Abstract
Human albumin is thought to hydrolyze esters because multiple equivalents of product are formed for each equivalent of albumin. Esterase activity with p-nitrophenyl acetate has been attributed to turnover at tyrosine 411. However, p-nitrophenyl acetate creates multiple, stable, acetylated adducts, a property contrary to turnover. Our goal was to identify residues that become acetylated by p-nitrophenyl acetate and determine the relationship between stable adduct formation and turnover. Fatty acid-free human albumin was treated with 0.5 mm p-nitrophenyl acetate for 5 min to 2 weeks, or with 10 mm p-nitrophenyl acetate for 48 h to 2 weeks. Aliquots were digested with pepsin, trypsin, or GluC and analyzed by mass spectrometry to identify labeled residues. Only Tyr-411 was acetylated within the first 5 min of reaction with 0.5 mm p-nitrophenyl acetate. After 0.5–6 h there was partial acetylation of 16–17 residues including Asp-1, Lys-4, Lys-12, Tyr-411, Lys-413, and Lys-414. Treatment with 10 mm p-nitrophenyl acetate resulted in acetylation of 59 lysines, 10 serines, 8 threonines, 4 tyrosines, and Asp-1. When Tyr-411 was blocked with diisopropylfluorophosphate or chlorpyrifos oxon, albumin had normal esterase activity with β-naphthyl acetate as visualized on a nondenaturing gel. However, after 82 residues had been acetylated, esterase activity was almost completely inhibited. The half-life for deacetylation of Tyr-411 at pH 8.0, 22 °C was 61 ± 4 h. Acetylated lysines formed adducts that were even more stable. In conclusion, the pseudo-esterase activity of albumin is the result of irreversible acetylation of 82 residues and is not the result of turnover.
Human albumin has been reported to have esterase activity with p-nitrophenyl acetate (1, 2), α-naphthyl acetate, phenyl acetate, 1-naphthyl N-methylcarbamate (3), β-naphthyl acetate (4), aspirin (5), ketoprofen glucuronide (6), carprofen acylglucuronide (7), cyclophosphamide (8), nicotinate esters (9), long- and short-chain fatty acid esters (10), octanoyl ghrelin (11), organophosphorus pesticides (12), carbaryl (13), o-nitrotrifluoroacetanilide (14), o-nitroacetanilide (15), and nerve agents (16).
One site in albumin is rapidly acetylated by p-nitrophenyl acetate, showing a burst of product, but up to 5.2 molar equivalents are incorporated when albumin is treated with a 9-fold excess of p-nitrophenyl [14C]acetate (1). The 5 equivalents of label are not removable by extensive dialysis. The esteratic site has been identified as Tyr-411 based on site-directed mutagenesis studies (17). Mass spectrometry (MS)2 has identified Tyr-411 as the residue labeled by organophosphorus esters including diisopropylfluorophosphate (DFP), soman, sarin, dichlorvos, FP-biotin, and chlorpyrifos oxon (16, 18) confirming the report by Sanger that a tyrosine in albumin is labeled by DFP (19), and reports that a tyrosine in albumin is labeled by the nerve agents soman, sarin, cyclosarin, and tabun (20). When albumin is labeled with 1 mol of DFP, albumin loses the fast phase of its esterase activity (the burst), supporting the conclusion that DFP and p-nitrophenyl acetate bind to the same site, namely to Tyr-411 (21).
Our goal was to identify the additional sites labeled by p-nitrophenyl acetate. We wanted an explanation for the apparently inconsistent observation that albumin hydrolyzes p-nitrophenyl acetate at measurable rates, yet the 14C-acetylated albumin formed by p-nitrophenyl acetate is a stable adduct.
EXPERIMENTAL PROCEDURES
Materials—A 1 mg/ml solution of fatty acid-free human albumin (Fluka 05418, via Sigma) was dissolved in 10 mm Tris-Cl, pH 8.0. A 1 mg/ml solution of porcine pepsin (Sigma P6887) in 10 mm HCl was stored at -80 °C. Sequencing grade modified trypsin (Promega V5113) at a concentration of 20 μg in 50 μl of 50 mm acetic acid was stored at -80 °C. Endoproteinase GluC (Staphylococcus aureus protease V8) was from Worthington Biochemical Corp (Lakewood, NJ, LS003608). 1 mg of GluC powder was dissolved in 210 μl of water, aliquoted into microcentrifuge tubes each containing 10 μl (50 μg), dried in a vacuum centrifuge, and stored at -80 °C. A 10 mg/ml solution of α-cyano-4-hydroxycinnamic acid matrix (CHCA) (Applied Biosystems) in 50% acetonitrile, 0.1% trifluoroacetic acid was stored at room temperature. A 0.1 m solution of p-nitrophenyl acetate (Sigma N8130) in acetonitrile was stored at -20 °C. DFP, a liquid with a concentration of 5.73 m, was from Sigma (D0879). Chlorpyrifos oxon (ChemService Inc. West Chester, PA; MET-674B) was dissolved in methanol to make a 1 m solution and stored at -80 °C. β-naphthyl acetate, Fast Blue RR, and fluorodinitrobenzene (>99% pure) were from Sigma. Albumin samples were concentrated in a Microcon centrifugal device with a YM10 membrane whose molecular weight cutoff was 10,000 (Millipore 42406).
Pure Albumin Labeled with p-Nitrophenyl Acetate, Digested with Pepsin While Albumin Disulfides Were Intact—1 ml of a 1 mg/ml solution of fatty acid-free human albumin (15 μm) in 10 mm Tris-Cl, pH 8.0 was treated with a 33-fold molar excess of p-nitrophenyl acetate (5 μl of 0.1 m in acetonitrile) at 22 °C. 1 ml of control albumin solution was treated with 5 μl of acetonitrile. A 50-μl aliquot was removed into 50 μl of 1% trifluoroacetic acid containing 2 μl of 1 mg/ml pepsin after 5, 10, 15, 20, 30, 40, 50, 60 min, 6 h, 24 h, 9 days, and 14 days. The drop in pH to 1.4 stopped the reaction with p-nitrophenyl acetate. Digestion with pepsin was at 37 °C for 1 to 2 h. A 0.5-μl aliquot of the digest was spotted on a MALDI target plate, dried, and overlaid with 0.5 μl of CHCA matrix, for analysis in a MALDI-TOF/TOF 4800 mass spectrometer.
Absorbance at 400 nm to Measure p-Nitrophenolate Produced by Albumin Hydrolysis of p-Nitrophenyl Acetate—1 ml of 1 mg/ml human albumin in 10 mm Tris-Cl, pH 8.0 was incubated with 5 μl of 0.1 m p-nitrophenyl acetate at 22 °C. At various times after mixing, a 50-μl aliquot was withdrawn and diluted to 2 ml for measurement of absorbance at 400 nm. The reaction was followed until all of the p-nitrophenyl acetate was consumed. A control sample, without albumin, was followed in a similar manner to measure spontaneous hydrolysis of p-nitrophenyl acetate, in 10 mm Tris-Cl, pH 8.0. The amount of p-nitrophenylate ion formed in the albumin reaction was corrected for spontaneous hydrolysis.
The extinction coefficient for the p-nitrophenolate ion at pH 8.0 is 16,900 m-1 cm-1 at 400 nm (1). Therefore, the completely hydrolyzed 500 μm p-nitrophenyl acetate should have an absorbance at 400 nm of 8.45.
Albumin Incubated with Potassium Acetate—To determine whether acetylation of lysines was accomplished by reaction with p-nitrophenyl acetate or by reaction with free acetate, we measured acetylated lysines after incubation of albumin with potassium acetate. 1 ml of 1 mg/ml albumin in 10 mm Tris-Cl, pH 8.0 was incubated with 500 μm potassium acetate for 15 h at 22 °C. A 50-μl aliquot of the reaction mixture was removed into 50 μl of 1% trifluoroacetic acid containing 2 μl of 1 mg/ml pepsin and incubated for 2 h at 37 °C. The digest was analyzed with a MALDI-TOF-TOF 4800 mass spectrometer, with CHCA matrix.
HPLC Purification of the 1872-amu Acetylated Peptide LVRYTKKVPQVSTPTL—This peptide was purified to make it possible to get an MS/MS spectrum of acetylated YTK. The acetylated YTK peptide ionized in the mass spectrometer only when the number of ions in the mixture was minimal. 10 mg of human albumin in 1 ml of 10 mm Tris-Cl, pH 8.0 was treated with a 33-fold molar excess of p-nitrophenyl acetate for 5 min. The reaction was stopped by the addition of 1 ml of 1% trifluoroacetic acid and 0.1 ml of 1 mg/ml pepsin. After 1 h at 37 °C, the pepsin-digested albumin was purified on a Waters 625 LC system on a Phenomenex Prodigy 5 micron C18 100 × 4.60 mm column. Peptides were eluted with a 60-min gradient from 0.1% trifluoroacetic acid in water to 60% acetonitrile, 0.09% trifluoroacetic acid at a flow rate of 1 ml/min. An aliquot from each 1-min fraction was spotted on a MALDI target plate to identify the fractions that contained the 1872 mass. It was found that the 1872 mass eluted with 26% acetonitrile in fractions 26 and 27. These fractions were combined, dried, and injected into the same Phenomenex column, but this time the elution solvents were 10 mm potassium phosphate pH 7.0 and acetonitrile. In a 60-min gradient from 0 to 60% acetonitrile, the 1872 mass eluted in fractions 37–44. About 5000 pmol of the purified 1872-amu peptide in 225 μl of 50 mm ammonium bicarbonate were digested with 2 μg of Promega trypsin for 4 h at 37 °C, dried, dissolved in 50 μl of 5% acetonitrile, 0.1% formic acid, and infused into the QTRAP 4000 mass spectrometer.
Percent Acetylation Calculated from Cluster Areas in a MALDI-TOF-TOF 4800 (Applied Biosystems, Foster City) Mass Spectrometer—Essentially salt-free 0.5-μl samples were spotted on a MALDI target plate, air-dried, and overlaid with 0.5 μl of 10 mg/ml CHCA in 50% acetonitrile, 0.1% trifluoroacetic acid. MS spectra were acquired with laser power at 3000 volts in positive reflector mode. Each spectrum was the average of 500 laser shots. The percentage of acetylation was calculated by dividing the cluster area of the unlabeled peptide by the sum of the cluster areas for the unlabeled and labeled peaks. The mass spectrometer was calibrated against des-Arg-bradykinin (904.468 Da), angiotensin 1 (1296.685 Da), Glu-fibrinopeptide B (1570.677 Da), and neurotensin (1672.918 Da) (Cal Mix 1 from Applied Biosystems).
LC/MS/MS with the QTRAP 2000 and LCQ Deca XP Mass Spectrometers—Human albumin treated with p-nitrophenyl acetate was denatured by boiling 10 min in the presence of 10 mm dithiothreitol, carbamidomethylated with 90 mm iodoacetamide, and dialyzed against 2 × 4 liters of 10 mm ammonium bicarbonate. A 500-μg aliquot was digested with 10 μg of Promega trypsin overnight at 37 °C, or with 50 μg of GluC. The digest was dried in a vacuum centrifuge and dissolved in 5% acetonitrile, 0.1% formic acid to make 6.7 pmol/μl. A 10-μl aliquot was injected into the HPLC nanocolumn (218MS3.07515 Vydac C18 polymeric rev-phase, 75 μm i.d. × 150 mm long; P.J. Cobert Assoc, St. Louis, MO). Peptides were separated with a 90-min linear gradient from 0 to 60% acetonitrile at a flow rate of 0.3 μl/min and electrosprayed through a fused silica emitter (360 μm o.d., 75 μm i.d., 15 μm taper, New Objective) directly into the QTRAP 2000 (Applied Biosystems, Foster City, CA), a hybrid quadrupole linear ion trap mass spectrometer. An ion-spray voltage of 1900 V was maintained between the emitter and the mass spectrometer. Information-dependent acquisition was used to collect MS, enhanced MS, and MS/MS spectra for the three most intense peaks in each cycle, having a charge of +1 to +4, a mass between 200 and 1700 m/z, and an intensity >10,000 cps. All spectra were collected in the enhanced mode, using the trap function. Precursor ions were excluded for 30 s after one MS/MS spectrum had been collected. The collision cell was pressurized to 40 μTorr with pure nitrogen and collision energies between 20 and 40 eV were determined automatically by the software based on the mass and charge of the precursor ion. The mass spectrometer was calibrated on selected fragments from the MS/MS spectrum of Glu-fibrinopeptide B. The MS/MS data were processed using Analyst 1.4.1 software and submitted to Mascot for identification of peptide sequences (22).
A pepsin digest of the same carbamidomethylated preparation was analyzed on the LCQ Deca XP mass spectrometer (Thermo-Finnigan, San Jose, CA) coupled to the Ultimate 3000 HPLC system (Dionex, Sunnyvale, CA). The 75-μm i.d. C18 column was from LCPackings. HPLC solvents were: A) 0.1 m acetic acid and B) 80% acetonitrile, 0.1 m acetic acid. Peptides were eluted with a gradient from 0 to 55% B in 90 min. Data were collected with Xcalibur 2.0 and analyzed with OMMSA search engine (23).
Infusion in QTRAP 4000 Mass Spectrometer—Peptides dissolved in 50% acetonitrile, 0.1% formic acid were infused into the QTRAP 4000 (Applied Biosystems, Foster City, CA) mass spectrometer at a flow rate of 0.3 μl/min through an 8-μm emitter (FS360-50-8-D, New Objective) via a 25-μl Hamilton syringe mounted on a Harvard syringe pump. 500 MS/MS spectra were accumulated for each parent ion.
Deacetylation of Tyr-411—The production and disappearance of acetylated Tyr-411 was followed by MALDI-TOF mass spectrometry. A 15 μm solution of human albumin in 10 mm Tris-Cl, pH 8.0 was treated with an equimolar concentration of p-nitrophenyl acetate at 22 °C. After various times, a 10-μl aliquot was mixed with 10 μl of 1% trifluoroacetic acid and 1 μl of 1 mg/ml pepsin. After 1–2 h at 37 °C a 0.5 μl aliquot of the digest was spotted on a MALDI target plate. Cluster areas for the unlabeled peptides at 1717 and 1830 amu and the labeled peptides at 1759 and 1872 amu were used to calculate % labeling on Tyr-411.
Percentage of Acetylated Lysines Calculated from Amino Acid Composition Analysis—Quantitation of the acetylated lysine was obtained by the use of fluorodinitrobenzene (Allfrey et al., Ref. 30). Fluorodinitrobenzene reacts with un-acetylated lysine to form ε-N-dinitrobenzylated lysine. Acid hydrolysis, in preparation for amino acid composition analysis hydrolyzes the ε-N-acetyl lysines to yield free lysine, but the dinitrobenzylated lysines are relatively stable to acid hydrolysis. The amount of free lysine appearing in the amino acid composition analysis, corrected for hydrolysis of dinitrobenzylated lysine, represents the amount of lysine that was originally acetylated.
Fluorodinitrobenzylation employs the method originally described by Sanger (24). Acetylated and control albumin were dialyzed into NaHCO3 (20 mg/ml). Two hundred micrograms of dialyzed albumin were adjusted to 1 ml with NaHCO3 and mixed with 2 ml of absolute ethanol. The mixture was shaken at room temperature for 1 h, at which time 30 μl of fluorodinitrobenzene was added and that mixture shaken for an additional 2 h. The insoluble yellow product was washed twice with water, twice with absolute ethanol, twice with diethyl ether, air-dried, and submitted for amino acid composition analysis.
Nondenaturing Gel Electrophoresis—Albumin esterase activity was demonstrated on a nondenaturing 4–30% polyacrylamide gradient gel, stained for esterase activity with β-naphthyl acetate and Fast Blue RR (25). Gels were shaken in 100 ml of 0.05 m Tris-Cl, pH 7.4 containing 50 mg of β-naphthylacetate in 1 ml ethanol, and 50 mg of solid Fast Blue RR. Though most of the Fast Blue RR does not dissolve, pink bands of esterase activity appear within 15–30 min. The gel was counterstained with Coomassie Blue to show protein concentration.
The positive control samples, not acetylated on any residues, were 5 μl of human serum and 200 μg of fatty acid-free human albumin. The negative control was albumin that had been denatured in 8 m urea, reduced with 10 mm dithiothreitol, carbamidomethylated with iodoacetamide, dialyzed to remove salts, and concentrated to 10 μg/μl. Albumin acetylated on 100% of Tyr-411 only, was prepared by incubating 10 mg/ml albumin with 5 mm p-nitrophenyl acetate (33-fold excess) in 10 mm Tris-Cl, pH 8.0 for 5 min, followed immediately by gel electrophoresis. Albumin acetylated on 100% Tyr-411 and 20% of the lysines was prepared by incubating 1 mg/ml albumin with 0.5 mm p-nitrophenyl acetate for 6 h at 22°C. Albumin with free Tyr-411 and 20% acetylated lysines was prepared by incubating 1 mg/ml albumin with 0.5 mm p-nitrophenyl acetate, 0.01% sodium azide for 2 weeks at 22 °C, during which time Tyr-411 was completely deacetylated as shown by mass spectrometry. Albumin with 100% acetylated Tyr-411 and 60–100% acetylated lysines was prepared by incubating 1 mg/ml albumin with 10 mm p-nitrophenyl acetate for 48 h. Percent acetylation of Tyr-411 and lysines was determined by mass spectrometry as described above, and by amino acid composition analysis after reaction with fluorodinitrobenzene.
Albumin labeled with DFP on 80% of Tyr-411 was prepared by treating 2 mg/ml albumin with a 20-fold excess of DFP in 10 mm Tris-Cl, pH 8.0 for 2 h at 22 °C. Albumin labeled with chlorpyrifos oxon on 95% of Tyr-411 was prepared by treating 1 mg/ml albumin in 10 mm Tris-Cl, pH 8.0 with a 7.5-fold excess of chlorpyrifos oxon for 48 h at 22 °C. The percent free and modified Tyr-411 was determined by mass spectrometry. No lysines were labeled by DFP or chlorpyrifos oxon.
RESULTS
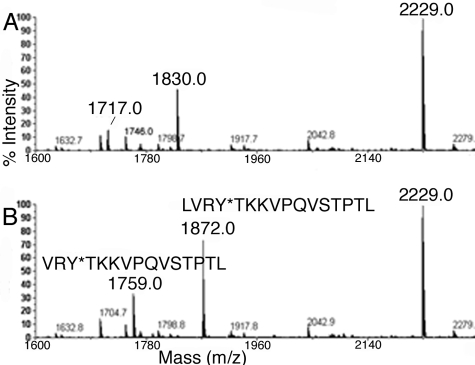
Tyrosine 411 Is Rapidly Acetylated by p-Nitrophenyl Acetate—A 5-min reaction of albumin (15 μm) with p-nitrophenyl acetate (500 μm) results in complete acetylation of Tyr-411. This is indicated by the mass shift of +42 amu for peptic peptides VRYTKKVPQVSTPTL and LVRYTKKVPQVSTPTL (missed cleavage), which have masses of 1717 and 1830 amu in the control albumin digest (Fig. 1A), but masses of 1759 and 1872 amu after treatment with p-nitrophenyl acetate (Fig. 1B). No unlabeled masses at 1717 and 1830 amu remain after treatment with p-nitrophenyl acetate, indicating 100% labeling. No other peptides were found to have a mass shift at this time point.
FIGURE 1.
MALDI-TOF mass spectra of peptic peptides of human albumin before (A) and after (B) 5 min reaction with 0. 5 mmp-nitrophenyl acetate. The mass of peptides at 1717 and 1830 amu increased by +42 amu due to acetylation of Tyr-411.
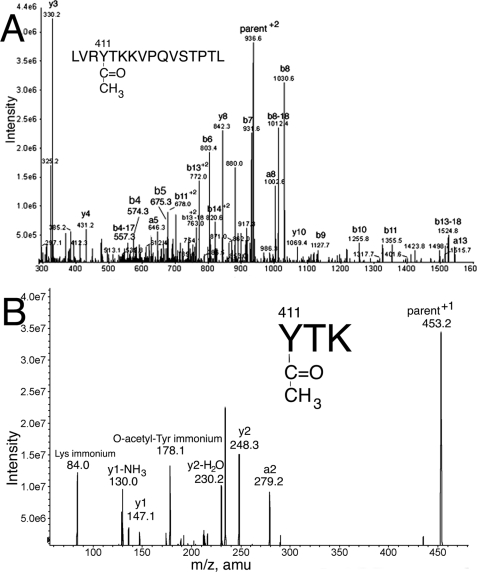
The labeled residue was identified as tyrosine 411 by fragmentation of the doubly charged parent ion 936.5 m/z (corresponding to the singly charged 1872 amu ion) in the QTRAP mass spectrometer. Fig. 2A shows fragmentation of parent ion 936.5 m/z. All major peaks are assigned to fragments of LVRYTKKVPQVSTPTL. The presence of the b4 ion (representing residues LVRY* plus acetyl) at 574.3 amu indicates acetylation of that fragment. The most likely candidate for acetylation is tyrosine. The b4–17 ion at 557.3 amu, and the b-ion fragments b5 ion at 675.3 amu through b13–18 at 1524.8 amu are all 42 amu larger than predicted from the amino acid sequence, further supporting acetylation of tyrosine in LVRYTKKVPQVSTPTL.
FIGURE 2.
MS/MS spectra to identify the residue acetylated after 5 min reaction of 1 mg/ml albumin with 0.5 mm p-nitrophenyl acetate. A, fragmentation of the doubly charged form of peptide 1872 identifies acetylated Tyr 411 in LVRY*TKKVPQVSTPTL. B, fragmentation of the singly charged peptide of mass 453.2 identifies acetylated Tyr-411 in Y*TK. The peak at 178.1 is the O-acetyl-tyrosine immonium ion.
To obtain additional support for acetylation on tyrosine, peptide LVRYTKKVPQVSTPTL from Fig. 2A was purified by HPLC, digested with trypsin, and analyzed by mass spectrometry. Fig. 2B shows fragmentation of the 453.2 amu singly charged parent ion. The labeled peaks in the spectrum support the sequence YTK where the acetyl group is on the phenolic oxygen of Tyr-411. The mass at 178.1 is the O-acetyl-tyrosine immonium ion. The 178.1 mass was found in other peptides that had an acetylated tyrosine. A peak at 178.1 can be used as a marker for O-acetylated tyrosine.
There is no possibility that the acetyl group is on the amino group of tyrosine, because the reaction with p-nitrophenyl acetate was performed while the amino group of Tyr-411 was in a peptide bond and therefore unavailable for modification. Taken together, the data indicate that Tyr-411 is the first albumin residue to be acetylated by p-nitrophenyl acetate.
Lysine 413 and Lysine 414 Are Slowly Acetylated by p-Nitrophenyl Acetate—After 6 h of reaction of 15 μm albumin with 500 μm p-nitrophenyl acetate, a second and third site on peptides VRYTKKVPQVSTPTL and LVRYTKKVPQVSTPTL were acetylated, as indicated by peaks with mass shifts of +42, +84, and +126 amu. The 1717 amu peak (supplemental Fig. S1A) shifted to 1759 amu, 1801 amu, and 1843 amu (supplemental Fig. S1B), while the 1830 amu peak shifted to 1872, 1914, and 1956 amu (supplemental Fig. S1B). At 6 h, about 50% of peptides VRYTKKVPQVSTPTL and LVRYTKKVPQVSTPTL were labeled with one acetate, about 50% with 2 acetates, and 1–2% with 3 acetates (percentage estimates are based on the cluster areas of the peaks). The percentage of acetylated peptides did not increase after 6 h, because 90% of the p-nitrophenyl acetate had been hydrolyzed by that time. The possibility that acetate (rather than p-nitrophenyl acetate) might be acetylating lysines was ruled out by the finding that no mass shifts occurred when 1 mg/ml albumin was treated with 0.5 mm potassium acetate.
Peptides VRYTKKVPQVSTPTL and LVRYTKKVPQVSTPTL with a mass shift of +84 amu were acetylated on Tyr-411 and Lys-413, or on Tyr-411 and Lys-414. Supplemental Fig. S2 shows the MS/MS spectrum for doubly charged parent ion 957.5 of peptide LVRYTKKVPQVSTPTL at the 6-h time point. The b4 ion at 574.1 amu (representing residues LVRY* plus 1 acetyl) and the b5 ion at 675.7 amu (representing residues LVRY*T plus 1 acetyl) support acetylation of Tyr-411. The b6 ion at 844.3 amu (representing residues LVRY*TK* plus 2 acetyls) supports acetylation of Lys-413 in addition to Tyr-411. The existence of the b6 ion mass at 803.3 amu (representing residues LVRY*TK plus 1 acetyl) suggests that Lys-413 can appear without being acetylated. The appearance of an un-acetylated form of Lys-413 together with acetylated b7 to b11 ions is consistent with the second acetyl group on Lys-414. Other +84 ions were found (data not shown) where the second acetyl group was exclusively on Lys-413 or on Lys-414. Based on the relative intensities of the peaks at 803.3 and 844.3 amu, acetylated Lys-414 was more abundant than acetylated Lys-413. From the above analysis, it follows that triply acetylated peptides, with a mass shift of +126 amu, were acetylated on Tyr-411, Lys-413, and Lys-414.
The N Terminus of Albumin, Lysine 4, Lysine 12, and Serine 5 Are Slowly Acetylated by p-Nitrophenyl Acetate—Another prominent peptide in pepsin-digested albumin was DAHKSEVAHRFKDLGEENF at 2229 amu (supplemental Fig. S1A). After 6 h of reaction of 15 μm albumin with 500 μm p-nitrophenyl acetate, 20% of this peak had acquired a mass of +42 amu (supplemental Fig. S1B), 2% had acquired a mass of +84 amu, and about 0.5% had increased in mass by +126 amu. Incubation with 10 mm p-nitrophenyl acetate for 48 h resulted in complete disappearance of the 2229 and 2271 amu peaks and appearance of +84, +126, and +168 amu ions at 2313, 2355, and 2397 amu (data not shown). MS/MS spectra showed that the +42 amu parent ion was acetylated on Asp-1, the N terminus of albumin, the +84 amu parent ion was acetylated on Asp-1 and Lys-12, and the +126 amu ion was acetylated on Asp-1, Lys-12, and Lys-4 (data not shown).
The +168 amu ion was acetylated on Asp-1, Lys-4, Ser-5, and Lys-12 (supplemental Fig. S3). Most of the major peaks in the spectrum could be assigned to the DAHKSEVAHRFKDLGEENF peptide. The 366.1 amu mass was consistent with the b3 ion (representing D*AH plus 1 acetyl). Though histidine is a potential candidate for acetylation, N-acetylhistidine is unstable (26). Therefore, acetylation is on the N terminus. The mass at 536.2 amu is consistent with the b4 ion (representing D*AHK* plus 2 acetyls). The 665.2 amu mass is consistent with the b5 ion (representing D*AHK*S* plus three acetyls). Additional b-ions at 794.4 amu (b6), 893.0 amu (b7), and 964.3 amu (b8) support the presence of 3 acetyls. The mass at 1296.5 amu is consistent with the y10 ion (representing RFK*DLGEENF plus 1 acetyl) where acetylation is on the lysine.
Analysis of cluster areas in the MALDI-TOF spectra showed that the N-terminal Asp-1 and Lys-12 were 100% acetylated, consistent with the disappearance of peaks at 2229 and 2271 amu. About 65% of Lys-4 and 6% of Ser-5 residues were acetylated, indicating that serine is not nearly as reactive with p-nitrophenyl acetate as lysine.
Residues Acetylated by 0.5 mm p-Nitrophenyl Acetate—Within the first 5 min of reaction of 15 μm albumin with 0.5 mm p-nitrophenyl acetate, Tyr-411 was acetylated 99–100%. No other residue was significantly acetylated within 5 min. Six hours after addition of p-nitrophenyl acetate, residues Asp-1, Lys-12, Lys-413, and Lys-414 were acetylated about 20–25% while Tyr-411 was still acetylated 100%. An additional 19 peptides had a mass shift of +42 amu as observed by MALDI-TOF, however peak intensities were low, and the peptide sequences could not be determined. After 50 h, all of the p-nitrophenol had been exhausted, and half of the Tyr-411 was free. By 2 weeks, none of the Tyr-411 was labeled. The albumin still carried a lot of label, but the label was on lysines and on the N-terminal Asp-1.
Residues Acetylated by 10 mm p-Nitrophenyl Acetate—Supplemental Table S1 lists the residues in human albumin acetylated by treatment of 15 μm albumin with 10 mm p-nitrophenyl acetate at pH 8.0, 22 °C for 48 h. The Mascot search engine matched 215 peptides to albumin in accession gi:3212456, yielding 76% coverage in the tryptic digest, and matched 331 peptides for 85% coverage in the GluC digest. Three peptides from a pepsin digest are included in supplemental Table S1. MS/MS spectra were manually evaluated before a labeled peptide was included in supplemental Table S1. MS/MS spectra positively identified acetylation of 59 lysines, 10 serines, 8 threonines, 4 tyrosines, and the N-terminal aspartate. Albumin has 59 lysines; every lysine was at least partially acetylated. The 10 acetylated serines were Ser-5, Ser-65, Ser-192, Ser-202, Ser-287, Ser-312, Ser-419, Ser-427, Ser-435, and Ser-454. The 8 acetylated threonines were Thr-68, Thr-76, Thr-79, Thr-83, Thr-467, Thr-474, Thr-527, and Thr-540. The four acetylated tyrosines were Tyr-84, Tyr-161, Tyr-401, and Tyr-411. The N-terminal aspartate Asp-1 was acetylated. Tyrosine 138, the ligand binding site in subdomain IB, which is modified by metabolites of polycyclic hydrocarbons (27) and by nitric oxide (28), was not modified by p-nitrophenyl acetate.
Both chemical and in vivo ε-N-acetylation of lysines has precedent (26, 29–32). Chemical O-acetylation of tyrosines also has precedent (33). Reports on acetylation of serines and threonines are more rare, though indirect evidence has been reported for bovine growth hormone-treated with acetic anhydride (34).
The peptides in supplemental Table S1 do not have an acetylated Lys at the C terminus, with the exception of Lys-162. The vast majority of acetylated lysines appear as missed cleavages within the peptide. This confirms reports in the literature (29) that trypsin generally does not recognize acetylated lysine as a cleavage site.
Carbamylation by ammonium cyanate, a degradation product of urea, would add a mass of +43, a value close to the mass of +42 from acetylation. To avoid carbamylation artifacts, we did not use urea in our protocol.
Lysines Are Stably Acetylated—An estimate of the stability of acetylated lysines was obtained by measuring % acetylation of Lys-225 in peptide SQRFPK*AEF with time. After 6 h of reaction of 15 μm albumin with 0.5 mm p-nitrophenyl acetate, 9% of Lys-225 was acetylated. No change in % acetylation of this peptide was observed for up to 9 days. The lack of an increase in % acetylation with time is explained by the fact that 90% of the p-nitrophenyl acetate had been consumed after 6 h. The absence of a loss in % acetylation supports the conclusion that acetylated lysine is stable at pH 8.0. A peptide with 9% acetylated lysines was chosen for this example to make the point that a particular residue may be only partially acetylated.
A second example of stable acetylation was obtained by MALDI-TOF analysis of peptide D*AHK*S*EVAHRFK*DLGEENF. After 48 h reaction of 15 μm albumin with 10 mm p-nitrophenyl acetate, followed by dialysis to remove excess reagent, 100% of the N terminus (Asp-1), 100% of Lys-12, 78% of Lys-4, and 5% of Ser-5 were acetylated. After 18 days, the % acetylations were unchanged. We conclude that acetylated lysines, as well as the acetylated N terminus and acetylated serine, are stable at pH 8.0 and 22 °C.
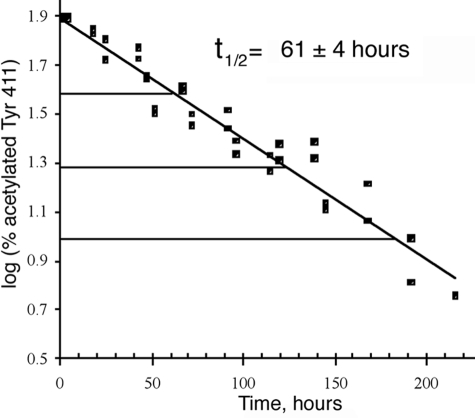
Deacetylation of Tyr-411—Treatment of 15 μm albumin with 15 μm p-nitrophenyl acetate in 10 mm Tris-Cl, pH 8.0 resulted in exclusive acetylation of Tyr-411. No other residues were significantly acetylated. Tyr-411 was maximally acetylated after 1 h. Fig. 3 shows the maximum acetylation level of 80% and loss of the acetyl group with time. The half-life for deacetylation of Tyr-411 at pH 8.0, 22 °C was 61 ± 4 h. This very slow deacetylation rate (k = 0.0002 min-1) confirms a previous report that deacetylation is the rate-limiting step for reaction of human albumin with p-nitrophenyl acetate (14). On the other hand, turnover with o-nitrotrifluoroacetanilide is explained by the instability of the trifluoroacetyl adduct due to the electronic effect of the fluorine atoms (14).
FIGURE 3.
Deacetylation rate of Tyr-411. At time 0, Tyr-411 was acetylated 80%. After 61 h at pH 8.0, 22 °C, Tyr-411 was acetylated 40%. Deacetylation was monitored for 3.5 half-lives until only 5% of Tyr-411 was still acetylated. The three horizontal lines mark 3 half-lives. % acetylation was calculated from cluster areas in the MALDI-TOF mass spectrometer for peptides VRYTKKVPQVSTPTL and LVRYTKKVPQVSTPTL.
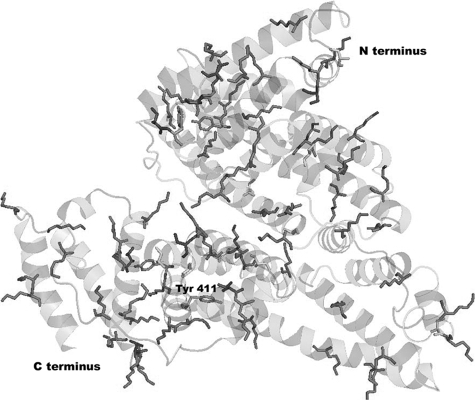
Location of Ester-reactive Residues—The location of the reactive residues in the crystal structure of human albumin is shown in Fig. 4. All acetylated lysines, serines, and threonines are exposed to the surface. Tyr-411 is in a pocket 4.5 Å from Arg-410. Acetylated Tyr-84, -161, and -401 may also have been activated by interaction with nearby arginines.
FIGURE 4.
Surface location of acetylated residues in human albumin. The crystal structure of human albumin (Protein Data Bank code 1bm0) shows side-chains for acetylated lysines, tyrosines, serines, and threonines. Asp-1 and Lys-4 are missing from the structure (41).
Time to Complete Hydrolysis of 0.5 mm p-Nitrophenyl Acetate—Six hours after addition of p-nitrophenyl acetate to 15 μm albumin in 10 mm Tris-Cl, pH 8.0, 22 °C, 90% of the 0.5 mm p-nitrophenyl acetate had been consumed, and the rate of p-nitrophenol production by albumin had slowed so it was indistinguishable from the rate by buffer alone. By 24 h, the end point absorbance of 8.42, at 400 nm, was reached, and no further change in absorbance was obtained. The theoretical end point calculated from the extinction coefficient of 16,900 m-1 cm-1 for p-nitrophenolate ion at pH 8.0 was 8.45 (1), a value in close agreement with the observed end point.
Percent Acetylated Lysines—About 50% of the hydrolysis of 0.5 mm p-nitrophenyl acetate was due to reaction with 15 μm albumin, and about 50% to reaction with pH 8.0 buffer. Assuming that the 250 μm p-nitrophenyl acetate that reacted with 15 μm albumin resulted in stable acetylation of albumin, one can calculate about 16–17 molar equivalents of acetate bound per mol of albumin. If 16 of 59 lysines are acetylated, then on average 27% of each lysine was acetylated. This value is close to the 20–25% acetylation calculated for Lys-414 by MALDI-TOF analysis.
A third method was used to calculate % acetylated lysines. This method is based on the principle that lysines labeled with fluorodinitrobenzene are relatively stable to acid hydrolysis, whereas acetylated lysines are deacetylated to free lysines. Standard acid hydrolysis followed by amino acid composition analysis allows an estimate of the number of acetylated lysines (30). It was found that 27% of the lysines were acetylated in albumin treated with 0.5 mm p-nitrophenyl acetate, while 67% of the lysines were acetylated in albumin treated with 10 mm p-nitrophenyl acetate.
Esterase Activity of Albumin—The esterase activity of human albumin can be visualized on a nondenaturing gel stained for esterase activity with β-naphthyl acetate and Fast Blue RR. The β-naphthol reacts with the diazonium salt of Fast Blue RR to produce a pink, insoluble azodye, which precipitates at the site where naphthol is released.
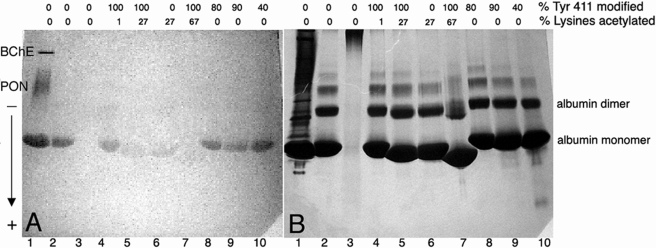
The gel in Fig. 5A shows naphthol production by the albumin present in 5 μl of human plasma (lane 1), as well as by 200 μg of pure human albumin (lane 2). Additional esterase bands in lane 1 are from butyrylcholinesterase and paraoxonase. The question of interest was how much of the apparent esterase activity of albumin was due to Tyr-411 and how much to irreversible acetylation of lysines? To answer this question, albumin preparations with various percent acetylation of Tyr-411 and lysines were loaded on the gel. The esterase activity in lane 4 where 100% of Tyr-411 was acetylated, was similar to that in lane 2 where none of the Tyr-411 was acetylated. The esterase activity in lane 5 where 100% of the Tyr-411 was acetylated, and 27% of the lysines were acetylated, was substantially decreased. The albumin in lane 6 was acetylated only on lysines, because Tyr-411 had completely deacetylated during 2 weeks incubation; the esterase activity in lane 6 was similar to that in lane 5, showing that lysines contributed more to albumin esterase activity than Tyr-411. The albumin in lane 7 was maximally acetylated by 10 mm p-nitrophenyl acetate, corresponding to the 82 residues acetylated in supplemental Table S1. The esterase activity in lane 7 was nearly abolished. Blocking Tyr-411 by covalent modification with DFP (lane 8) or chlorpyrifos oxon (lanes 9 and 10) had little effect on the apparent esterase activity.
FIGURE 5.
Nondenaturing gel stained for esterase activity (A) and counterstained with Coomassie Blue (B). Lane 1, 5 μl of human serum where the esterase bands are butyrylcholinesterase (BChE), paraoxonase (PON), and albumin. Lane 2, 200 μg of 99% pure fatty acid-free human albumin. Lane 3, 200 μg of denatured, carbamidomethylated albumin. Lane 4, 200 μg of albumin treated 5 min with 0.5 mm p-nitrophenyl acetate to acetylate 100% of Tyr-411. Lane 5, 200 μg of albumin treated with 0.5 mm p-nitrophenyl acetate for 6 h. Lane 6, 200 μg of albumin treated with 0.5 mm p-nitrophenyl acetate for 2 weeks. Lane 7, 200 μg of albumin treated with 10 mm p-nitrophenyl acetate for 48 h. Lane 8, 200 μg of albumin treated with DFP to label 80% of Tyr-411. Lane 9, 200 μg of albumin treated with chlorpyrifos oxon to label 90% of Tyr-411. Lane 10, 200 μg of albumin treated with chlorpyrifos oxon to label 40% of Tyr-411. The percent labeling of Tyr-411 was estimated from cluster areas of peaks in the MADLI-TOF mass spectrometer. The percent labeling of lysines was estimated from amino acid composition analysis. The arrow indicates the direction of migration of proteins on the gel.
The gel was counterstained with Coomassie Blue in Fig. 5B to show that protein loading per lane was equivalent. Slower migrating bands are consistent with higher molecular weight forms of albumin. Pure albumin is predominantly monomeric, but also forms dimers, and higher multimers. Fig. 5B clearly shows that the highly acetylated albumin (67%) in lane 7 migrated substantially further than native albumin in lane 2. Albumin that was 27% acetylated migrated slightly further than native. This behavior is consistent with elimination of positive charge from the protein by acetylation of the ε-N-amino groups of the lysines, thus giving the acetylated protein a greater net negative charge so that it would be attracted more readily to the positively charged electrode at the bottom of the gel.
The esterase activity of albumin is characterized by an initial burst of product formation that is equal to one equivalent of albumin, followed by slower formation of multiple equivalents of product (1). The fast phase has been attributed to initial acetylation of Tyr-411 (21), which is consistent with the rapid labeling of Tyr-411 described in Fig. 1. From the activity shown in Fig. 5, it can be concluded that the apparent slow phase esterase activity of albumin is due to release of p-nitrophenol upon acetylation of 59 lysines, and to some extent the surface accessible serines, threonines, and other tyrosines. This myriad of acetylations creates the appearance of enzymatic turnover, but it is not a true turnover process. Lysine, serine, and threonine are acetylated but do not release the acetate. Acetylated Tyr-411 does release the bound acetate (that is, it turns over), but the release rate is too slow to account for a significant part of the apparent esterase activity of albumin in a 30-min assay. Albumin must therefore be regarded as a pseudo-esterase, not an enzymatic esterase.
DISCUSSION
Albumin Esterase Activity—In our previous reports we had identified Tyr-411 as the residue in albumin that is labeled by soman, sarin, DFP, chlorpyrifos oxon, FP-biotin, and dichlorvos (16, 18). Others have also identified tyrosine as the site of covalent binding of soman, sarin, cyclosarin, and tabun to albumin (20). Organophosphorus agents inhibit the esterase activity of butyrylcholinesterase and other serine esterases by covalent binding to the active site serine (35). By analogy, we had expected that OP binding to albumin would inhibit the esterase activity of albumin because it is commonly assumed that Tyr-411 is the active site residue on albumin that is responsible for esterase activity. The burst activity of albumin with p-nitrophenyl acetate is indeed inhibited by labeling Tyr-411 with an organophosphorus agent (21). However our results show that the slow steady state esterase activity of albumin is not inhibited by binding OP to Tyr-411, but instead is inhibited by acetylation of lysines. These results lead to a revised model of the esterase activity of albumin.
In this revised model of the esterase activity of albumin up to 82 residues participate. The most reactive residue is Tyr-411. Deacetylation of Tyr-411 occurs with a half-life of 61 ± 4 h, which means the p-nitrophenolate product formed in a 30-min reaction cannot come from turnover on Tyr-411. The majority of the “esterase” activity of albumin is due to a half-reaction with lysines, serines, threonines, and tyrosines. These acetylated residues do not turnover but form stable adducts. At high p-nitrophenyl acetate concentration a total of 59 lysines, 10 serines, 8 threonines, 4 tyrosines, and the N-terminal aspartate are acetylated.
The lysines of albumin are acetylated by p-nitrophenyl acetate, but are not labeled by organophosphorus esters. We have found no evidence for labeling of lysines by organophosphorus agents.
Confirmation of Means and Bender—When Means and Bender (1) found stable incorporation of [14C]acetate into albumin they concluded that accelerated formation of p-nitrophenolate ion in the presence of serum albumin was not due to increased hydrolysis, but to rapid acetylation of serum albumin by p-nitrophenyl acetate. Means and Bender did not identify the residues involved and therefore their conclusion has been overlooked during the past 30 years while investigators focused on Tyr-411 as the esteratic site (36). With the availability of mass spectrometry we have identified the specific residues in albumin that become acetylated by p-nitrophenyl acetate and therefore provide proof for the concept introduced by Means and Bender to explain albumin esterase activity. Our results support their conclusion that the overall reaction rate, as reflected by appearance of p-nitrophenolate ion, corresponds to the sum of a large number of simultaneous reactions at different sites on the protein plus spontaneous hydrolysis.
Additional support for the conclusion that lysines and tyrosine participate in the esterase activity of albumin comes from studies of chemically modified albumin (7). Modification of tyrosine and lysine suppressed hydrolysis of carprofen glucuronide, leading to the conclusion that Tyr and Lys have roles in hydrolysis.
Significance—It is expected that other esters will also acetylate albumin. Acetyl salicylic acid (aspirin) has been shown to acetylate up to 3 residues on human albumin, though only one acetylation site has been identified to date, namely Lys-199 (37–39). Though acetylation rates must be very slow, the high concentration of albumin in plasma (0.6 mm) makes these reactions pharmacologically relevant.
Albumin is unusual in the family of plasma proteins because it has no carbohydrates. This makes the amino acids of albumin more accessible to acetylation than those of a protein like butyrylcholinesterase whose surface is sugar-coated. Nevertheless, it is possible that other proteins including butyrylcholinesterase are acetylated by carboxylic acid esters on multiple sites. Several lysines on ubiquitin are acetylated by aspirin (26). In some cases chemical acetylation has an important physiological consequence. For example, the anti-inflammatory action of aspirin is explained by acetylation of the N-terminal serine of prostaglandin synthetase (40).
Supplementary Material
Acknowledgments
Mass spectra and amino acid composition analysis were obtained with the support of the Mass Spectrometry and Proteomics core facility and the Protein Structure core facility at the University of Nebraska Medical Center.
This work was supported, in whole or in part, by the National Institutes of Health NIH CounterACT U01 NS058056-02 (to O. L.) and NIH Eppley Cancer Center Grant P30CA36727. This work was also supported by U.S. Army Medical Research and Materiel Command W81XWH-07-2-0034 (to O. L.), W81XWH-06-1-0102 (to S. H. H.), and DGA Grant 03co010-05/PEA01 08 7 (to P. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
Footnotes
The abbreviations used are: MS, mass spectrometry; DFP, diisopropylfluorophosphate; amu, atomic mass unit; CHCA, α-cyano-4-hydroxycinnamic acid; LC/MS/MS, liquid chromatography tandem mass spectrometry; MALDI-TOF, matrix-assisted laser desorption ionization time of flight mass spectrometry; i.d., inner diameter.
References
- 1.Means, G. E., and Bender, M. L. (1975) Biochemistry 14 4989-4994 [DOI] [PubMed] [Google Scholar]
- 2.Tildon, J. T., and Ogilvie, J. W. (1972) J. Biol. Chem. 247 1265-1271 [PubMed] [Google Scholar]
- 3.Casida, J. E., and Augustinsson, K. B. (1959) Biochim. Biophys. Acta 36 411-426 [DOI] [PubMed] [Google Scholar]
- 4.Morikawa, M., Inoue, M., Tsuboi, M., and Sugiura, M. (1979) Jpn. J. Pharmacol. 29 581-586 [DOI] [PubMed] [Google Scholar]
- 5.Rainsford, K. D., Ford, N. L., Brooks, P. M., and Watson, H. M. (1980) Eur. J. Clin. Investig. 10 413-420 [DOI] [PubMed] [Google Scholar]
- 6.Dubois-Presle, N., Lapicque, F., Maurice, M. H., Fournel-Gigleux, S., Magdalou, J., Abiteboul, M., Siest, G., and Netter, P. (1995) Mol. Pharmacol. 47 647-653 [PubMed] [Google Scholar]
- 7.Georges, H., Presle, N., Buronfosse, T., Fournel-Gigleux, S., Netter, P., Magdalou, J., and Lapicque, F. (2000) Chirality 12 53-62 [DOI] [PubMed] [Google Scholar]
- 8.Kwon, C. H., Maddison, K., LoCastro, L., and Borch, R. F. (1987) Cancer Res. 47 1505-1508 [PubMed] [Google Scholar]
- 9.Salvi, A., Carrupt, P. A., Mayer, J. M., and Testa, B. (1997) Drug Metab. Dispos. 25 395-398 [PubMed] [Google Scholar]
- 10.Tove, S. B. (1962) Biochim. Biophys. Acta 57 230-235 [DOI] [PubMed] [Google Scholar]
- 11.De Vriese, C., Hacquebard, M., Gregoire, F., Carpentier, Y., and Delporte, C. (2007) Endocrinology 148 2355-2362 [DOI] [PubMed] [Google Scholar]
- 12.Sogorb, M. A., Diaz-Alejo, N., Escudero, M. A., and Vilanova, E. (1998) Arch. Toxicol. 72 219-226 [DOI] [PubMed] [Google Scholar]
- 13.Sogorb, M. A., Carrera, V., and Vilanova, E. (2004) Arch. Toxicol. 78 629-634 [DOI] [PubMed] [Google Scholar]
- 14.Masson, P., Froment, M. T., Darvesh, S., Schopfer, L. M., and Lockridge, O. (2007) J. Enzyme Inhib. Med. Chem. 22 463-469 [DOI] [PubMed] [Google Scholar]
- 15.Manoharan, I., and Boopathy, R. (2006) Arch. Biochem. Biophys. 452 186-188 [DOI] [PubMed] [Google Scholar]
- 16.Li, B., Nachon, F., Froment, M. T., Verdier, L., Debouzy, J. C., Brasme, B., Gillon, E., Schopfer, L. M., Lockridge, O., and Masson, P. (2008) Chem. Res. Toxicol. 21 421-431 [DOI] [PubMed] [Google Scholar]
- 17.Watanabe, H., Tanase, S., Nakajou, K., Maruyama, T., Kragh-Hansen, U., and Otagiri, M. (2000) Biochem. J. 349 813-819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, B., Schopfer, L. M., Hinrichs, S. H., Masson, P., and Lockridge, O. (2007) Anal. Biochem. 361 263-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanger, F. (1963) Proc. Chem. Soc. 5 76-83 [Google Scholar]
- 20.Williams, N. H., Harrison, J. M., Read, R. W., and Black, R. M. (2007) Arch. Toxicol. 81 627-639 [DOI] [PubMed] [Google Scholar]
- 21.Means, G. E., and Wu, H. L. (1979) Arch. Biochem. Biophys. 194 526-530 [DOI] [PubMed] [Google Scholar]
- 22.Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999) Electrophoresis 20 3551-3567 [DOI] [PubMed] [Google Scholar]
- 23.Geer, L. Y., Markey, S. P., Kowalak, J. A., Wagner, L., Xu, M., Maynard, D. M., Yang, X., Shi, W., and Bryant, S. H. (2004) J. Proteome Res. 3 958-964 [DOI] [PubMed] [Google Scholar]
- 24.Sanger, F. (1945) Biochem. J. 39 507-515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, B., Sedlacek, M., Manoharan, I., Boopathy, R., Duysen, E. G., Masson, P., and Lockridge, O. (2005) Biochem. Pharmacol. 70 1673-1684 [DOI] [PubMed] [Google Scholar]
- 26.Macdonald, J. M., LeBlanc, D. A., Haas, A. L., and London, R. E. (1999) Biochem. Pharmacol. 57 1233-1244 [DOI] [PubMed] [Google Scholar]
- 27.Brunmark, P., Harriman, S., Skipper, P. L., Wishnok, J. S., Amin, S., and Tannenbaum, S. R. (1997) Chem. Res. Toxicol. 10 880-886 [DOI] [PubMed] [Google Scholar]
- 28.Jiao, K., Mandapati, S., Skipper, P. L., Tannenbaum, S. R., and Wishnok, J. S. (2001) Anal. Biochem. 293 43-52 [DOI] [PubMed] [Google Scholar]
- 29.Violand, B. N., Schlittler, M. R., Lawson, C. Q., Kane, J. F., Siegel, N. R., Smith, C. E., Kolodziej, E. W., and Duffin, K. L. (1994) Protein Sci. 3 1089-1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allfrey, V. G., Di Paola, E. A., and Sterner, R. (1984) Methods Enzymol. 107 224-240 [DOI] [PubMed] [Google Scholar]
- 31.Lapko, V. N., Smith, D. L., and Smith, J. B. (2001) Protein Sci. 10 1130-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gershey, E. L., Vidali, G., and Allfrey, V. G. (1968) J. Biol. Chem. 243 5018-5022 [PubMed] [Google Scholar]
- 33.Riordan, J. F., and Vallee, B. L. (1972) Methods Enzymol. 25 500-506 [DOI] [PubMed] [Google Scholar]
- 34.Oikawa, A., Dellacha, J. M., and Sonenberg, M. (1967) Biochem. J. 104 947-952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nachon, F., Asojo, O. A., Borgstahl, G. E., Masson, P., and Lockridge, O. (2005) Biochemistry 44 1154-1162 [DOI] [PubMed] [Google Scholar]
- 36.Sakurai, Y., Ma, S. F., Watanabe, H., Yamaotsu, N., Hirono, S., Kurono, Y., Kragh-Hansen, U., and Otagiri, M. (2004) Pharm. Res. 21 285-292 [DOI] [PubMed] [Google Scholar]
- 37.Hawkins, D., Pinckard, R. N., and Farr, R. S. (1968) Science 160 780-781 [DOI] [PubMed] [Google Scholar]
- 38.Walker, J. E. (1976) FEBS Lett. 66 173-175 [DOI] [PubMed] [Google Scholar]
- 39.Yang, F., Bian, C., Zhu, L., Zhao, G., Huang, Z., and Huang, M. (2007) J. Struct. Biol. 157 348-355 [DOI] [PubMed] [Google Scholar]
- 40.Roth, G. J., and Siok, C. J. (1978) J. Biol. Chem. 253 3782-3784 [PubMed] [Google Scholar]
- 41.Sugio, S., Kashima, A., Mochizuki, S., Noda, M., and Kobayashi, K. (1999) Protein Eng. 12 439-446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.