is higher in shPDK-1 compared with shCt cells (n = 4, p < 0.001). Total activity was determined as activity following PDP-1 treatment of cell extracts. C, 14CO2 captured from uniformly labeled glucose is higher in shPDK-1 compared with shCt cells (n = 4, p < 0.01). D, extracellular lactate release is decreased in shPDK-1 cells compared with shCt (n = 3, p < 0.001). E, 1H NMR spectroscopic analysis of extracts from shCT cells (blue trace, n = 6) and shPDK-1 cells (red trace, n = 2) shows most prominent change in the lactate peaks.
Comparative analysis of the 1H NMR spectra generated for the culture medium revealed elevated levels of glucose in the medium that contained the PDK-1 shRNA cell lines compared with the controls (p < 0.01), where glucose to lactate ratios were also significantly higher (p < 0.05). The data also suggested elevated levels of branched chain amino acids in the same medium, although this was not statistically significant (p = 0.085).6
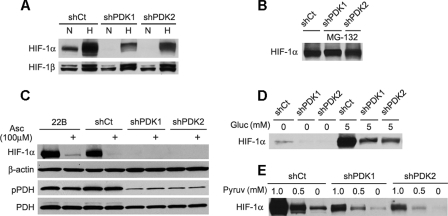
Decreased HIF-1α with PDK Inhibition—Many cancer cells are known to have a high level of HIF-1α accumulation under basal as well as hypoxic conditions (9, 25). Prior work has shown that glucose metabolism contributes to this normoxic HIF-1 activation in cancer cells. Removal of medium glucose or its substitution with nonglycolytic fuel sources has been shown to diminish hypoxia-independent basal HIF-1α accumulation and also to lower HIF-1α stabilization by hypoxia (9, 25), suggesting a key role for glycolytic metabolism in regulating both normoxic and hypoxic HIF-1 activation. To examine whether altering the ratio of glycolysis to glucose oxidation via PDC activation had an impact on HIF-1α stabilization, we measured nuclear HIF-1α accumulation in cells following 16 h of culture with or without exposure to hypoxia (1% O2) for the final 4 h (Fig. 3A). We found that the normoxic and hypoxia-dependent HIF-1α accumulation seen in the parent 22B cells was retained in the shCt cells. However, PDK-1 knockdown eliminated normoxic and reduced hypoxic accumulation of HIF-1α. A similar effect was seen in 22B cells stably transfected with shRNA targeting PDK-2, which has been proposed to form functional heterodimers with PDK-1 (26). However, HIF-1α accumulation following treatment with the proteasome inhibitor MG-132 was similar in all transfectants (Fig. 3B), suggesting that HIF-1α reduction by PDK knockdown was not because of decreased HIF-1α protein production. Normoxic or glycolytic HIF-1α accumulation also differs from hypoxic HIF-1α accumulation in that it is reduced by ascorbate (9, 27, 28). The basal HIF-1α accumulation seen in the parent 22B and the shCt cells following 24 h of culture in Krebs buffer was indeed abolished by 100 μm ascorbate (Fig. 3C). Ascorbate did not, however, affect the phosphorylation status of PDHα, suggesting that the reduction of normoxic HIF-1α accumulation by ascorbate occurs independently from PDC activity.
FIGURE 3.
HIF-1α expression is reduced in stably transfected PDK shRNA 22B cell lines. A, Western blot of nuclear extracts shows loss of normoxic (N) and decreased hypoxic (H) HIF-1α expression in shPDK-1 and shPDK-2 cells compared with shCt. B, basal HIF-1 Western blot of normoxic HIF-1α expression following treatment with the 26 S proteosome inhibitor MG132 (15 μm) shows equivalent HIF-1α expression between the cell lines. C, ascorbate treatment (100 μm for 24 h) decreases normoxic HIF-1α expression independently of PDH phosphorylation demonstrated by Western blot of nuclear (HIF-1α and β-actin) and cytoplasmic (pPDHα and PDHα) cell extracts. Cells in A–C were cultured for 24 in complete medium. D, Western blot of nuclear extracts from cells cultured in Krebs buffer shows higher glucose-dependent HIF-1α expression in shCt cells as compared with shPDK-1 and shPDK-2 cells. E, dose-response curves for pyruvate induction of HIF-1 in glucose-free Krebs buffer show higher pyruvate sensitivity in shCt cells than shPDK-1 or shPDK-2 cells. Incubation time for experiments in D and E was 4 h.
The direct regulation of HIF-1 by glucose metabolism is most readily and specifically demonstrated using short term culture of cells in Krebs buffer rather than in complete medium, which contains added serum factors and amino acids (9, 25, 27). Under these conditions, the shCt 22B cells showed a much stronger HIF-1α accumulation in the presence of 5 mm glucose than the shPDK-1 and shPDK-2 cells (Fig. 3D). Elimination of glucose from the Krebs buffer prevented HIF-1α accumulation in all cells. HIF-1α stabilization by glucose is also known to require the metabolism of glucose to lactate and pyruvate (9). Both of these metabolites can substitute for glucose in promoting normoxic HIF-1α accumulation in many cell lines, but lactate requires conversion to pyruvate for its action (9, 25). We observed that substitution of pyruvate for glucose in the Krebs buffer produced a more robust, dose-dependent HIF-1α accumulation over 4 h in the shCt compared with the shPDK cells (Fig. 3E). The basal HIF-1α accumulation in 22B cells was lost following PDK-1 and PDK-2 knockdown. These observations suggested that reduction of PDC activity via PDK-mediated phosphorylation of PDHα can sensitize cells toward HIF-1α stabilization by glucose or glycolytic metabolites.
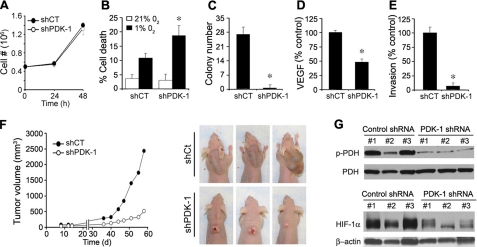
PDK Inhibition Reduces Expression of a Malignant Phenotype—We next determined whether several phenotypic biological changes relevant to malignancy were altered by enhancement of PDC activity in 22B cells. No difference was observed in the anchorage-dependent growth of 22B cells following knock-down of PDK-1 (Fig. 4A). However, the shPDK-1 cells showed significantly higher cell death under prolonged hypoxia (Fig. 4B), as well as a profound reduction in their ability to form colonies in soft agar (Fig. 4C). Moreover, the shPDK-1 cells also displayed a 50% decrease in VEGF secretion as assessed by VEGF enzyme-linked immunosorbent assay analysis (Fig. 4D) and showed a loss of invasiveness in Matrigel™-coated Boyden chambers (Fig. 4E). These in vitro results suggested that PDC activity may be an important determinant of aggressive tumor growth. To examine the impact of PDK knockdown on in vivo tumor growth, we performed xenograft studies using nude mice. After injecting 1 × 107 cells into the posterior neck scruff of 10 mice for each group, palpable tumors were seen at 1 week and subsequently measured bi-weekly for 2 months. Nine of 10 mice formed tumors in each group. Fig. 4F (left panel) shows that PDK-1 knockdown led to a dramatic reduction of tumor growth. Three examples of representative tumors show the marked differences seen in gross tumor morphology (Fig. 4F, right panel). The shPDK-1 tumors remained white with the development of externally visible erosive areas. The much larger shCt tumors exhibited no externally visible erosion and appeared to be well vascularized based on their dark color and subsequent histology.7 Western blot analysis of three separate tumors from each group showed that the differential PDHα phosphorylation state and HIF-1α accumulation observed in vitro was maintained in vivo (Fig. 4G).
FIGURE 4.
In vitro and in vivo reversal of malignant phenotype by PDK inhibition in 22B cells. A, shPDK-1 and shCt cells demonstrate similar anchorage-dependent growth rates. B, hypoxia treatment increases shPDK-1 cell death 2-fold greater than 22B shCt (n = 3, p < 0.01). C, shPDK-1 cells demonstrate decreased capacity to form colonies in soft agar compared with shCt (n = 3, p < 0.001). D, VEGF production, assayed by enzyme-linked immunosorbent assay, is reduced in shPDK-1 cells compared with shCt cells (n = 6, p < 0.001). E, invasiveness through Matrigel™-coated Boyden chambers is decreased in shPDK-1 cells compared with 22B shCt cells (n = 6, p < 0.001). F, left panel, xenograft tumor growth in nude mice is reduced for shPDK-1 cells compared with shCt cells (n = 9, p < 0.05). F, right panel, three representative images of shCt and shPDK-1 tumors are shown. G, Western blot analysis of cytoplasmic (pPDHα and PDHα) and nuclear (HIF-1α and β-actin) cell extracts from three separate xenograft tumors from each group shows that the decreased pPDHα and HIF-1α expression in shPDK-1 compared with shCt cells observed in vitro is maintained in vivo.
DISCUSSION
Altered Glucose Metabolism in Cancer—Aerobic glycolysis occurs transiently in normal cells, but has been known for more than 80 years as a persistent feature of malignant cells. Commonly referred to as the Warburg effect, this metabolic pattern reflects a relative imbalance between the glycolytic and oxidative phases of glucose metabolism. There are likely to be many prerequisites of cell transformation that occur before aerobic glycolysis becomes associated with malignancy, including the loss of cell cycle regulation. However, several of the oncogenes commonly dysregulated in cancer, including Akt (29), Myc (30), and Ras (31), also appear to up-regulate overall glycolysis. HIF-1 also directly enhances glycolysis by increasing the transcription of glycolytic enzyme genes, and HIF stabilization is a prominent feature of many cancers (3). This is in part due to the above oncogenic dysregulations but also due to the diffusion-limited and hypoxic microenvironment of solid tumors (3, 5). In addition to up-regulated glycolysis, decreased glucose oxidation in cancer cells can result from mitochondrial DNA mutations (27), p53 mutations (32, 33), and rare familial mutations in the tricarboxylic acid cycle enzymes succinate dehydrogenase and fumarate hydratase (7, 8, 34). Thus, many molecular features of cancer cells can increase the ratio of glycolytic to oxidative glucose metabolism and lead to the buildup of glycolytic metabolites.
Altered lactate dehydrogenase activity and decreased PDC activity have also gain attention as contributors to the aerobic glycolysis pattern in cancer cells (19, 20, 35, 36). These enzymatic activities are positioned at the interface of glycolytic and oxidative glucose metabolism and normally serve to regulate the transient imbalances between glycolytic and oxidative glucose metabolism in normal tissues. PDC activity is regulated by reversible inhibitory phosphorylation as a normal homeostatic regulatory mechanism and is used by organs, such as the liver, and muscle to help adjust metabolic fuel routing in response to changes in blood nutrient levels (10). These physiological changes in PDC phosphorylation are normally transient in nature. However, the identification of PDK-1 as an HIF-1 regulated gene has suggested that the PDC may become persistently inhibited in cancer cells and may be a key factor in producing the Warburg aerobic glycolysis metabolic pattern (11, 12).
Novel Signaling Actions of Altered Metabolism in Cancer—Given the above links between molecular dysregulation and metabolic phenotype in cancer cells, it is not surprising that the accumulation of glycolytic metabolites in human tumors is strongly associated with poor clinical outcome (5). That the accumulation of glycolytic and tricarboxylic acid cycle metabolites may actually enhance malignancy in cancer is, however, a more recent and surprising realization. Promotion of angiogenesis (37, 38), metastasis (5), and immune evasion (39) are among the relevant biological actions recently attributed to glycolytic metabolites. The seminal finding of HIF-1 regulation by glycolytic metabolites (25) has provided one possible mechanism for these novel biological effects. Lactate, pyruvate, and several tricarboxylic acid cycle intermediates have been shown to stabilize HIF-1 under normoxic conditions and by direct or indirect inhibition the prolyl hydroxylase (PHD) enzymes, which target HIF-1α for degradation. Both succinate and fumarate have been reported to stabilize HIF-1 in a manner reversible by 2-oxoglutarate, the natural substrate of the PHDs (7, 8). On the other hand, pyruvate, oxaloacetate (9), and malate (40) have been reported to inhibit PHD activity in a 2-oxoglutarate-independent manner. Because addition of ascorbate and Fe2+, the other required PHD cofactors, reverses their inhibitory effect, these latter metabolites have been proposed to promote uncoupled enzyme reactions that lock PHDs into inactive states (9, 34). Some of the discrepancies reported in the literature regarding these phenomena may reflect differences between experimental conditions used with in vitro and cell-based assays, potential inter-conversion of metabolites, and cell type-specific metabolic phenotypes (7–9, 25, 27, 34, 41, 42). Nevertheless, enhanced HIF-1 stabilization by glycolytic metabolites represents an important concept for understanding the close association of aerobic glycolysis with malignancy. We have previously shown that even in normal human primary cells sustained elevation of lactate and pyruvate can indeed increase HIF-1 stabilization (25). We thus believe that HIF-1 regulation by glycolytic metabolites may represent a widespread signaling mechanism akin to hypoxic regulation of HIF-1. As with hypoxic activation of HIF-1, regulation of this transcription factor by glycolytic metabolites is likely to be beneficial for normal tissues (37). This is because HIF-1 activates genes governing angiogenesis, erythropoiesis, and enhanced cell survival (3). Unfortunately, this same response is believed to also be beneficial for the survival of cancer cells, much to the detriment of the organism. Thus, although sustained HIF-1 activation does not cause cancer, it can amplify malignant progression by enhancing survival of cancer cells, particularly under hypoxia, as demonstrated in our paper. This capability is essential for formation of solid tumor, in which hypoxia is a key selection pressure for cell survival.
Reciprocal Interactions between PDK-1 and HIF-1 Activity—Here we have demonstrated that inhibition of PDK-1 expression in HNSCC leads to a dramatic reduction in lactate levels, HIF-1α expression, and attenuation of the expression of a malignant phenotype. Thus, restoration of PDC activity via inhibition of PDHα phosphorylation in cancer cells not only blunts the Warburg phenotype but also reduces in vivo malignant potential. The reduction in expression of a malignant phenotype following PDK knockdown may in part be due to the reduced HIF-1α expression. Although PDK-1 is known to be an HIF-1-regulated protein (11, 12), this work is the first to demonstrate that expression of PDK-1 can in turn regulate HIF-1. Moreover, our data suggest that the reduction in HIF-1α stability seen following PDK knockdown may in fact be due to decreased buildup of glycolytic metabolites, as reflected by lactate. The approaches used in this study were not sensitive enough to resolve changes in and interconversions between various glycolytic metabolites. However, regulation of HIF-1α accumulation by glucose and pyruvate was observed to be dependent on the state of PDC inhibition and was also sensitive to ascorbate but not 2-oxoglutarate.8
In addition to changes in cellular energy metabolism, redox status has been shown to modulate hypoxia-inducible factor prolyl hydroxylase activity in vivo and in vitro (9, 34). Following PDK-1 knockdown, we observed a prominent change in the glucose metabolism profile and in the ability of glucose metabolites to regulate HIF-1. We cannot rule out that this represents adaptations in redox buffering systems in the stably transfected shPDK-1 and -2 cells. Regardless, the identification of PDK-1 as an HIF-1-regulated protein along with our findings showing that inhibition of PDK-1 and -2 expression reduces basal HIF-1α suggests the existence of important signaling loops between cell metabolism and HIF-1. It also suggests that mutations in cancer that result in either increased HIF-1 or PDK expression can activate a malignancy-promoting feed-forward loop.
Glucose Metabolites and Anabolic Support—HIF-1 is known to induce transcription of most glycolytic genes and glucose transporters. By also enhancing the phosphorylation of PDHα via PDK-1 induction, sustained HIF-1 activation may help ensure that dividing cells get enough ATP from glycolysis, whereas sparing glycolytic metabolites for anabolic purposes. The routing of glucose-derived and other metabolic intermediates toward the synthesis of macromolecules instead of toward oxidation supports the high rate of cell division and anabolic increase in cancer cells (2). The reduction in lactate accumulation and enhancement of glucose oxidation in the shPDK-1 cells was accompanied by a significant drop in glycerophosphocholine levels and a rise in total CH2 and CH3 lipids. This suggests that the routing of glucose metabolism by PDC activity may also impact lipid oxidation and membrane lipid turnover in cancer cells. Of note, membrane lipid turnover is a key feature of dividing cells and is also involved in growth factor signaling (2).
The PDC as a Therapeutic Target for Cancer—Pharmacological inhibition of PDK with DCA was reported recently to inhibit xenograft tumor growth in a variety of carcinoma and glioblastoma cancer cell lines (19). DCA treatment was also shown to increase the effects of cytotoxic therapy (20). However, DCA is a weak PDK inhibitor and may have differential potency for distinct PDK isoenzyme combinations (26). These studies did not clearly demonstrate reduced PDHα phosphorylation and PDC activity as the mechanism of the DCA effects. In our study the effectiveness of DCA in lowering PDHα phosphorylation and lactate generation was much greater for the primary 22A cells than for the metastatic 22B cells. Although we do not yet know whether this differential sensitivity is because of differential activity of PDK and PDP homologues in the two cell types, our results clearly show heterogeneous DCA sensitivity in human cancer cells. DCA also produced cytotoxicity in the 22B cells even in the absence of evidence for inhibition of PDHα phosphorylation. Thus, in addition to providing support for PDK isoforms as potential therapeutic targets for killing cancer cells, our study suggests a need for developing better PDK inhibitors. PDK activity can also be enhanced allosterically by the intramitochondrial accumulation of NADH or acetyl-CoA (10), which may allow downstream mitochondrial molecular lesions, such as tricarboxylic acid cycle enzyme defects, electron transport chain defects, and cytochrome c oxidase defects to also inhibit PDC activity and produce the Warburg metabolic phenotype in cancer cells. Besides PDK inhibitors, there may be other approaches for correcting the metabolic milieu within tumors, including the optimization of vitamins and co-enzymes required for PDC activity. Recent advances in the ability to quantitatively image tumor lactate and pyruvate in biopsies (5, 43), as well as in vivo (44, 45), now offer an ideal opportunity to test therapeutic approaches aimed at reverting the Warburg effect in cancer. Our results also stress the need for a better understanding of cell and tumor type-specific metabolic responses to therapy.
Acknowledgments
We thank Jake Friedman for assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants NS73814 and CA113506 (to A. V.), DK47844 (to R. H.), P50 CA96784 and R01DE015939 from NIDCR (to J. C.) and SPORE grant from NCI. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HIF-1, hypoxia-inducible factor; PDC, pyruvate dehydrogenase complex; PDH, pyruvate dehydrogenase; PDK, pyruvate dehydrogenase kinase; PDP, pyruvate dehydrogenase phosphatase; PHD, HIF prolyl hydroxylase; HNSCC, human head and neck squamous cell carcinoma; DCA, dichloroacetate; shRNA, short hairpin ribonucleic acid; VEGF, vascular endothelial growth factor; DMEM, Dulbecco's modified Eagle's medium; PBS, phosphate-buffered saline; shRNA, short hairpin RNA.
N. D. Halim, T. McFate, A. Mohyeldin, P. Okagaki, L. G. Korotchkina, M. S. Patel, N. H. Jeoung, R. A. Harris, M. J. Schell, and A. Verma, manuscript in preparation.
N. H. Jeoung, unpublished data.
O. Teahan and T. M. Tseng, unpublished data.
T. McFate and A. Mohyeldin, unpublished data.
A. Verma, unpublished data.
References
- 1.Warburg, O. (1930) The Metabolism of Tumors, Constable Press, London, UK
- 2.Deberardinis, R. J., Lum, J. J., Hatzivassiliou, G., and Thompson, C. B. (2008) Cell Metab. 7 11-20 [DOI] [PubMed] [Google Scholar]
- 3.Harris, A. L. (2002) Nat. Rev. Cancer 2 38-47 [DOI] [PubMed] [Google Scholar]
- 4.Mabjeesh, N. J., and Amir, S. (2007) Histol. Histopathol. 22 559-572 [DOI] [PubMed] [Google Scholar]
- 5.Walenta, S., Schroeder, T., and Mueller-Klieser, W. (2004) Curr. Med. Chem. 11 2195-2204 [DOI] [PubMed] [Google Scholar]
- 6.Jaakkola, P., Mole, D. R., Tian, Y. M., Wilson, M. I., Gielbert, J., Gaskell, S. J., von Kriegsheim, A., Hebenstreit, H. F., Mukherji, M., Schofield, C. J., Maxwell, P. H., Pugh, C. W., and Ratcliffe, P. J. (2001) Science 292 468-472 [DOI] [PubMed] [Google Scholar]
- 7.Isaacs, J. S., Jung, Y. J., Mole, D. R., Lee, S., Torres-Cabala, C., Chung, Y. L., Merino, M., Trepel, J., Zbar, B., Toro, J., Ratcliffe, P. J., Linehan, W. M., and Neckers, L. (2005) Cancer Cell 8 143-153 [DOI] [PubMed] [Google Scholar]
- 8.Selak, M. A., Armour, S. M., Mackenzie, E. D., Boulahbel, H., Watson, D. G., Mansfield, K. D., Pan, Y., Simon, M. C., Thompson, C. B., and Gottlieb, E. (2005) Cancer Cell 7 77-85 [DOI] [PubMed] [Google Scholar]
- 9.Lu, H., Dalgard, C. L., Mohyeldin, A., McFate, T., Tait, A. S., and Verma, A. (2005) J. Biol. Chem. 280 41928-41939 [DOI] [PubMed] [Google Scholar]
- 10.Roche, T. E., and Hiromasa, Y. (2007) Cell. Mol. Life Sci. 64 830-849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, J. W., Tchernyshyov, I., Semenza, G. L., and Dang, C. V. (2007) Cell Metab. 3 177-185 [DOI] [PubMed] [Google Scholar]
- 12.Papandreou, I., Cairns, R. A., Fontana, L., Lim, A. L., and Denko, N. C. (2007) Cell Metab. 3 187-197 [DOI] [PubMed] [Google Scholar]
- 13.Jeoung, N. H., Sanghani, P. C., Zhai, L., and Harris, R. A. (2006) Anal. Biochem. 356 44-50 [DOI] [PubMed] [Google Scholar]
- 14.Itoh, Y., Esaki, T., Shimoji, K., Cook, M., Law, M. J., Kaufman, E., and Sokoloff, L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4879-4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, E., Tsang, T. M., Huang, J. T., Leweke, F. M., Koethe, D., Gerth, C. W., Nolden, B. M., Gross, S., Schreiber, D., Nicholson, J. K., and Bahn, S. (2006) PLoS Med. 3 e327, 1420-1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieterle, F., Ross, A., Schlotterbeck, G., and Senn, H. (2006) Anal Chem. 78 4281-4290 [DOI] [PubMed] [Google Scholar]
- 17.Zou, C. P., Youssef, E. M., Zou, C. C., Carey, T. E., and Lotan, R. (2001) Oncogene 20 6820-6827 [DOI] [PubMed] [Google Scholar]
- 18.Patel, M. S., and Korotchkina, L. G. (2006) Biochem. Soc. Trans. 34 217-222 [DOI] [PubMed] [Google Scholar]
- 19.Bonnet, S., Archer, S. L., Allalunis-Turner, J., Haromy, A., Beaulieu, C., Thompson, R., Lee, C. T., Lopaschuk, G. D., Puttagunta, L., Bonnet, S., Harry, G., Hashimoto, K., Porter, C. J., Andrade, M. A., Thebaud, B., and Michelakis, E. D. (2007) Cancer Cell 11 37-51 [DOI] [PubMed] [Google Scholar]
- 20.Cairns, R. A., Papandreou, I., Sutphin, P. D., and Denko, N. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9445-9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tzeng, H. F., Blackburn, A. C., Board, P. G., and Anders, M. W. (2000) Chem. Res. Toxicol. 13 231-236 [DOI] [PubMed] [Google Scholar]
- 22.Stacpoole, P. W., and Greene, Y. J. (1992) Diabetes Care 15 785-791 [DOI] [PubMed] [Google Scholar]
- 23.Hassoun, E. A., and Ray, S. (2003) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 135 119-128 [DOI] [PubMed] [Google Scholar]
- 24.Laughter, A. R., Dunn, C. S., Swanson, C. L., Howroyd, P., Cattley, R. C., and Corton, J. C. (2004) Toxicology 203 83-98 [DOI] [PubMed] [Google Scholar]
- 25.Lu, H., Forbes, R. A., and Verma, A. (2002) J. Biol. Chem. 277 23111-23115 [DOI] [PubMed] [Google Scholar]
- 26.Boulatnikov, I., and Popov, K. M. (2003) Biochim. Biophys. Acta 1645 183-192 [DOI] [PubMed] [Google Scholar]
- 27.Zhou, S., Kachhap, S., Sun, W., Wu, G., Chuang, A., Poeta, L., Grumbine, L., Mithani, S. K., Chatterjee, A., Koch, W., Westra, W. H., Maitra, A., Glazer, C., Carducci, M., Sidransky, D., McFate, T., Verma, A., and Califano, J. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7540-7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowles, H. J., Raval, R. R., Harris, A. L., and Ratcliffe, P. J. (2003) Cancer Res. 63 1764-1768 [PubMed] [Google Scholar]
- 29.Elstrom, R. L., Bauer, D. E., Buzzai, M., Karnauskas, R., Harris, M. H., Plas, D. R., Zhuang, H., Cinalli, R. M., Alavi, A., Rudin, C. M., and Thompson, C. B. (2004) Cancer Res. 64 3892-3899 [DOI] [PubMed] [Google Scholar]
- 30.Shim, H., Dolde, C., Lewis, B. C., Wu, C. S., Dang, G., Jungmann, R. A., Dalla-Favera, R., and Dang, C. V. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6658-6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blum, R., Jacob-Hirsch, J., Amariglio, N., Rechavi, G., and Kloog, Y. (2005) Cancer Res. 65 999-1006 [PubMed] [Google Scholar]
- 32.Zhou, S., Kachhap, S., and Singh, K. K. (2003) Mutagenesis 18 287-292 [DOI] [PubMed] [Google Scholar]
- 33.Matoba, S., Kang, J. G., Patino, W. D., Wragg, A., Boehm, M., Gavrilova, O., Hurley, P. J., Bunz, F., and Hwang, P. M. (2006) Science 312 1650-1653 [DOI] [PubMed] [Google Scholar]
- 34.Gottlieb, E., and Tomlinson, I. P. (2005) Nat. Rev. Cancer 5 857-866 [DOI] [PubMed] [Google Scholar]
- 35.Fantin, V. R., St-Pierre, J., and Leder, P. (2006) Cancer Cell 9 425-434 [DOI] [PubMed] [Google Scholar]
- 36.Koukourakis, M. I., Giatromanolaki, A., Bougioukas, G., and Sivridis, E. (2007) Cancer Biol. Ther. 6 1476-1479 [DOI] [PubMed] [Google Scholar]
- 37.Hunt, T. K., Aslam, R. S., Beckert, S., Wagner, S., Ghani, Q. P., Hussain, M. Z., Roy, S., and Sen, C. K. (2007) Antioxid. Redox. Signal. 9 1115-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, M. S., Moon, E. J., Lee, S. W., Kim, M. S., Kim, K. W., and Kim, Y. J. (2001) Cancer Res. 61 3290-3293 [PubMed] [Google Scholar]
- 39.Fischer, K., Hoffmann, P., Voelkl, S., Meidenbauer, N., Ammer, J., Edinger, M., Gottfried, E., Schwarz, S., Rothe, G., Hoves, S., Renner, K., Timischl, B., Mackensen, A., Kunz-Schughart, L., Andreesen, R., Krause, S. W., and Kreutz, M. (2007) Blood 109 3812-3819 [DOI] [PubMed] [Google Scholar]
- 40.Pan, Y., Mansfield, K. D., Bertozzi, C. C., Rudenko, V., Chan, D. A., Giaccia, A. J., and Simon, M. C. (2007) Mol. Cell. Biol. 27 912-925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koivunen, P., Hirsilä, M., Remes, A. M., Hassinen, I., E., Kivirikko, K. I., and Myllyharju, J. (2007) J. Biol. Chem. 282 4524-4532 [DOI] [PubMed] [Google Scholar]
- 42.Hewitson, K. S., Liénard, B. M., McDonough, M. A., Clifton, I. J., Butler, D., Soares, A. S., Oldham, N. J., McNeill, L. A., and Schofield, C. J. (2007) J. Biol. Chem. 282 3293-3301 [DOI] [PubMed] [Google Scholar]
- 43.Sattler, U. G., Walenta, S., and Mueller-Klieser, W. (2007) Lab. Investig. 87 84-92 [DOI] [PubMed] [Google Scholar]
- 44.Golman, K., Zandt, R. I., Lerche, M., Pehrson, R., and Ardenkjaer-Larsen, J. H. (2006) Cancer Res. 66 10855-10860 [DOI] [PubMed] [Google Scholar]
- 45.Day, S. E., Kettunen, M. I., Gallagher, F. A., Hu, D. E., Lerche, M., Wolber, J., Golman, K., Ardenkjaer-Larsen, J. H., and Brindle, K. M. (2007) Nat. Med. 13 1382-1387 [DOI] [PubMed] [Google Scholar]