Abstract
Riboswitches are metabolite-sensitive elements found in mRNAs that control gene expression through a regulatory secondary structural switch. Along with regulation of lysine biosynthetic genes, mutations within the lysine-responsive riboswitch (L-box) play a role in the acquisition of resistance to antimicrobial lysine analogs. To understand the structural basis for lysine binding, we have determined the 2.8Å resolution crystal structure of lysine bound to the Thermotoga maritima asd lysine riboswitch ligand-binding domain. The structure reveals a complex architecture scaffolding a binding pocket completely enveloping lysine. Mutations conferring antimicrobial resistance cluster around this site as well as highly conserved long range interactions, indicating that they disrupt lysine binding or proper folding of the RNA. Comparison of the free and bound forms by x-ray crystallography, small angle x-ray scattering, and chemical probing reveals almost identical structures, indicating that lysine induces only limited and local conformational changes upon binding.
Small non-protein-coding RNAs and mRNA sequences play a central role in cellular regulatory processes and are involved in virtually every aspect of the maintenance and transmission of genetic information. One prevalent form of riboregulation in bacteria is the riboswitch; at least 4% of all genes in Bacillus subtilis and related species are controlled in this fashion (1). This non-protein-coding element exerts genetic control in a cis-fashion by interacting with a cellular metabolite, thereby directing formation of one of two mutually exclusive mRNA secondary structures (reviewed in Ref. 2). Depending upon their placement within the mRNA, riboswitches control transcription or translation in bacteria (3) and alternative splicing or mRNA stability in eukarya (4, 5).
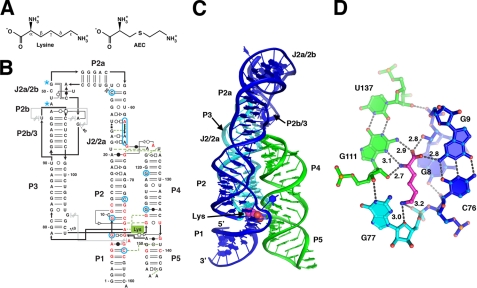
Currently, there at least 20 distinct families of riboswitches that recognize a diverse set of metabolites including nucleo-bases, sugars, vitamin cofactors, amino acids, and metal ions (2). The lysine-binding riboswitch is of particular interest for several reasons. Although in vitro selection methods are capable of generating aptamers to an equally diverse set of compounds (6), one of the few molecules for which an aptamer has failed to be raised is lysine (7). This suggests that RNA may require a complex architecture for recognition of this otherwise simple amino acid. Second, the lysine riboswitch has been the focus of studies involving the potential of riboswitches as targets of antimicrobial agents (8–10) because resistance to lysine analogs such as S-(2-aminoethyl)-l-cysteine (AEC2, see Fig. 1A) in Escherichia coli and B. subtilis is the result of mutations within the lysine riboswitch that regulates the lysC gene (see Fig. 1B, highlighted in blue) (11, 12).
FIGURE 1.
A, the chemical structure of lysine and the antimicrobial compound AEC. The carbon atom designation is shown for lysine. B, secondary structure of the T. maritima lysine riboswitch, reflecting the tertiary structure of the RNA. Base-pairing interactions are shown using the nomenclature of Leontis and Westhof (42). Nucleotides shown in red are >90% conserved across phylogeny, and positions where mutations confer resistance to AEC are circled in blue (blue asterisks denote approximate positions). C, schematic diagram of the tertiary structure of the lysine riboswitch with each of the three stacks colored differently. Lysine is shown represented as van der Waals spheres. D, details of the hydrogen-bonding interactions between lysine and the RNA. The distances of the bonds between lysine and RNA are given in Ångströms.
A recent study of AEC resistance in E. coli uncovered a mechanism that implicates lysyl tRNA-synthetase (LysRS) as the primary target of this compound (10). The toxic effects of AEC are a result of its incorporation into proteins in the place of lysine due to the inability of LysRS to discriminate between the two compounds. Mutations in the lysine riboswitch confer resistance to AEC because they result in a loss of lysine-dependent regulation of key lysine biosynthetic enzymes, increasing the intracellular lysine concentration. This allows lysine to effectively outcompete AEC for binding to LysRS, alleviating its toxic effects (10). Thus, development of new effective lysine analog antimicrobials will require targeting both LysRS and the lysine riboswitch (10). In the current study, we have solved the crystal structure of the lysine riboswitch in complex with lysine, revealing the basis for recognition of both the cognate ligand and the antimicrobial analogs, and provided insights into how mutations in the riboswitch might confer AEC resistance through two different means.
EXPERIMENTAL PROCEDURES
RNA Preparation and Crystallization—A 161-nucleotide double-stranded DNA coding for the Thermotoga maritima lysine riboswitch aptamer domain controlling the asd gene was constructed by PCR using overlapping oligonucleotides. The RNA was transcribed and purified using previously published techniques (13). The refolded RNA was then exchanged into 10 mm Na-HEPES, pH 7.0, 5 mm MgCl2, and 2 mm lysine before storage at 4 °C. For the free state, RNA was refolded in the lysine supplemented buffer, exchanged three times into 10 mm Na-HEPES, pH 7.0, 5 mm MgCl2 followed by overnight dialysis into 1 liter of lysine-free buffer. The final concentration was determined by absorbance at 260 nm (ε = 1,570,000 m–1 cm–1, molecular weight = 52,433 g mol–1). RNA was stored at 4 °C until use.
The riboswitch was crystallized by the hanging drop vapor diffusion method in the presence of 1 mm lysine or in the absence of lysine for the free state crystals. Drops were set up by mixing 1 μl of RNA with 1μl of a mother liquor solution consisting of 2 m Li2SO4, 5 mm MgCl2, and 10 mm Na-HEPES, pH 7.0, and 60 mm iridium hexammine to obtain the heavy atom derivative crystals. Identical conditions were used to grow the free state crystals except that no iridium hexammine was used in the mother liquor. Crystals were obtained within 24 h and required no additional cryoprotection agent; they were looped with 0.2–0.3-mm loops and flash-frozen in liquid nitrogen before data collection.
Data Collection—Data for the bound state iridium hexammine derivative crystal were collected on beamline X29A at the Brookhaven National Synchrotron Light Source X-rays at the iridium absorption peak. These data were integrated and scaled using HKL2000 (14). All data used in phasing and refining came from a single crystal. Data for the unliganded structure were collected using CuKα wavelength (1.5418 Å) radiation on an R-AXIS IV++ home source (Riguaku MSC), and the data were indexed and scaled using D*TREK (15).
Phasing and Structure Determination—Phases were determined by single wavelength anomalous diffraction using data extending to 2.8 Å. SHELXD (16) was used to find three iridium heavy atom sites within the asymmetric unit that had reasonably high occupancy. These heavy atom sites were used to calculate phases in SHELXE (17). The resulting experimental density map, following density modification (0.5 solvent fraction), displayed clear features corresponding to RNA backbone and base pairing (supplemental Fig. S1). This map was used for initial building of the model.
The model was built in Coot (18) and refined in PHENIX (19) using iterative rounds of building and refinement. The RNA nucleotides were initially built along with four iridium hexammine molecules. This model was brought through multiple rounds of simulated annealing and atomic displacement factor refinement before building lysine into the model. At this point, the density for the entire ligand was clearly visible and was validated by inspection using a simulated annealing omit map in which the ligand and a few surrounding nucleotides were omitted from the model (supplemental Fig. S2). One round of water picking was carried out by the PHENIX-ordered solvent protocol; waters were chosen based on peak size in an Fo – Fc map. Rfree was monitored in each round to ensure that it was dropping. Figures were prepared using PyMOL (20).
The unliganded RNA model was built using the bound form as a molecular replacement solution using only the RNA and re-refined using iterative rounds of simulated annealing and atomic displacement factor refinement. The final refinement statistics are shown in supplemental Table S1, and the structure factors and models have been deposited in the Protein Data Bank (accession codes 3D0U and 3D0X).
Chemical Probing Using Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension Chemistry—RNA sequences were constructed to correspond to the T. maritima RNA that was crystallized as well as the riboswitch controlling the B. subtilis lysC gene (21). The B. subtilis sequence was truncated in the P5 region to match the length of the T. maritima sequence to ensure that the RNAs are comparable, and the 5′- and 3′-structure cassettes were appended to these sequences as described previously (22). RNA was generated by run-off transcription and purified according to the same protocol used to generate RNA for the crystallographic studies.
RNA was prepared for modification by placing 1 μl of 2 μm RNA (2 pmol) into 11 μl of 0.5× Tris EDTA buffer. This sample was heated/cooled to allow the RNA to refold and then supplemented with 6 μl of buffer consisting of 333 mm K-HEPES, pH 8.0, and 333 mm NaCl. This buffer was supplemented with 2 mm lysine for the plus ligand reactions (final concentration of 667 μm), and MgCl2 was included in the folding buffer at concentrations ranging from 6.8 mm to 425 μm in 2-fold dilutions to yield the concentrations shown in the magnesium titration experiments. All reactions were supplemented with 1 μl of 3 mm Mg2+ to ensure proper extension by the polymerase during the reverse transcription step. Modifications were carried out using 30 mm NMIA for the recommended five half-lives at 20 °C; reverse transcription and gel running procedures were performed as described previously (22).
Small Angle X-ray Scattering (SAXS) Data Collection and Analysis—The B. subtilis lysC riboswitch was prepared for SAXS analysis using an Ettan LC liquid chromatography system configured with a Superose 6 PC 3.2 size-exclusion column (GE Healthcare), an in-line vacuum degasser, and a 0.01-μm filter. Three distinct sample buffer conditions were used and are referred to as native-bound, bound, and unfolded containing either 2 mm lysine and 5 mm MgCl2 or 2 mm EDTA, respectively. All sample buffer conditions contained 20 mm Na-HEPES, pH 6.5, and 50 mm KCl. Prior to gel filtration purification of the sample, the column was equilibrated with the appropriate buffer. Likewise, RNAs were refolded as described above at 5 μm and concentrated to 50 μl to a final concentration of 5 mg/ml. Each purification utilized 50 μl of sample, and a fraction corresponding to the major eluting peak was taken for direct SAXS experiments.
SAXS data were collected at the synchrotron beamline 12.3.1 of the Advanced Light Source (Berkeley, CA). All scattering data were collected at 1.0332 Å using a minimal sample volume of 18 μl. A full scattering curve was measured as two separate exposures at 6 and 60 s for the sample and buffer. The x-ray scattering due to the riboswitch RNA was determined by subtracting the background x-ray scattering of the gel filtration buffer from that containing the RNA and buffer. Data were reduced and combined with Primus to produce the final x-ray scattering curve (23). Data were collected over a range of RNA concentrations (0.7–0.1 mg/ml). No concentration-dependent changes were observed in the lowest scattering angles.
The radius of gyration (Rg), which describes the mass distribution of a particle around its rotational center of mass, was determined either by using the Guinier approximation (Guinier Rg) (24) within the angular range (q) of q·Rg < 1.3 or by calculating the electron pair-distribution function (Real Space Rg) in Gnom (23, 25). All final plots were prepared with KaleidaGraph.
RESULTS AND DISCUSSION
Crystallization and Structural Determination—To understand the basis for lysine recognition and AEC resistance, we have solved the structure of a riboswitch that controls the expression of the T. maritima β-aspartate semialdehyde dehydrogenase gene (asd). This RNA, which is centered about a conserved five-way junction motif, contains all of the nucleotides whose identity is >90% conserved across phylogeny (Fig. 1B, red) (3). An iridium hexammine derivative yielded sufficiently high quality data from which an experimental electron density map could be calculated (supplemental Fig. S1). Data collection and refinement statistics for both liganded and unliganded structures that include all 161 nucleotides are presented in supplemental Table S1 (for the bound structure, final Rwork = 18.2%, Rfree = 20.9%). The 2.8 Å resolution structure of the RNA-lysine complex agrees well with previous genetic, biochemical, and phylogenetic analysis of the RNA (21, 26, 27). The global architecture of the RNA comprises three sets of coaxially stacked helices (P1-P2/2a, P2b-P2b/3-P3, and P4-P5) arranged roughly parallel to one another (Fig. 1C), a mode of helical organization common in larger RNAs (28). At the center of this fold is the five-way junction containing the majority of the nucleotides with >90% phylogenetic conservation, in which a single lysine is observed wedged between helix P1 and the J2/3 joining region.
Overall Structure—The tertiary architecture of the RNA is dominated by formation of a three-helix bundle structure composed of the P2, P3, and P4 helices (Fig. 1C) stabilized via interactions mediated by their terminal loops. A kissing loop interaction is observed between the terminal loops of P2 and P3 that was identified as important for the ability of the B. subtilis lysC riboswitch to efficiently terminate transcription (29). Unlike other structurally characterized kissing loop interactions, there is a stacking interaction between G40 and U91, which are oriented perpendicular to the P2b/3 helical axis. These two bases form hydrogen-bonding interactions with the major groove of the central four base pairs of the P2b/3 helix. This additional dinucleotide “staple” may constitute an adaptation for function at elevated physiological temperatures; similar observations were made in the selection of thermophilic ribozymes, where mutations that add new tertiary interactions or further stabilize existing ones are responsible for adaptation to elevated temperatures (30).
The ability of the two terminal loops to interact is achieved by a ∼120° bend at J2a/2b using a novel internal loop motif. In the majority of other lysine riboswitches, this turn is achieved by the structurally similar canonical kink-turn motif (31). Thus, although the majority of the aptamer domain is highly conserved, some elements of the peripheral region of the lysine riboswitch have evolved unique solutions to the stabilization of a common global architecture, reflecting the modular nature of RNA structure in general (32).
The second element stabilizing the three-helix bundle is an interaction between the terminal pentaloop of P4 and an internal loop motif adjacent to the sarcin/ricin motif between P2 and P2a (33). The pentaloop of P4 forms a structure similar to a standard GNRA tetraloop motif by flipping out a uridine residue (U125). Rather than docking with another helix using the sugar edge of the three stacked adenosine residues, as commonly observed for most tetraloop-mediated interactions (34), the adenine bases interact with the minor groove of P2 using their Watson-Crick faces. Unusually, A123 forms the central base of a U21·A123·G65 base triple that anchors the interaction.
Lysine Recognition—The ligand-binding pocket is contained within the core of the five-way junction motif, sitting between the P1 helix and J2/3 and is flanked by the first base pairs of the P2 and P4 helices (Fig. 1D). The carboxylate group of lysine forms a set of hydrogen bonds with the N2 amino groups of the G111·U137 wobble pair, the G9-C76 Watson-Crick pair and the 2′-hydroxyl group of G8. Further contacts to the N3 and O2′ atoms of G111 are made by the α-amino group of lysine. The ε-amino group of lysine is recognized by a combination of electrostatic and hydrogen-bonding interactions within a pocket that places it close to the non-bridging phosphate oxygen of G77 along with the O4 oxygen atom of the ribose sugar. The relatively small size of the ε-amino pocket near G77 precludes efficient recognition by homoarginine and N6-trimethyl-l-lysine (8, 21). This is likely the basis for discrimination between the related metabolites lysine and diaminopimelate.
Discrimination between lysine and other closely related compounds is further achieved through indirect recognition of the methylene linker of the side chain. The lysine side chain is bound in an extended conformation that allows it to span the two sites of interaction of the polar atoms, consistent with the ability of a lysine analog that contains a trans-double bond between the γ- and δ-carbons to productively bind this riboswitch (8). As a result, compounds containing shorter or longer side chains (l-ornithine and l-α-homolysine, respectively) are not efficiently bound because their side chain is of the incorrect length to allow the proper contacts between all of the polar atoms of lysine and the RNA. The hydrophobic methylene groups are primarily contacted through stacking interactions with G77, A78, and the G8·G152 pair.
Despite being a critical component of proper recognition, the methylene groups of lysine are not tightly packed against the RNA. The loose packing around lysine explains the ability of antimicrobial lysine analogs containing modifications at the γ-position (Fig. 1A), such as l-3-[(2-aminoethyl)-sulfonyl]-alanine, l-4-oxalysine, and AEC, to bind reasonably well to the riboswitch (8, 21). Similarly, the thiamine pyrophosphate riboswitch only moderately contacts the central thiazole ring of thiamine pyrophosphate (35, 36). In each case, moieties recognized through indirect readout, which are generally the hydrophobic groups, are modified to yield riboswitch-binding antimicrobial agents (37).
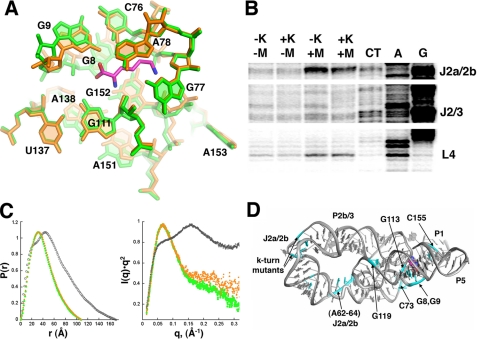
Ligand-dependent Conformational Changes in the RNA—Lysine is completely buried within the five-way junction (100% solvent-inaccessible), indicating that there is some form of folding event concurrent with binding. To test the hypothesis that lysine-dependent conformational changes occur within the riboswitch, we crystallized the RNA in the absence of lysine and determined its structure. The RNA crystallized under the same conditions and in the same space group. The resulting structure is nearly identical to the complexed form (supplemental Fig. S3A), with only minor differences between the two structures in the positioning of the 5′-side of the P1 helix (supplemental Fig. S3B). This finding reveals that the global architecture can be formed in the absence of ligand. Close examination of the binding pocket shows that positioning of some of the nucleotides is perturbed by 2–3 Å, but the overall pattern of base interactions remains the same (Fig. 2A).
FIGURE 2.
A, closeup of the lysine-binding pocket with the superposition of the free (orange) and bound (green; lysine in magenta) RNA. B, chemical modification of the T. maritima and B. subtilis lysine riboswitch emphasizing two key regions of each RNA that undergoes strong magnesium- and lysine-dependent changes. The full annotated gels are shown in supplemental Fig. S3. C, small angle x-ray data corresponding to free (EDTA, gray; magnesium, orange) and lysine-bound (green). The left panel shows the experimental electron pair distribution plot where the x-intercept reflects the most likely maximum intermolecular scattering distance (Dmax). The right panel is a Kratky plot that reflects the extent of unfoldedness of the macromolecule (40). D, map of mutations (cyan) conferring AEC resistance onto the lysine riboswitch structure.
The above result, however, could potentially be an artifact of the crystal lattice where crystallization induces a conformation that does not exist in solution. To further explore the potential similarity between the bound and unbound lysine riboswitch in solution, we probed backbone flexibility using selective 2′-hydroxyl acylation analyzed by primer extension chemistry to monitor local changes in the RNA (22) and SAXS to monitor global changes. To ensure that the observed behavior is a general feature of the L-box, we probed both the T. maritima asd and the B. subtilis lysC RNAs at varying magnesium concentrations in the absence and presence of 667 μm lysine (Fig. 2B, and supplemental Figs. S4 and S5). In particular, we wished to probe these RNAs under near physiological salt concentrations (100 mm NaCl, 0.5–2 mm MgCl2), as opposed to the high salt conditions of the crystals (2 m Li2SO4, 5 mm MgCl2, 60 mm iridium hexammine). These experiments reveal two general trends in the response of the L-box to magnesium and ligand. First, both RNAs exhibit clear changes in reactivity upon the addition of magnesium. These changes are centered on J2a/2b and tertiary interactions involving the terminal loops of P2, P3, and P4. For example, the conserved uridine in L4 becomes reactive in both RNAs at 2 mm magnesium (Fig. 2B), reflecting its exposure to the solvent in the folded structure. Second, both RNAs show limited lysine-dependent changes localized principally to the five-way junction, primarily J2/3, the base of P5, and 3′-side of the P1 helix (Fig. 2B). It is noteworthy that at high magnesium concentrations (>1 mm), lysine does not appear to significantly affect the structure in the B. subtilis variant; lysine-dependent effects are most pronounced near physiological magnesium concentrations (0.2–0.5 mm). These data are in agreement with both in-line probing of the B. subtilis lysC riboswitch (8, 21) and an observed magnesium-dependent fluorescence change of a 2-aminopurine label-incorporated riboswitch at the equivalent position to A153 in the five-way junction (38). Together, these data indicate that the structure of the L-box can be induced by magnesium and nearly native in the absence of lysine.
To further validate the chemical probing data, we analyzed the B. subtilis lysC riboswitch by SAXS. This solution method is sensitive to the global conformation of a macromolecule and provides a direct measure of the radius of gyration (Rg), maximum dimension (Dmax), and electron-pair distribution function of the macromolecule (39). The addition of 5 mm magnesium in the absence of lysine induces a significant compaction of the RNA (Fig. 2C, left; supplemental Fig. S6). This compact form is not significantly altered by the inclusion of 2 mm lysine, as anticipated. More importantly, the electron-pair distribution plots describing both the bound and the unbound forms of the riboswitch in 5 mm magnesium are identical. The maximum dimension of the RNA in the presence of magnesium, regardless of lysine (108–110 Å; supplemental Table S2), is consistent with the crystal structure after taking into account the first solvation shell (∼108 Å). In the absence of magnesium, the riboswitch is weakly folded, as illustrated by the hyperbolic feature of the Kratky plot (Fig. 2C, right). In the presence of magnesium, the Kratky plot converges rapidly at small scattering angles, reflecting a well folded molecule (40). Together with the chemical probing data, these results indicate that this riboswitch can adopt a near native form without lysine in solution with the aid of magnesium. Thus, lysine likely gains access to the binding pocket through small, localized fluctuations in the RNA structure, mostly centered within J2/3.
Resistance Mutations in the Lysine Riboswitch—Resistance to the antimicrobial lysine analog AEC is conferred in both E. coli and B. subtilis via mutations within the lysine riboswitch (Fig. 2D, cyan) (8, 11, 12). Some of these mutations map (e.g. G8 and G9) around the binding pocket, abrogating direct contacts with lysine. More interestingly, others are observed in the distal regions of the P2 and P4 helix (nucleotides A62–A64 and G129), disrupting formation of key tertiary interactions. Notably, some of these mutants bind lysine with nearly the same affinity as the wild type B. subtilis lysC RNA (8), suggesting that AEC resistance is gained by decreasing the rate at which the RNA folds into a binding-competent structure. As transcriptional regulation has a short temporal window in which to direct the downstream secondary structural switch (41), a lowered folding rate would lead to loss of regulatory control. In this fashion, riboswitch-mediated regulation is governed by the rates at which individual elements are able to fold rather than interplay between the thermodynamic stabilities of each structure in the switch. Further studies will be needed to address the role of the ligand in the kinetic folding pathway of the riboswitch and its relation to efficient genetic regulation.
Supplementary Material
Acknowledgments
We thank Francis Reyes for assistance with model building and Jeffrey Kieft, Rob Knight, Quentin Vicens, Sunny Gilbert, and the Batey laboratory for insightful discussions and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM073850 (to R. T. B.). R. T. B. is a paid consultant of BioRelix, a company dedicated to developing antibiotic therapeutics targeted against riboswitches. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The atomic coordinates and structure factors (code 3D0U, 3D0X) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains two supplemental tables and six supplemental figures.
Footnotes
The abbreviations used are: AEC, S-(2-aminoethyl)-l-cysteine; SAXS, small angle x-ray scattering; LysRS, lysyl tRNA-synthetase.
References
- 1.Irnov, Kertsburg, A., and Winkler, W. C. (2006) Cold Spring Harbor Symp. Quant. Biol. 71 239–249 [DOI] [PubMed] [Google Scholar]
- 2.Winkler, W. C., and Breaker, R. R. (2005) Annu. Rev. Microbiol. 59 487–517 [DOI] [PubMed] [Google Scholar]
- 3.Barrick, J. E., and Breaker, R. R. (2007) Genome Biol. 8 R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheah, M. T., Wachter, A., Sudarsan, N., and Breaker, R. R. (2007) Nature 447 497–500 [DOI] [PubMed] [Google Scholar]
- 5.Wachter, A., Tunc-Ozdemir, M., Grove, B. C., Green, P. J., Shintani, D. K., and Breaker, R. R. (2007) Plant Cell 19 3437–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson, D. S., and Szostak, J. W. (1999) Annu. Rev. Biochem. 68 611–647 [DOI] [PubMed] [Google Scholar]
- 7.Famulok, M. (1994) J. Am. Chem. Soc. 116 1698–1706 [Google Scholar]
- 8.Blount, K. F., Wang, J. X., Lim, J., Sudarsan, N., and Breaker, R. R. (2007) Nat. Chem. Biol. 3 44–49 [DOI] [PubMed] [Google Scholar]
- 9.Blount, K. F., and Breaker, R. R. (2006) Nat. Biotechnol. 24 1558–1564 [DOI] [PubMed] [Google Scholar]
- 10.Ataide, S. F., Wilson, S. N., Dang, S., Rogers, T. E., Roy, B., Banerjee, R., Henkin, T. M., and Ibba, M. (2007) ACS Chem. Biol. 2 819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu, Y., Shevtchenko, T. N., and Paulus, H. (1992) FEMS Microbiol. Lett. 71 23–27 [DOI] [PubMed] [Google Scholar]
- 12.Patte, J. C., Akrim, M., and Mejean, V. (1998) FEMS Microbiol. Lett. 169 165–170 [DOI] [PubMed] [Google Scholar]
- 13.Montange, R. K., and Batey, R. T. (2006) Nature 441 1172–1175 [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307–326 [DOI] [PubMed] [Google Scholar]
- 15.Pflugrath, J. W. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 1718–1725 [DOI] [PubMed] [Google Scholar]
- 16.Schneider, T. R., and Sheldrick, G. M. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1772–1779 [DOI] [PubMed] [Google Scholar]
- 17.Sheldrick, G. M. (2002) Z. Kristallographie 217 644–650 [Google Scholar]
- 18.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 19.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948–1954 [DOI] [PubMed] [Google Scholar]
- 20.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, 0.99 Ed., DeLano Scientific, Palo Alto, CA
- 21.Sudarsan, N., Wickiser, J. K., Nakamura, S., Ebert, M. S., and Breaker, R. R. (2003) Genes Dev. 17 2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson, K. A., Merino, E. J., and Weeks, K. M. (2006) Nat. Protoc. 1 1610–1616 [DOI] [PubMed] [Google Scholar]
- 23.Svergun, D. I. (1992) J. Appl. Crystallogr. 25 495–503 [Google Scholar]
- 24.Guinier, A., and Fournet, G. (1955) Small Angle Scattering of X-rays, Wiley, New York
- 25.Putnam, C. D., Hammel, M., Hura, G. L., and Tainer, J. A. (2007) Q. Rev. Biophys. 40 191–285 [DOI] [PubMed] [Google Scholar]
- 26.Grundy, F. J., Lehman, S. C., and Henkin, T. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12057–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodionov, D. A., Vitreschak, A. G., Mironov, A. A., and Gelfand, M. S. (2003) Nucleic Acids Res. 31 6748–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laederach, A., Chan, J. M., Schwartzman, A., Willgohs, E., and Altman, R. B. (2007) RNA (Cold Spring Harbor) 13 643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blouin, S., and Lafontaine, D. A. (2007) RNA (Cold Spring Harbor) 13 1256–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, F., Gooding, A. R., and Cech, T. R. (2006) RNA (Cold Spring Harbor) 12 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein, D. J., Schmeing, T. M., Moore, P. B., and Steitz, T. A. (2001) EMBO J. 20 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leontis, N. B., Lescoute, A., and Westhof, E. (2006) Curr. Opin. Struct. Biol. 16 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Correll, C. C., Munishkin, A., Chan, Y. L., Ren, Z., Wool, I. G., and Steitz, T. A. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13436–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B., and Steitz, T. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 4899–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serganov, A., Polonskaia, A., Phan, A. T., Breaker, R. R., and Patel, D. J. (2006) Nature 441 1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thore, S., Leibundgut, M., and Ban, N. (2006) Science 312 1208–1211 [DOI] [PubMed] [Google Scholar]
- 37.Edwards, T. E., and Ferre-D'Amare, A. R. (2006) Structure (Camb.) 14 1459–1468 [DOI] [PubMed] [Google Scholar]
- 38.Mulhbacher, J., and Lafontaine, D. A. (2007) Nucleic Acids Res. 35 5568–5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipfert, J., and Doniach, S. (2007) Annu. Rev. Biophys. Biomol. Struct. 36 307–327 [DOI] [PubMed] [Google Scholar]
- 40.Russell, R., Millett, I. S., Tate, M. W., Kwok, L. W., Nakatani, B., Gruner, S. M., Mochrie, S. G., Pande, V., Doniach, S., Herschlag, D., and Pollack, L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4266–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickiser, J. K., Winkler, W. C., Breaker, R. R., and Crothers, D. M. (2005) Mol. Cell 18 49–60 [DOI] [PubMed] [Google Scholar]
- 42.Leontis, N. B., and Westhof, E. (2001) RNA (Cold Spring Harbor) 7 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.