Abstract
Lipoproteins are emulsion particles that consist of lipids and apolipoproteins. Their natural function is to transport lipids and/or cholesterol to different tissues. We have taken advantage of the hydrophobic interior of these natural emulsions to solubilize DNA. Negatively charged DNA was first complexed with cationic lipids containing a quaternary amine head group. The resulting hydrophobic complex was extracted by chloroform and then incorporated into reconstituted chylomicron remnant particles (≈100 nm in diameter) with an efficiency ≈65%. When injected into the portal vein of mice, there were ≈5 ng of a transgene product (luciferase) produced per mg of liver protein per 100 μg injected DNA. This level of transgene expression was ≈100-fold higher than that of mice injected with naked DNA. However, such a high expression was not found after tail vein injection. Histochemical examination revealed that a large number of parenchymal cells and other types of cells in the liver expressed the transgene. Gene expression in the liver increased with increasing injected dose, and was nearly saturated with 50 μg DNA. At this dose, the expression was kept at high level in the liver for 2 days and then gradually reduced and almost disappeared by 7 days. However, by additional injection at day 7, gene expression in the liver was completely restored. By injection of plasmid DNA encoding human α1-antitrypsin, significant concentrations of hAAT were detected in the serum of injected animals. This is the first nonviral vector that resembles a natural lipoprotein carrier.
Keywords: gene transfer, cationic lipid, emulsion, apolipoprotein, human α1-antitrypsin
The liver is an important target organ for gene therapy, because this organ plays a central role in the metabolism and production of serum proteins. There are many lethal metabolic diseases resulting from the defect or deficiency of hepatocyte-derived gene products (1–5). Acquired diseases such as hepatoma and viral hepatitis are also likely to be a target of hepatic gene therapy. Advancements in hepatic gene therapy depend to a large degree on the development of a delivery system capable of efficiently introducing genes into the hepatocytes.
Although a variety of techniques have been developed to introduce genetic materials into the liver, each has its distinct problem. Since retroviral vectors have no infectivity to quiescent cells (6), they have been used mainly for ex vivo gene therapy, i.e., transduction of cultured hepatocytes and their subsequent reimplantation into the host liver (7, 8). Direct introduction requires enhanced proliferation activity of the hepatocytes by means of a partial hepatectomy (9, 10). Injection of adenoviral vectors into the portal vein or systemic circulation leads to high levels of foreign gene expression (11, 12). However, the expression is usually transient and repetitive injection is unsuccessible because of high immunogenicity of the vector (13, 14). Nonviral gene transfer methods, including injection of plasmid DNA alone (15, 16) or plasmid DNA complexed with polylysine (17, 18) or liposomes (19–21), have also been developed, but the level of gene expression is usually not high enough for therapeutic purposes.
Lipoproteins are naturally occurring biological emulsions and serve as carriers of cholesterol and other lipids in the systemic circulation (22). Dietary lipids absorbed by the intestine are packed into triglyceride-rich lipoproteins termed chylomicrons (23). In the blood circulation, chylomicrons are remodeled to chylomicron remnants by hydrolysis of the core triglycerides by lipoprotein lipase and by accepting apolipoproteins (24). Finally, the remnants are taken up by liver parenchymal cells via apolipoprotein-specific receptors (25). Recently, reconstituted chylomicron remnants (RCR) were established using commercially available lipids and shown to be taken up by hepatocytes following intravenous injection (26). If the therapeutic DNA can be incorporated into the RCR, the resulting nonviral particles could be an efficient vector for hepatic gene therapy.
We report here the incorporation of DNA into RCR by means of the formation of a hydrophobic DNA complex with a quaternary ammonium derivative of cholesterol. Intraportal injection of the resulting particles leads to highly efficient expression of foreign genes in the livers of mice. The therapeutic potential of the novel vector is demonstrated using human α1-antitrypsin (hAAT) gene and its production in the liver.
MATERIALS AND METHODS
Materials.
Cholesterol (Chol), cholesteryl oleate, l-α-lysophosphatidylcholine, olive oil, and dimethyldioctadecylammonium bromide were obtained from Sigma. l-α-phosphatidylcholine were from Avanti Polar Lipid, luciferase from Calbiochem and 5-bromo-4-chloro-3-indoyl-β-d-galactoside (X-Gal) from GIBCO/BRL. Luciferase assay system and Coomassie Plus Protein Assay Reagent were from Promega and Pierce, respectively. Calibrator 4 as a standard for hAAT and goat anti-hAAT were purchased from Incstar (Stillwater, MN) and Vectastain ABC kit were from Vector.
Plasmids.
Plasmid pCMVL and pCMVLacZ contain a fire fly luciferase gene and a bacterial β-galactosidase (β-gal) gene, respectively, and both of them are driven by a human cytomegalovirus (CMV) immediate early promoter. pRSVhAAT (27) contains a hAAT gene driven by a Raus sarcoma virus (RSV) promoter. pAAVCMVhAAT was reconstructed from pAAVlac.26 (28) and pRSVhAAT. Briefly, pAAVlac.26 was digested with XbaI followed by ligation with double-stranded oligodeoxynucleotide (5′-CTAGACTCGAGT-3′) to insert a XhoI site. After digestion of both plasmids by HindIII and XhoI, the 4,510-bp fragment from pAAVlac.26 and the 1680-bp fragment from pRSVhAAT were isolated and ligated together. The resulting pAAVCMVhAAT contains two AAV inverted terminal repeat sequences flanking hAAT gene driven by the CMV promoter. These plasmids were amplified in DH5α strain of Escherichia coli, isolated by alkaline lysis and purified by cesium chloride gradient centrifugation (29).
Synthesis of Cationic Derivatives of Cholesterol.
3-β-[N-(N′,N′-dimethylethane)-carbamoyl]cholesterol (DC-Chol), a tertiary ammonium derivative of Chol, was synthesized as described (30). 3-β-[N-(N′,N′,N′-trimethylethane)carbamoyl]cholesterol (TC-Chol), a quaternary ammonium derivative of Chol, was synthesized by methylation of DC-Chol by iodomethane.
Formation of Hydrophobic DNA/Cationic Lipid Complex.
Formation of hydrophobic complex of DNA and cationic lipids was carried out according to the method of Reimer et al. (31). Plasmid DNA (4 μg) containing trace amount of 125I-labeled DNA was incubated with various amount of cationic lipids in 410 μl of Bligh and Dyer monophase (chloroform:methanol:water = 1:2.1:1) at room temperature for 30 min. Subsequently, the monophase was partitioned into a two-phase system by the addition of 100 μl each of chloroform and water. The sample was mixed by vortexing, and the separation of the upper aqueous and lower organic phases was facilitated by centrifugation at 2,000 × g for 10 min at room temperature. Two hundred microliters of the aqueous phase and 100 μl of the organic phase were collected separately and their radioactivities were measured using a gamma-counter (Gamma 5500B, Beckman). Radioactivity in the interface was calculated by subtracting the radioactivities of aqueous and organic phases from the total radioactivity.
Incorporation of Hydrophobic DNA/TC-Chol Complex into RCR.
Hydrophobic DNA/TC-Chol complexes was incorporated into RCR composed of olive oil:l-α-phosphatidylcholine:l-α-lysophosphatidyl choline:cholesteryl oleate: Chol (70:22.7:2.3:3.0:2.0, weight ratio), as follows. The complex prepared from 400 μg of DNA and 1.25 mg of TC-Chol in 4.1 ml of the monophase was extracted in 2 ml of chloroform The complex was mixed with 40 mg of lipids dissolved in chloroform and evaporated under a stream of nitrogen. Following vacuum desiccation for 1 h, 1.6 ml of water was added and left at room temperature for 1 h. The hydrated mixture of DNA/TC-Chol complex and lipids was emulsified by vortexing for 2 min and homogenization by a Tissue Tearor (Model 985-370, Biospec Products, Bartlesville, OH) for 30 sec at 65°C. Under this condition, no degradation of the plasmid DNA was found by agarose gel electrophoresis. Finally, the emulsion were extruded 20 times through a polycarbonate membrane filter with 100 nm pore size (Nuclepore).
Incorporation of DNA into the RCR was determined by its flotation on centrifugation in the absence or presence of a density gradient. After extrusion, a half ml of the emulsion was mixed with 4 ml of water and then centrifuged at 16,000 × g for 20 min at room temperature using a SW50.1 rotor (Beckman). In the case of the density gradient centrifugation, the emulsion (0.5 ml) was mixed with 2-fold volume of NaCl solution with a density of 1.346 and discontinuous gradient was then formed using 1 ml each of NaCl solution with a density of 1.065, 1.020 and 1.006 (32). Fractions of a half ml each were collected from the top to the bottom of the centrifuge tube and the radioactivity of 125I-DNA in each fraction was measured in a gamma counter.
Incorporation of DNA into RCR was also tested as protection of DNA from DNase I digestion. Empty RCR was prepared in the same concentration as RCR containing DNA. 40 ng plasmid DNA was simply mixed with 10 μl empty RCR with or without TC-Chol or 10 μl of the emulsion was mixed with 0.1U DNase I and incubated at 37°C for 10 min. After enzyme digestion, samples were incubated at 65°C for 10 min with or without addition of 1 μl 10% SDS. The samples were then run on a 0.8% agarose gel in TAE buffer and DNA was visualized by ethidium bromide staining.
Intraportal Injection.
Six weeks old female CD1 mice were anesthetized with intramuscular injection of ketamine hydrochloride (1 mg/20 g of body weight). After the liver was exposed through a ventral midline incision, various amounts of DNA/TC-Chol complex-incorporated RCR (DNA/TC-Chol-RCR), which was dispersed in 1 ml of 5.2% (isotonic) mannitol, were injected into the portal vein using a 1/2-inch 30-gauge needle and 1 ml syringe.
Measurement of Luciferase Activity.
At indicated days after injection, mice were sacrificed and liver, spleen, lung, kidney, and heart were collected. The organs were homogenized with lysis buffer (0.05% Triton X-100, 2 mM EDTA, 0.1 M Tris, pH 7.8) using a Tissue Tearor for 1 min. After 2 cycles of freeze in liquid nitrogen and thaw at 37°C, the homogenates were centrifuged at 14,000 × g for 10 min at 4°C. Supernatant (20 μl) was mixed with 100 μl of Luciferase assay system and relative light unit was measured with a luminometer (AutoLumant LB 953, EG & G, Salem, MA) for 20 sec. Conversion from relative light unit to luciferase protein mass was calculated from a standard curve (1 to 10,000 pg; pg luciferase = −5.0 + 8.3 × 10−3 × relative light unit, r2 = 0.99) based on purified luciferase protein as a standard. Protein concentration of the supernatant was also determined by a Coomassies Protein Assay Reagent, using BSA as a standard.
X-Gal Staining.
Forty-eight hours after injection, the liver was perfused in situ with 5 ml of 1.25 mM EGTA in PBS (pH 7.5) to remove the blood. The liver was dissected, cut into small blocks and frozen in OCT embedding compound (Miles Scientific). Cryosections of 10 μm in thickness were sampled and placed on polylysine-coated glass slides. Following fixation with 2% glutaraldehyde in PBS containing 0.04% Nonidet P-40, the liver sections were incubated in a staining solution (0.08% X-Gal), 5 mM each of K3Fe(CN)6 and K4Fe(CN)6, and 2 mM MgCl2 in PBS) at 37°C for 24 h. The sections were then rinsed three times with PBS and lightly counter stained with hematoxylin.
ELISA for hAAT.
ELISA method for measuring blood level of hAAT was modified from a published procedure (33), except that an ABC kit containing avidin and biotinylated peroxidase was used.
RESULTS
Preparation of Hydrophobic DNA/TC-Chol Complex.
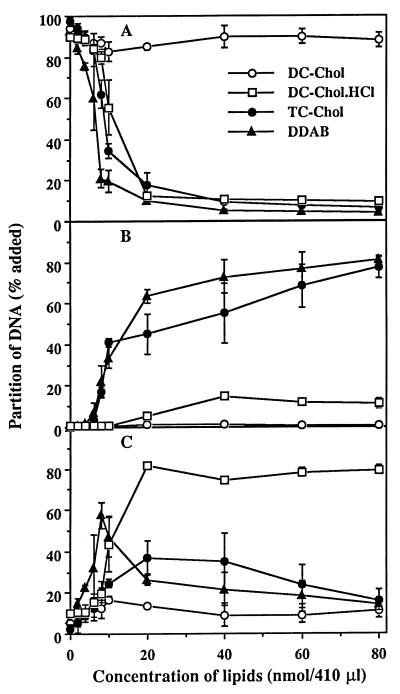
Since RCR is an emulsion containing no aqueous phase in its interior, native DNA cannot be incorporated into RCR because of its high degree of hydrophilicity. It has been reported that cationic lipids form a hydrophobic complex with DNA by an electrostatic interaction (34, 35). We have previously described the complex of DNA and cationic liposomes containing DC-Chol, which transfects a variety of cultured cell lines with high efficiency (36, 37). We attempted to make a hydrophobic complex of DNA with DC-Chol, but the DC-Chol did not increase the solubility of DNA in the organic phase (Fig. 1A). Cationic charge of DC-Chol does not seem strong enough to form a hydrophobic complex with DNA, probably due to incomplete ionization of DC-Chol in the monophase. Although DC-Chol hydrochloride, a salt form of DC-Chol, drastically decreased the solubility of DNA in the aqueous phase, the resulting hydrophobic complex was not soluble in the organic phase (Fig. 1B) and collected from the interface (Fig. 1C). TC-Chol, a quaternary ammonium derivative of DC-Chol, formed a hydrophobic complex with DNA that was extractable with chloroform (Fig. 1B). At lower concentrations of TC-Chol, the amount of hydrophobic complex in the interface increased and reached the maximum level at 10 nmol of TC-Chol at which the ratio of cationic charge of TC-Chol to anionic charge of DNA was ≈1 (Fig. 1C). After that, the complex in the interface (Fig. 1C) was quantitatively transferred to the organic phase (Fig. 1B). The results with TC-Chol were almost the same as that obtained with dimethyldioctadecylammonium bromide, a cationic lipid having a quaternary amine head group (38). In the preparation of DNA/TC-Chol complex for in vivo study, the concentration of both DNA and TC-Chol was almost 30× higher than the above condition. However percentile collection of the complex was not changed.
Figure 1.
Effect of cationic lipids on the partition of DNA into organic phase. 125I-DNA (4 μg) was incubated with indicated amounts of cationic lipids in the Bligh and Dyer monophase (410 μl) at room temperature for 30 min. The monophase was separated into two phases by the addition of chloroform and water (100 μl each). Partition of DNA into the aqueous phase (A) and organic phase (B) were determined by measuring their radioactivities. Partition of DNA into the interface (C) was determined by subtracting the amount of DNA in the aqueous and organic phases from total DNA added. Each point presents the mean from three experiments ± SD.
Incorporation of Hydrophobic DNA/TC-Chol Complex into RCR.
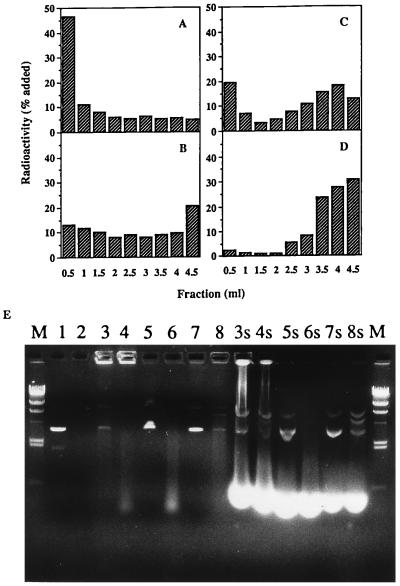
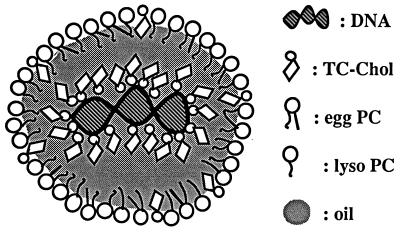
The hydrophobic DNA/TC-Chol complex extracted with chloroform was emulsified together with commercially available lipids by homogenization, and incorporation of DNA into RCR was confirmed by means of its flotation by centrifugation. More than 65% of DNA added as the complex floated up with the emulsions into the top three fractions (Fig. 2A), whereas no flotation of DNA was observed when free DNA was used (Fig. 2B). This means that more than two-thirds of DNA/TC-Chol complexes were incorporated into the RCR. It has been reported that hydrophobic complexes of DNA and cationic lipids reversibly dissociate depending on the concentration of inorganic salt in the solvent (35). DNA/TC-Chol-RCR was mixed with NaCl solution and then density gradient centrifugation was carried out. About 32% of DNA was recovered from the upper emulsion fractions (Fig. 2C). When free DNA was mixed with empty RCR containing TC-Chol in its lipid composition, majority of DNA associated with the RCR, resulting in flotation after the centrifugation in the water (data not shown). However, no flotation of DNA was observed after the NaCl density gradient centrifugation (Fig. 2D). Moreover, most DNA incorporated into RCR is protected from DNase I digestion (Fig. 2E, lanes 3, 4), while simple mixture of free DNA and empty RCR with or without TC-Chol has no or weaker protection against DNase I (Fig. 2E, lanes 5–8). One percent SDS can partially break down RCR and more DNA can migrate into the gel (Fig. 2E, lanes 3s and 4s). This result confirmed that most DNA in RCR is completely neutralized by TC-Chol and stays in the wells of the agarose gel (lanes 3, 4), while small amount of DNA that runs as free plasmid is not protected and is probably attached on the outer surface of RCR. Based on these two methods, it is estimated that at least 60–80% of DNA incorporated into RCR is likely to localize in the internal oil space of RCR. Resulting DNA/TC-Chol-RCR has a homogeneous particle distribution with a mean diameter of 107 ± 16 nm (average ± SD, n = 3) measured by light scattering using a submicron particle analyzer (N4 Plus, Coulter). A proposed structure of the DNA/TC-Chol-RCR is given in Fig. 3.
Figure 2.
(A–D) Determination of DNA-incorporation into RCR by centrifugation. DNA/TC-Chol complex (A, C) or free DNA (B, D) supplemented with a trace amount of 125I-DNA was incorporated into RCR (A–C) or mixed with empty RCR containing TC-Chol (D). The samples (0.5 ml) were centrifuged in 4.5 ml (final) of water (A, B) or NaCl density gradient (C, D). Fractions (0.5 ml) were collected from the top to the bottom of centrifuge tubes and the radioactivities were measured. (E) Determination of DNA incorporation into RCR by DNase I digestion assay. DNA-TC-Chol complex was incorporated into RCR (lanes 3, 4, 3s, 4s) or free DNA was mixed with empty RCR without (lanes 5, 6, 5s, 6s) or with (lanes 7, 8, 7s, 8s) TC-Chol. Lane 1 and 2 are DNA only. 0.1 unit DNase I was added to each even numbered sample and incubated at 37°C for 10 min. The samples were incubated at 65°C for 10 min after treatment with (lanes 3s–8s) or without (lanes 1–8) 1% SDS and then run on a 0.8% agarose gel. M is λ DNA HindIII fragments. The brightly stained lower band in lanes 3s to 8s is due to SDS solubilized lipids.
Figure 3.
A proposed structure of the DNA/TC-Chol-RCR.
In Vivo Gene Expression in Mice.
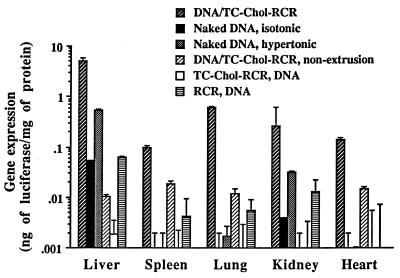
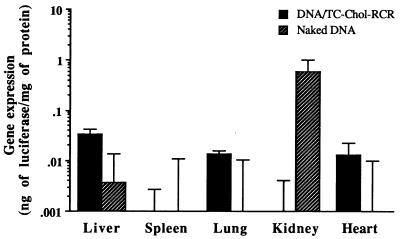
Recently, it was reported that naked DNA delivered intraportally efficiently expressed the gene in hepatocytes (39). We injected our new formulation by the same manner and compared their gene expression efficiency with that of naked DNA two days after injection. As shown in Fig. 4, a high amount of luciferase protein was produced in the liver of mice injected with 100 μg DNA of pCMVL DNA/TC-Chol-RCR. The level was almost 100-fold higher than that of naked DNA-injected mice (isotonic). Although the level of gene expression in the liver by naked DNA was significantly increased when a hypertonic solution (15% mannitol, 0.9% NaCl) was used for injection (hypertonic) as reported (39), the level was still 10-fold lower than that of the DNA/TC-Chol-RCR. Luciferase protein was also produced in the spleen, lung, and heart by intraportal injection of pCMVL DNA/TC-Chol-RCR, but the levels were 25- to 800-fold lower than that in the liver. The particle size of the DNA/TC-Chol-RCR was important for an efficient gene delivery into hepatocytes. Injection of DNA/TC-Chol-RCR without extrusion (DNA/TC-Chol-RCR, nonextrusion), which had a mean diameter of 352 ± 135 nm, resulted in a low level of the luciferase protein production in the liver, spleen, lung, and heart. We also examined the necessity of DNA incorporation into the interior of RCR. First, we prepared empty RCRs with or without TC-Chol, and then mixed with 100 μg of pCMVL DNA and injected. The RCR containing TC-Chol formed aggregations immediately after the addition of DNA (TC-Chol-RCR, DNA), and showed no gene expression in any organs. In the case of the mixture of DNA and empty RCR without TC-Chol (RCR, DNA), gene expression in each organ was almost same as that of the mice injected with only naked DNA (isotonic). These results indicate that high level of gene expression requires the location of DNA in the interior of RCR.
Figure 4.
Gene expression in organs of mice following intraportal injection. CD1 mice were intraportally injected with 100 μg of plasmid pCMVL DNA in the various formulations. Two days after injection, mice were sacrificed and luciferase activity and protein concentration of tissue extracts were analyzed. Each column presents the mean from three animals ± SD.
Distribution and strength of gene expression after tail vein injection were also examined in mice (Fig. 5). Two days after injection of 100 μg DNA of pCMVL DNA-TC-Chol-RCR, gene expression was detected, being the highest in the liver and also in the lung and heart. The level of gene expression in the liver was significantly higher as compared with naked DNA, but was almost 100-fold lower than that of the mice injected intraportally (Fig. 4).
Figure 5.
Gene expression in organs of mice following intravenous injection. Mice were injected from the tail vein with naked pCMVL DNA or pCMVL DNA/TC-Chol-RCR at the dose of 100 μg of DNA. Two days after injection, mice were sacrificed and luciferase activity in the various organ extracts was measured. Each column presents the mean from three animals ± SD.
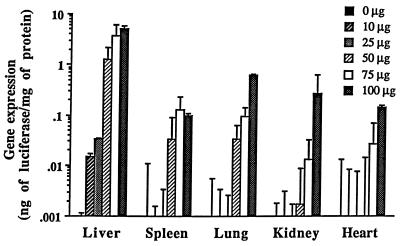
The effect of injected dose on the gene expression in mice following intraportal injection was examined (Fig. 6). Even by injection of 10 μg of DNA, DNA/TC-Chol-RCR induced a measurable level of gene expression in the liver. The level of gene expression in each organ increased with increasing injected dose. In the liver, the gene expression jumped up between 25 and 50 μg of DNA and was almost saturated with 100 μg of DNA.
Figure 6.
Dose dependence of gene expression by intraportal injection of DNA/TC-Chol-RCR. Various amounts of pCMVL/TC-Chol-RCR were intraportally injected into mice. Luciferase activity in the extracts from various organs was measured on day 2. Each column presents the mean from three animals ± SD.
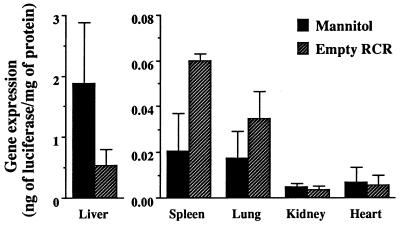
To reveal whether the gene expression in the liver results from uptake of DNA/TC-Chol-RCR particles themselves or not, the effect of empty RCR preinjection on the gene expression was investigated (Fig. 7). Gene expression in the liver by injection of 50 μg DNA of DNA/TC-Chol-RCR was reduced to about one-third by preinjection of empty RCR at the dose corresponding to 100 μg DNA in DNA/TC-Chol-RCR. However, the effect on the spleen and lung was opposite to that on the liver, i.e., 3-fold increase in the spleen and 2-fold increase in the lung. No effect was observed in the kidney and heart.
Figure 7.
Effect of preinjection of empty RCR on gene delivery by DNA/TC-Chol-RCR. Fifteen minutes after intraportal injection of 0.6 ml of isotonic mannitol with or without empty RCR (5.0 mg of total lipids), mice were intraportally injected with 50 μg DNA of pCMVL/TC-Chol-RCR dispersed in 0.6 ml of isotonic mannitol. Two days after injection, mice were sacrificed and luciferase activity in the various organ extracts was measured. Each column presents the mean from three animals ± SD.
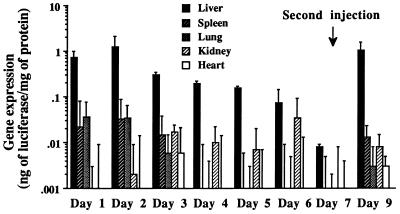
Fig. 8 shows the time course of gene expression at the dose of 50 μg of pCMVL DNA. High level of gene expression was observed at day 1 and day 2 in the liver, but there was a significant reduction at day 3 (Fig. 8). After that, gene expression in the liver gradually decreased and almost disappeared by day 7. Other organs except the kidney showed similar time course of change in gene expression. The kidney retained a constant level of gene expression from day 2 to day 6. Fig. 8 also shows the possibility of repetitive injection of DNA/TC-Chol-RCR. Two days after the second injection at day 7, comparable gene expression to that obtained after the first injection was observed in the liver.
Figure 8.
Time course of gene expression after the first and second injection. Mice were intraportally injected with 50 μg DNA of pCMVL/TC-Chol-RCR and sacrificed on the indicated day after injection. Some of them were injected again at day 7 by the same method as the first injection and were sacrificed 2 days after the second injection. Luciferase activity in the various organ extracts were measured. Each column presents the mean from three animals ± SD.
Histochemical Analysis of Gene Expression in the Liver.
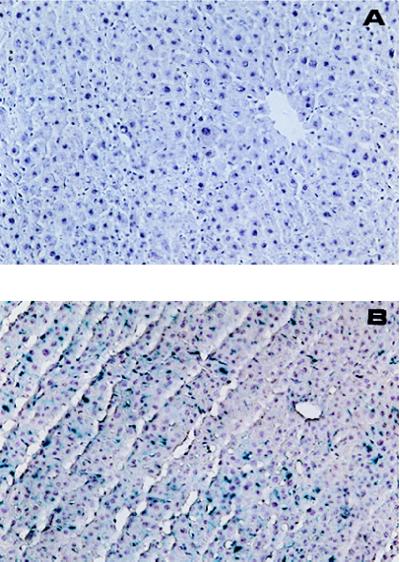
To elucidate the population and localization of cells in the liver that are transfected by intraportal injection of DNA/TC-Chol-RCR, pCMVLacZ DNA was complexed with TC-Chol, incorporated into RCR (pCMVLacZ DNA/TC-Chol-RCR) and injected into a mouse. Liver cells expressing β-gal activity were visualized by X-Gal staining (Fig. 9). Liver sections from a control (isotonic mannitol-injected) mouse or mouse injected with naked DNA did not show any change in color, while the color of liver sections from a mouse injected with DNA/TC-Chol-RCR turned to blue. By microscope examination, in the liver cryosections from a control mouse injected with empty RCR, no β-gal positive cell was observed (A). However, ≈10% of the cell population in the sections from the DNA/TC-Chol-RCR injected mouse was stained by X-Gal and these blue cells were found all over the section (B). It appeared at higher magnification that these cells were not only hepatocytes, which have a polygonal shape and round nuclei, but also nonparenchymal cells (photo not shown).
Figure 9.
X-Gal staining of the liver sections from a mouse injected with pCMVLacZ/TC-Chol-RCR. A mouse was intraportally injected with empty RCR (A) or 100 μg DNA of pCMVLacZ/TC-Chol-RCR (B). Two days after injection, liver cryosections of 10 μm in thickness from the mouse were stained with X-Gal for 24 h. After counter staining, the sections were examined under a light microscopy (Diaphot, Nikon) at an original magnification of ×100.
Secretion of Therapeutic Protein into the Blood Circulation.
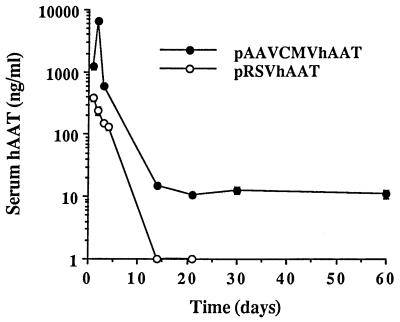
Secretion of gene products into the blood circulation and its time course were examined using two different plasmids containing therapeutic hAAT gene driven by different promoters (Fig. 10). In both cases, significant amount of hAAT was produced and secreted into the blood circulation. pAAVCMVhAAT, however, produced the highest serum level (6 μg/ml) of hAAT at day 2, which was 17-fold higher than the highest serum hAAT level achieved at day 1 after injection of the same dose of pRSVhAAT. Moreover, although the serum level of hAAT rapidly decreased by 7 days after injection of both DNA, detectable concentration of hAAT was constantly observed until 60 days after injection of pAAVCMVhAAT.
Figure 10.
Delivery and expression of hAAT gene by DNA/TC-Chol-RCR. Mice were intraportally injected with 50 μg DNA of pRSVhAAT/TC-Chol-RCR or pAAVCMVhAAT/TC-Chol-RCR. At the indicated days after injection, the blood was collected from the tail vein of mice and serum concentration of hAAT was determined by ELISA. Each point presents the mean from three animals ± SD.
DISCUSSION
One of the most promising nonviral gene delivery systems is cationic liposomes (37, 38, 40–46). In a variety of cationic lipids synthesized, DC-Chol (37), N-[1-(2, 3-dimyristyloxy)propyl]-N, N-dimethyl-N-(2-hydroxyethyl)ammonium bromide (46) and N,N,N-trimethyl-2-bis[(1-oxo-9-octadecenyl)oxy]-(Z, Z)-1-propanaminium methyl sulfate (47) have been used in human clinical trials by local injection. However, the DNA/liposome complex is inactive or less effective in the presence of serum probably due to dissociation of DNA from the complex or formation of large aggregates. Our novel formulation seems to offer serum resistance that is critical for in vivo systemic application. DNA/TC-Chol complex becomes hydrophobic when the net charge of the complex is neutralized, and a portion of those complexes is apparently solubilized into the oil core of RCR as shown in Fig. 3. This is supported by the fact that more than half of DNA in DNA/TC-Chol-RCR were not dissociated by the addition of high concentration of NaCl (Fig. 2C) that could readily dissociate DNA from its cationic lipid complex in a monophase even at low concentrations (31). It is also supported by the fact that most DNA in RCR was protected from DNase I digestion (Fig. 2E). Therefore, it is unlikely to form aggregates in the presence of serum proteins and DNA could be protected from digestion by serum nucleases by the lipid in the RCR.
One of the major obstacles to using particulate carriers for systemic gene delivery is a limited extravasation of the particles through the vascular wall. The liver is a favorable target organ for this kind of carriers because the sinusoidal wall of the liver lacks a basement membrane and possesses a fenestrated (≈100 nm) endothelium (48). Therefore, the size of the particle is an important factor for efficient delivery (49). By extrusion through polycarbonate membranes, our formulation became small enough to pass through the fenestrae and get into the space of Disse that is in direct contact with the hepatocytes. This was clearly shown by the result that gene expression in the liver by injection of extruded DNA/TC-Chol-RCR was almost 500-fold higher than that of unextruded one (Fig. 4). After extravasation of the particles with the help of high injection pressure, the particles could be retained in the space of Disse and directly taken up by the hepatocytes, or free DNA could be taken up following its release from the particles. The former is more likely to occur, because gene expression in the liver was almost saturated with a high dose (Fig. 6) and was inhibited competitively by preinjection of empty RCR (Fig. 7).
Intraportal injection seems to be less invasive than reimplantation of hepatocytes or partial hepatectomy, but intravenous injection is more applicable for not only liver but also other organs or tissues. DNA/TC-Chol-RCR led to a certain level of gene expression in the liver by single intravenous injection (Fig. 5). However, the level was significantly lower as compared with the intraportal injection (Fig. 4) and the expression was not limited in the liver. This is likely because we did not incorporate any apolipoproteins into the surface of the RCR in this work. Protein-free emulsions can pick up appropriate apolipoproteins in the systemic circulation (32) and still have the possibility to target to the receptor-bearing cells. However, their colloidal stability may not be as good as the ones containing apolipoproteins. By the addition of apolipoprotein E, RCR should be stabilized and its efficiency and targetability could be further increased because chylomicron remnants are efficiently taken up by hepatocytes via specific recognition of apolipoprotein E (24, 25). Moreover, chylomicron is only one type of lipoprotein that transports lipids and cholesterol. Other lipoproteins that have been reconstituted for drug delivery include low density lipoprotein (50) and high density lipoprotein (51). It is conceivable that they could also be used for gene delivery to cells that express a specific receptor. The low density lipoprotein receptor is wide spread; many cell types contain high levels of receptors including cancer cells, hepatocytes, fibroblasts, endothelial cells, etc. It may also be possible to prepare emulsions with conjugated glycoproteins, antibodies and other ligands. Thus, gene delivery system using emulsions should not be limited to the liver.
It has been reported that multiple direct injection of plasmid DNA into liver led to a significant gene expression, but only for cells surrounding the injection site and not for more distant tissue (52). As apparent by the X-Gal staining (Fig. 9), ≈10% of the liver cells were transfected by intraportal injection of DNA/TC-Chol-RCR. While the transfected cells were not only hepatocytes (Fig. 9), the total amount of transgene products produced in the liver exceeded 800 ng per liver (Fig. 4).
Long-term expression is usually required for successful gene therapy of hereditary diseases. In this regard, retroviral vectors exceed all other vectors, but their application for hepatic gene therapy is obliged to use an invasive method such as reimplantation or partial hepatectomy. Adenoviral vectors, that offer an excellent transfection efficiency to the liver in vivo, usually result in no significant gene expression after the second injection (14, 53). Recently, new types of adenoviral vector (54) and oral tolerization (55) have been developed to reduce the immunogenicity. Our novel vector also led to only a transient gene expression (Figs. 8 and 10) but are probably able to be injected repetitively because it contains neither protein nor peptide that acts as an antigen. Recovery of gene expression in the liver was achieved following the second injection of DNA/TC-Chol-RCR (Fig. 7). It might be possible to maintain a high level of gene expression by repetitive injection of RCR formulation by using a catheter method that has been established for multiple portal vein infusion (53).
The strength and extension of gene expression can also be controlled by using transcriptional regulatory elements. The transcriptional activity of CMV promoter was significantly higher than that of RSV promoter, whereas the serum level of hAAT was still subtherapeutic at 0.5% of normal at its peak expression time (20, 52). It has been reported that AAV inverted terminal repeat, that is necessary for the site specific integration of AAV to the host genome, seems to allow long-term persistence of foreign DNA (28, 56, 57). As shown in Fig. 8, integration of the hAAT gene into the mouse hepatocyte genome probably did not occur because the serum hAAT concentration rapidly decreased and became <1% of the maximum concentration within a week. The integration of gene through inverted terminal repeat may require another factors. Another possible way to prolong gene expression is to use immunosuppressive reagents. It has been reported that gene expression by naked DNA injection was increased and/or prolonged by coinjection of cyclosporine A or dexamethasone (39). The only vector that can achieve high enough serum hAAT concentration is an adenoviral vector (27). However, the same adenoviral vector used by another investigator produced serum hAAT levels similar to what is reported here (58). Although the efficiency of transgene expression should depend on the structure of the plasmid DNA including the selection of promoter and/or enhancer, a relatively high gene transfer activity of our novel vector is likely to find therapeutic uses in treating diseases such as hepatoma, viral hepatitis and many other liver disorders.
Acknowledgments
We thank Dr. H. Deshmukh for synthesis of TC-Chol, to Dr. M. Kay for pRSVhAAT, and to Dr. M. During for pAAVlac.26. This work was supported by National Institutes of Health Grants CA59327, CA64654, CA71731, and DK 44935.
ABBREVIATIONS
- RCR
reconstituted chylomicron remnants
- hAAT
human α1-antitrypsin
- Chol
cholesterol
- X-Gal
5-bromo-4-chloro-3-indolyl-β-d-galactoside
- β-gal
β-galactosidase
- DC-Chol
3-β-[N-(N′,N′-dimethyl-ethane)carbamoyl]cholesterol
- TC-Chol
3-β-[N-(N′,N′,N′-trimethylethane)carbamoyl]-cholesterol
- DNA/TC-Chol-RCR
DNA/TC-Chol complex-incorporated reconstituted chylomicron remnants
- RSV
Rous sarcoma virus
- AAV
adeno-associated virus
References
- 1.Wilson J M. Nat Genet. 1996;12:232–233. doi: 10.1038/ng0396-232. [DOI] [PubMed] [Google Scholar]
- 2.Wilson J M, Grossman M, Wu C H, Chowdhury N R, Wu G Y, Chowdhury J R. J Biol Chem. 1992;267:963–967. [PubMed] [Google Scholar]
- 3.Ferkol T, Lindberg G L, Ghen J, Perales J C, Crawford D R, Ratnoff O D, Hanson R W. FASEB J. 1993;7:1081–1091. doi: 10.1096/fasebj.7.11.8370479. [DOI] [PubMed] [Google Scholar]
- 4.Kay M A, Baley P, Rothemberg S, Leland F, Fleming L, Ponder K P, Liu T, Finegold M, Darlington G, Pokorny W, Woo S L C. Proc Natl Acad Sci USA. 1992;89:89–93. doi: 10.1073/pnas.89.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao J, Jin L, Chen L-M, Chen V C, Chao L. Hum Gene Ther. 1996;7:901–911. doi: 10.1089/hum.1996.7.8-901. [DOI] [PubMed] [Google Scholar]
- 6.Miller D G, Adam M A, Miller A D. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossman M, Raper S E, Kozarsky K, Stein E A, Engelhardt J F, Muller D, Lupien P J, Wilson J M. Nat Genet. 1994;6:335–341. doi: 10.1038/ng0494-335. [DOI] [PubMed] [Google Scholar]
- 8.Ledley F, Darlington G, Hahn T, Woo S. Proc Natl Acad Sci USA. 1987;84:5335–5339. doi: 10.1073/pnas.84.15.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferry N, Duplessis O, Houssin D, Danos O, Heard J-L. Proc Natl Acad Sci USA. 1991;88:8377–8381. doi: 10.1073/pnas.88.19.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kay M A, Li Q T, Liu T J, Leland F, Finegold M, Woo S L C. Hum Gene Ther. 1992;3:641–647. doi: 10.1089/hum.1992.3.6-641. [DOI] [PubMed] [Google Scholar]
- 11.Rosefeld M A, Siegfried W, Yoshimura K, Yoneyama K, Fukayama M, Stier L E, Paakko P K, Gilardi P, Stratford-Perricaudet L D, Perricaudet M, Jallat S, Pavirani A, Lecocq J-P, Crystal R G. Science. 1991;252:431–434. doi: 10.1126/science.2017680. [DOI] [PubMed] [Google Scholar]
- 12.Li Q T, Kay M A, Finegold M, Stratford-Perricaudet L D, Woo S L C. Hum Gene Ther. 1993;4:403–409. doi: 10.1089/hum.1993.4.4-403. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Ertl H C J, Wilson J M. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 14.Barr D, Tubb J, Ferguson D, Lieber A, Perkins A J, Kay M A. Gene Ther. 1995;2:151–155. [PubMed] [Google Scholar]
- 15.Malone R W, Hickman M A, Lehmann-Bruinsma K, Sih T R, Walzem R, Carlson D M, Powell J S. J Biol Chem. 1994;269:29903–29907. [PubMed] [Google Scholar]
- 16.Hickman M A, Malone R W, Lehmann-Bruinsma K, Sih T R, Knoell D, Szoka F C, Walzem R, Carlson D M, Powell J S. Hum Gene Ther. 1994;5:1477–1483. doi: 10.1089/hum.1994.5.12-1477. [DOI] [PubMed] [Google Scholar]
- 17.Wu G Y, Wu C H. J Biol Chem. 1988;263:14621–14624. [PubMed] [Google Scholar]
- 18.Wu G Y, Wilson J M, Shalaby F, Grossman M, Shafritz D A, Wu C H. J Biol Chem. 1991;266:14338–14342. [PubMed] [Google Scholar]
- 19.Kaneda A, Iwai K, Uchida T. Science. 1989;243:375–378. doi: 10.1126/science.2911748. [DOI] [PubMed] [Google Scholar]
- 20.Alino S F, Crespo J, Bobadilla M, Lejarreta M, Blaya C, Crespo A. Biochem Biophys Res Commun. 1994;204:1023–1030. doi: 10.1006/bbrc.1994.2565. [DOI] [PubMed] [Google Scholar]
- 21.Hara T, Aramaki Y, Takada S, Koike K, Tsuchiya S. Gene Ther. 1995;2:784–788. [PubMed] [Google Scholar]
- 22.Havel R J, Eder H A, Bragdon J H. J Clin Chem. 1955;34:1345–1353. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Windler E, Chao Y-S, Havel R J. J Biol Chem. 1980;255:8303–8307. [PubMed] [Google Scholar]
- 24.Huettinger M, Retzek H, Eder M, Goldenberg H. Clin Biochem. 1988;21:87–92. doi: 10.1016/s0009-9120(88)80093-6. [DOI] [PubMed] [Google Scholar]
- 25.Mahley R W, Hussain M M. Curr Opin Lipidol. 1991;2:170–176. [Google Scholar]
- 26.Rensen P C N, Van Dijk M C R, Havenaar E C, Bijsterbosch M K, Kruijt J K, Van Berkel T J C. Nat Med. 1995;1:221–225. doi: 10.1038/nm0395-221. [DOI] [PubMed] [Google Scholar]
- 27.Guo Z S, Wang L H, Eisensmith R C, Woo S L C. Gene Ther. 1996;3:802–810. [PubMed] [Google Scholar]
- 28.Kaplitt M G, Leon P, Xiao X, Samulski R J, Pfaff D W, O’Malley K L, During M J. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Gao X, Huang L. Biochem Biophys Res Commun. 1991;179:280–285. doi: 10.1016/0006-291x(91)91366-k. [DOI] [PubMed] [Google Scholar]
- 31.Reimer D L, Zhang Y-P, Kong S, Wheeler J J, Graham R W, Bally M B. Biochemistry. 1995;34:12877–12883. doi: 10.1021/bi00039a050. [DOI] [PubMed] [Google Scholar]
- 32.Redgrave T G, Maranhao R C. Biochim Biophys Acta. 1985;835:104–112. doi: 10.1016/0005-2760(85)90036-0. [DOI] [PubMed] [Google Scholar]
- 33.Kay M A, Graham F, Leland F, Woo S L C. Hepatology. 1995;21:815–819. [PubMed] [Google Scholar]
- 34.Ruysschaert J, El Ouahabi A, Willeaume V, Huez G, Fuks R, Vandenbranden M, Di Stefano P. Biochem Biophys Res Commun. 1994;203:1622–1628. doi: 10.1006/bbrc.1994.2372. [DOI] [PubMed] [Google Scholar]
- 35.Wong F M P, Reimer D L, Bally M B. Biochemistry. 1996;35:5756–5763. doi: 10.1021/bi952847r. [DOI] [PubMed] [Google Scholar]
- 36.Farhood H, Serbina N, Huang L. Biochim Biophys Acta. 1995;1235:289–295. doi: 10.1016/0005-2736(95)80016-9. [DOI] [PubMed] [Google Scholar]
- 37.Nabel G J, Nabel E G, Yang Z Y, Fox B A, Plautz G E, Gao X, Huang L, Shu S, Gordon D, Chang A E. Proc Natl Acad Sci USA. 1993;90:11307–11311. doi: 10.1073/pnas.90.23.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose J K, Buonocore L, Whitt M A. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 39.Budker V, Zhang G, Knechtle S, Wolff J A. Gene Ther. 1996;3:593–598. [PubMed] [Google Scholar]
- 40.Felgner P L, Gadek P R, Holm M, Roman R, Chan H W, Wenz M, Northrop J P, Ringold G M, Danielsen M. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koshizaka T, Hayashi Y, Yagi K. J Clin Biochem Nutr. 1989;7:185–192. [Google Scholar]
- 42.Ito A, Miyazoe R, Mitoma J, Akao T, Osaki T, Kunitake T. Biochem Int. 1990;22:235–241. [PubMed] [Google Scholar]
- 43.Leventis R, Silvius J R. Biochim Biophys Acta. 1990;1023:124–132. doi: 10.1016/0005-2736(90)90017-i. [DOI] [PubMed] [Google Scholar]
- 44.Felgner J H, Kumar R, Sridhar C N, Wheeler C J, Tsai Y J, Border R, Ramsey P, Martin M, Felgner P L. J Biol Chem. 1994;269:2550–2561. [PubMed] [Google Scholar]
- 45.Staedel C, Remy J, Hua Z, Broker T R, Chow L T, Behr J. J Invest Dermatol. 1994;102:768–772. doi: 10.1111/1523-1747.ep12377673. [DOI] [PubMed] [Google Scholar]
- 46.Nabel G J, Chang A E, Nabel E G, Plautz G E, Ensminger W, Fox B A, Felgner P, Shu S, Cho K. Hum Gene Ther. 1994;5:57–77. doi: 10.1089/hum.1994.5.1-57. [DOI] [PubMed] [Google Scholar]
- 47.Porteous D J, Dorin J R, McLachlan G, Davidson-Smith H, Davidson H, Stevenson B J, Carothers A D, Wallace W A H, Moralee S, Hoenes C, Kallmeyer G, Michaelis U, Naujoks K, Ho L-P, Samways J M, Imrie M, Greening A P, Innes J A. Gene Ther. 1997;4:210–218. doi: 10.1038/sj.gt.3300390. [DOI] [PubMed] [Google Scholar]
- 48.Nopanitaya W, Lamb J, Grisham J, Carson J. Br J Exp Pathol. 1976;57:604–609. [PMC free article] [PubMed] [Google Scholar]
- 49.Hara T, Aramaki Y, Tsuchiya S, Hosoi K, Okada A. Biopharm Drug Dispos. 1987;8:327–339. doi: 10.1002/bdd.2510080404. [DOI] [PubMed] [Google Scholar]
- 50.Bijsterbosch M K, Ziere G J, Van Berkel T J C. Mol Pharmacol. 1989;36:484–489. [PubMed] [Google Scholar]
- 51.Bijsterbosch M K, Schouten D, Van Berkel T J C. Biochemistry. 1994;33:14073–14080. doi: 10.1021/bi00251a016. [DOI] [PubMed] [Google Scholar]
- 52.Okuyama T, Huber R M, Bowling W, Pearline R, Kennedy S C, Flye M W, Ponder K P. Hum Gene Ther. 1996;7:637–645. doi: 10.1089/hum.1996.7.5-637. [DOI] [PubMed] [Google Scholar]
- 53.Marie-Jeanne T F D, Peeters V, Lieber A, Perkins J, Kay M A. BioTechniques. 1996;20:278–285. doi: 10.2144/96202rr05. [DOI] [PubMed] [Google Scholar]
- 54.Ilan Y, Droguett G, Chowdhury N R, Li Y, Sengupta K, Thummala N R, Davidson A, Chowdhury J R, Horwitz M S. Proc Natl Acad Sci USA. 1997;94:2587–2592. doi: 10.1073/pnas.94.6.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ilan Y, Parkash R, Davidson A, Jona V, Droguett G, Horwitz M S, Chowdhury N R, Chowdhury J R. J Clin Invest. 1997;99:1098–1106. doi: 10.1172/JCI119238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKeon C, Samulski R J. Hum Gene Ther. 1996;7:1615–1619. doi: 10.1089/hum.1996.7.13-1615. [DOI] [PubMed] [Google Scholar]
- 57.Koeberl D D, Alexander I E, Halbert C L, Russell D W, Miller A D. Proc Natl Acad Sci USA. 1997;94:1426–1431. doi: 10.1073/pnas.94.4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marie-Jeanne T F D, Peeters V, Patijn G A, Lieber A, Meuse L, Kay M A. Hum Gene Ther. 1996;7:1693–1699. doi: 10.1089/hum.1996.7.14-1693. [DOI] [PubMed] [Google Scholar]