Abstract
Human placentas are sources of cytokines, hormones and other substances that program receptive cells. One of these substances is HLA-G, which influences the functioning of both leukocytes and endothelial cells. In this study we investigated the possibility that these and/or other types of cells in extraembryonic fetal tissues might respond to HLA-G by interacting with one or another of the leukocyte immunoglobulin-like receptors (LILR). LILRB1 is expressed by most leukocytes and LILRB2 is expressed primarily by monocytes, macrophages and dendritic cells. Analysis of term placentas by immunohistochemistry and Real Time PCR demonstrated that LILRB1 and LILRB2 protein and specific messages are produced in the mesenchyme of term villous placenta but are differently localized. LILRB1 was abundant in stromal cells and LILRB2 was prominent perivascularly. Neither receptor was identified in trophoblast. Further investigation using double label immunofluorescence indicated that placental vascular smooth muscle but not endothelia exhibit LILRB2. Term umbilical cord exhibited the same LILRB2 patterns as term placenta. Samples obtained by laser capture dissection of vascular smooth muscle in umbilical cords demonstrated LILRB2 mRNA, and double labeling immunofluorescence showed that cord vascular smooth muscle but not endothelium exhibited LILRB2 protein. The presence of LILRB1 in placental stromal cells and LILRB2 in vascular smooth muscle strongly suggest that HLA-G has novel functions in these tissues that could include regulation of placental immunity as well as development and function of the extraembryonic vasculature.
1.0 Introduction
HLA-G, a major histocompatibility complex (MHC) class Ib molecule with multiple protein isoforms, is strongly expressed in cytotrophoblast (CTB) cells invading the decidualized endometrium (decidua) of the human uterus [1-3] as well as the villous CTB cells within the defined placenta [4]. Studies of HLA-G functions, which have been recently reviewed [5-8], provide substantial evidence for the ability of several HLA-G isoforms to drive immune cells into suppressive profiles. Binding to receptors such as the leukocyte immunoglobulin-like receptors (LILR)B1 (CD85j, ILT2, LIR-1), and LILRB2 (CD85d, ILT4, LIR-2), which interfere with immune cell activation, have been implicated in this process. Highly purified macrophages selected from maternal decidua are positive for both LILRB1 and LILRB2 [9]. Collectively, these findings have led to the postulate that HLA-G assists in developing the pregnant uterus as an immune privileged site
Regarding receptors, LILRB1 are found on many types of leukocytes whereas LILRB2 are generally believed to be restricted to the myeloid family, which includes monocytes, macrophages and dendritic cells [10,11]. Although few studies have addressed the question of cellular expression of LILRB in non-leukocytes, LILRB2 was recently reported by Manavalan et al. [12] on endothelial cells in biopsies from successfully transplanted human hearts. More recently, one of the soluble isoforms of HLA-G (HLA-G5, also known as soluble HLA-G1) was reported to target endothelial cells via CD160 [13-15]. Binding resulted in inhibition of certain functions, including the ability of the endothelial cells to proliferate and invade.
Because of the observations showing that HLA-G may not be restricted to modulation of immune cells but may instead also be involved in programming other cell lineages we designed the present study to determine whether cells in the placenta or umbilical cord might be targeted by HLA-G via binding to LILRB1 or LILRB2. The results indicate that in the villous placenta, LILRB1 expression is restricted to stromal cells, which are composed primarily of fibroblasts and macrophages. Unexpectedly, LILRB2 was poorly defined in villous placenta stromal cells but was highly expressed in vascular smooth muscle in both the placenta and the umbilical cord. The novel patterns of expression of these two receptors suggest that binding of HLA-G might drive specific immunological and vascular functions in human extraembryonic tissues.
2.0 Materials and methods
2.1 Tissues and cells
Tissue acquisitions were done in accordance with protocols approved by the institutional Human Subjects Committee. Term placentas and umbilical cords from normal cesarean deliveries performed to avoid fetal distress were used for the experiments. For immunohistologic studies, samples of villous placenta and umbilical cord were dissected manually into approximately 1 cm3 samples, then were embedded in tissue freezing medium and stored at -80°C until sectioned by cryostat (5 μm sections). For initial Real Time PCR experiments,samples were taken from term villous placenta, term umbilical cord, and manually dissected chorion membrane (n, 1 each). Three different placentas were used to prepare highly purified villous mesenchymal cells (MC) and villous cytotrophoblast (CTB) cells that were appropriate for Real Time PCR [16]. In brief, villous placenta was enzyme digested, cells were separated by gradient centrifugation, were washed and then were placed over a column to which W6/32, an anti-HLA class I monoclonal antibody (mAb) [17] was bound. HLA class I MC that had bound to the column were released and washed for use in Real Time PCR experiments and villous CTB cells in the flow-through from the column were treated similiarly. For laser capture microdissection, described in detail below, umbilical cords were fixed in 4% paraformaldehyde, embedded in paraffin, and stored at 4°C until sectioned. Captured mRNA in venous smooth muscle cells was evaluated by Real Time PCR.
2.2 Real Time PCR
Total RNA was isolated from a term placenta, a term umbilical cord and chorion membrane CTB cells (n, 1 each), all from normal term deliveries, as well as the JEG-3 human trophoblast cell line (American Type Culture Collection, Manassas, VA) and HUVEC, human venous endothelial cells (ATCC) (n, 1 sample each). Total RNA was also isolated from separated MC and CTB (n, 3 placentas, matching samples). These RNAs were obtained using TRIzol® reagent (Invitrogen) and were used to synthesize cDNA as previously described [18]. RNA from human umbilical vein smooth muscle cells isolated by laser capture microscopy as described below was purified using a Qiagen RNeasy Micro kit (Qiagen, Valencia, CA) and used for cDNA synthesis as described above. The levels of LILRB1 and LILRB2 messages in all of these samples were assessed by real time PCR using the TaqMan® Gene Expression Assay system (Applied Biosystems, Foster City, CA). The relative quantitation method was used to estimate the levels of LILRB in each sample, with glyceraldehyde-6-phosphate dehydrogenase (GAPDH) mRNA serving as the endogenous normalization control and peripheral blood mononuclear cell (PBMC) cDNA serving as the calibrator. Real Time PCR was performed in triplicate in 96-well MicroAmp™ optical reaction plates. Each 25 μl reaction contained 1× 6-carboxylfluorescein (FAM)-labeled LILRB1 or LILRB2 TaqMan® Gene Expression Assay or GAPDH endogenous control, TaqMan® Universal PCR Master Mix and cDNA diluted 1:25 in water. The PCR reaction was performed using a 7500 Real-Time PCR System with an initial AmpErase uracyl-N-glycosylase (UNG) activation step conducted at 50°C for 2 min followed by a10 min incubation at 95°C to activate the Amplitaq Gold® polymerase enzyme. This was followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for one min. The data were collected and analyzed using the ABI Sequence Detection Software, version 1.3.1 (Applied Biosystems).
2.3 Studies using immunohistochemistry
Five micrometer serial sections were cut from frozen third trimester placentas, fixed in acetone, and evaluated by immunohistochemical staining using mouse mAbs to LILRB1 (IgG1, clone M401) and LILRB2 (IgG1, clone M422), both provided by Amgen Inc., Thousand Oaks, CA. Mouse IgG1 (BD Pharmingen, San Jose, CA) was used as a negative control. Binding of primary antibodies was detected using biotinylated horse anti-mouse IgG (Vector, Burlingame, CA), followed by a streptavidin peroxidase conjugate (Zymed, South San Francisco, CA) and the 3-amino-9-ethylcarbozole in N,N-dimethylformamide (AEC) color development substrate (Zymed). Tissue sections were counterstained with Mayer’s hematoxylin and coverslipped for evaluation by light microscopy.
2.4 Double label immunofluorescence
Five micron sections were cut from frozen tissue blocks, dried then fixed in -20°C acetone for 10 min. The tissues were then washed for 5 min in phosphate buffered saline (PBS) with 0.2% Triton-X-100 (Sigma, Saint Louis, MO) containing 2% serum from the species in which secondary antibodies were generated. Blocking solution (PBS-Triton-X-100, 10% serum of secondary antibody, 20% human serum for FcR blocking) was applied to each sample for 1 hour (hr) at room temperature. Samples were washed 3 times for 5 min each and then evaluated by immunofluorescence staining. Primary mAb used included: anti-CD31 (R & D Systems, Minneapolis, MN, 10 μg/ml); anti-LILRB2 (provided by Amgen Inc., used at 10 μg/ml), and anti-LILRB2 (R & D Systems, Minneapolis, MN, used at 20 μg/ml); goat anti-smooth muscle actin (Everest Biotech, Oxfordshire, UK, 10 μg/ml). Isotype specific mouse IgG1 (BD Biosciences Pharmingen, San Jose, CA) was used as a negative control for the Amgen mAb, mouse IgG2a (BD Biosciences Pharmingen, San Jose, CA) was used as a negative control for the R & D mAb and goat IgG (Sigma) was used to control for the anti-smooth muscle actin. All control reagent concentrations were identical to the concentrations of the primary antibodies. Tissue sections were incubated with anti-LILRB2 or mouse IgG1 overnight at 4°C then washed with PBS. Double staining was achieved by applying sheep anti-CD31 or goat anti-smooth muscle actin (or appropriate control Ig) to the tissue section for 1 hr at 37° C followed by a second PBS wash. Sections were then incubated with fluorescent tagged secondary antibodies for 30 min at room temperature. Anti-CD31 mAb was detected using Alexa Fluor®-594-conjugated donkey anti-sheep IgG (Molecular Probes), anti-smooth muscle actin was detected using Alexa Fluor®-594-conjugated donkey anti-goat IgG and binding of the anti-LILRB mAbs was detected using Alexa Fluor®-488-conjugated donkey anti-mouse IgG (Molecular Probes, Eugene, OR). Sections were mounted using Prolong® Gold antifade reagent containing the nuclear stain DAPI (Molecular Probes). Fluorescent digital images were obtained using an Olympus IX71 inverted microscope equipped with a mercury arch UV lamp. UV light directed through an IX2-GS rotating filter turret allows each stain to be viewed according to its specific wavelength: DAPI stain (scan range: 320.0 nm to 520.0 nm), Alexa Fluor®-488 stain (FITC, 450.0 nm to 650.0 nm), and for the Alexa Fluor®-594 stain (TRITC, 500.0 nm to 700.0 nm). Images were taken using a Retiga 4000R digital camera by Q-imaging. Digital images were then uploaded using QCapture Pro software. Double stained images were obtained by superimposing multiple pictures from identical locations using combinations of the three different filters.
2.5 Laser microdissection
Paraffin-embedded umbilical cord tissue was cut into 10 μm sections and floated onto nuclease-free, polyethylene naphthalate (PEN)-membrane glass slides (Leica, North Central Instruments, Inc. Plymouth, MN). The smooth muscle layer of the umbilical vein was dissected away from stroma and endothelial cells using the Leica Laser Microdissection Microscope (AS LMD, North Central Instruments, Inc.) with a 337 nm laser using the 20× objective. Dissected smooth muscle was collected in nuclease-free tubes, and RNA was harvested immediately using the Qiagen RNeasy® Micro kit (Qiagen) and stored at -80°C until analysis.
3.0 Results
3.1 Localization of LILRB1 and LILRB2 proteins in placentas by immunohistochemistry
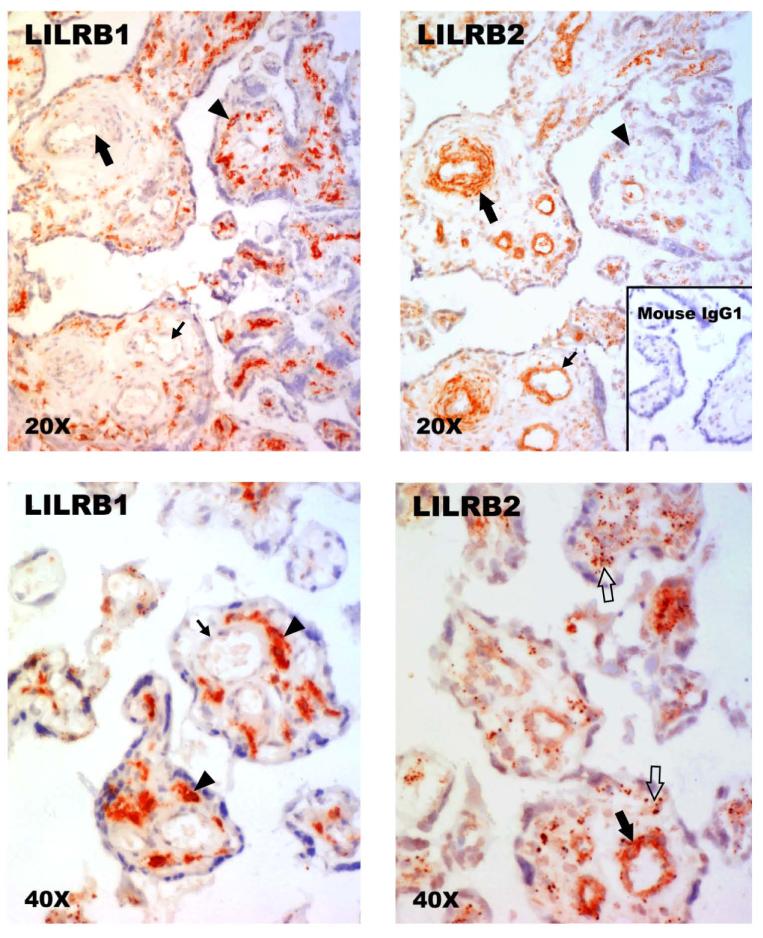
In the first set of experiments, we used standard immunohistochemistry with detection by a peroxidase signal to determine whether LILRB1 and/or LILRB2 proteins could be identified in human villous placental cells (n, 3 term placentas). Figure 1A shows that LILRB1 identified with the IgG1 mAb from Amgen was strongly localized to cells in the stroma of the villi. By contrast, as shown in Figure 1B, LILRB2, also identified with an IgG1 mAb from Amgen, was highly prominent in cells associated with fetal vessels, both arteries and veins. Syncytiotrophoblast and cells with the morphological characteristics of villous CTB cells were clearly negative with both anti-LILRB1 and anti-LILRB2 whereas immunostaining of endothelium with anti-LILRB2 was less well defined.
Fig. 1. Identification of LILRB1 and LILRB2 proteins in human term villous placentas by immunohistology.
(Upper left panel) Anti-LILRB1 (Amgen) fails to reveal signals in mesenchymal cells cuffing a small artery (large arrow) and a small vein (small arrow) but is strongly positive with pleiomorphic stromal cells (arrowhead). (Upper right panel) Anti-LILRB2 (Amgen) identifies positive cells cuffing a small artery (large arrow) and a small vein (small arrow) but does not yield readily identifiable signal in stromal cells (arrowhead). (Lower left panel) Anti-LILRB1 (Amgen) fails to reveal signals in cells cuffing a small vessel (large arrow) but is strongly positive with pleiomorphic stromal cells (arrowhead). (Lower right panel) Anti-LILRB2 (Amgen) identifies positive cells cuffing a small vessel (large filled arrow) and identifies diffuse, punctuate staining within the stroma (open arrows). Upper panels, original magnifications ×200; lower panels, original magnifications ×400. Positive stains in these experiments are identified by red signals. The slides were counterstained with Mayer’s hematoxylin.
The inset to Figure 1B illustrates lack of signals when the isotype-matched nonspecific IgG1 was substituted for the primary mAb provided by Amgen, Inc.
In Figure 1C a higher power magnification illustrates the intracellular staining pattern of anti-LILRB1 in an abundant population of pleomorphic stromal cells, which morphologically resembled fibroblasts and placental macrophages. In Figure 1D, staining of villous stromal cells by anti-LILRB2 was punctuate at the higher power magnification. This pattern did not permit assignment of LILRB2 positivity to any morphologically distinct stromal cell lineage.
Although the general patterns obtained using the anti-LILRB1 and anti-LILRB2 mAb from R & D, were the same as with the mAb from Amgen, staining was less distinct as background staining in placental villi was high (data not shown). No signal was observed when isotype-matched control IgG2a was substituted for the R & D mAbs nor was reactivity observed when tissue sections were treated only with secondary antibody.
The results of these experiments suggested that both LILRB1 and LILRB2 are present in human placental mesenchymal cells, but are differentially expressed according to cell type.
3.2 Double labeling immunofluorescence detects LILRB2 in vascular smooth muscle but not endothelia
Because distinctions between smooth muscle and endothelium in the display of LILRB2 remained unclear by light microscopy and endothelia had been reported as a site of HLA-G binding [12], we used double label immunofluorescence to investigate this question. Each of the double label immunofluorescence experiments was performed on a minimum of three different term placentas. Monoclonal antibodies against LILRB2, one from Amgen, Inc. and one from R & D were again used for each tissue.
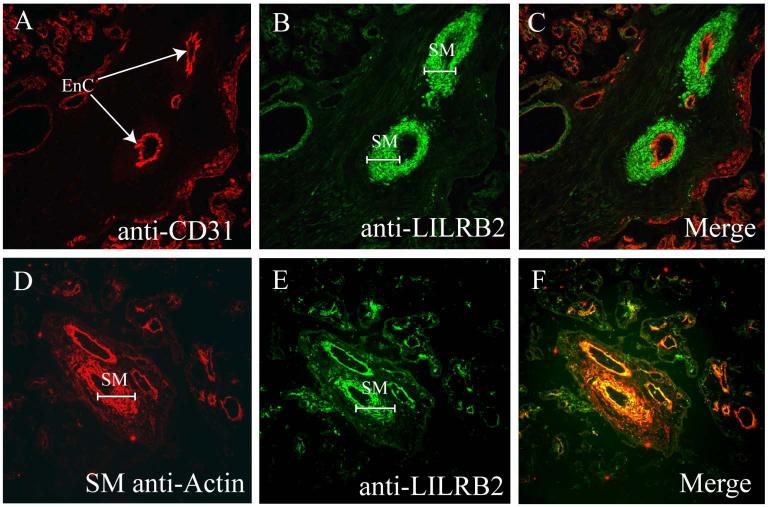
Figure 2A shows that vessels in the villi of a representative placenta were readily identified by the display of CD31 (red label) on villous endothelial cells. Arteries were identified by their deep cuffs of smooth muscle and constricted lumens. Veins were identified by their thinner smooth muscle cuffs and distended lumens. As illustrated in Figure 2B, cells cuffing arteries and veins were positive for LILRB2 (green label); syncytiotrophoblast was not. Merging of the two signals (Fig. 2C) showed that the anti-CD31 mAb (red) and the anti-LILRB2 (Amgen, green) mAb staining patterns did not overlap as no yellow signals were observed. Instead, areas of immunostaining were distinct from one another, indicating that smooth muscle but not endothelium exhibits the LILRB2 receptor for HLA-G. In Figures 2D (anti-smooth muscle actin, red), 2E (anti-LILRB2, Amgen, green) and 2F (merged images), the cuff of presumed smooth muscle was clearly identified as such using anti-smooth muscle actin. Note that overlapping signals yielding a bright yellow signal in Fig. 2F are concentrated in two or three layers of smooth muscle cells immediately beneath the endothelium.
Fig. 2. Identification of LILRB2 positive vascular smooth muscle cells and LILRB2 negative endothelial cells in term villous placenta by double label immunofluorescence.
(A) Anti-CD31. Arrows point to CD31+ arterial endothelial cells (EnC) in a term placental villus. (B) Anti-LILRB2. Brackets mark LILRB2+ smooth muscle (SM) cuffs around small arteries. (C) Merging of frames (A) and (B) reveals no double staining (yellow) in either endothelial cells or smooth muscle. (D) Anti-smooth muscle actin marks smooth muscle (SM) cuffs around villous placental vessels. (E) Anti-LILRB2. As in Panel (B), LILRB2 is prominent in smooth muscle (SM) cuffs around placental vessels. (F) Merging of frames (D) and (E) demonstrates bright yellow staining indicating that the vascular smooth muscle is LILRB2+. Images were captured at magnification ×200.
Controls consisting of isotype-specific mouse IgG (IgG1) substituted for the primary antibodies and controls where tissue sections received only secondary antibody were negative. As before, the anti-LILRB2 from R & D (data not shown) gave generally similar but less definitive results, with higher background staining in placental villi than the anti-LILRB2 mAb from Amgen.
Collectively, these results supplied evidence that LILRB2 is exhibited by vascular smooth muscle but not by vascular endothelium in the villous placenta.
3.3 LILRB1 and LILRB2 mRNAs are located in term villous placental mesenchymal cells
Since immunohistochemical studies do not identify the cells of origin of specific proteins and placental cell receptors may be soluble as well as cell-bound [19], studies of mRNAs in specific placental cells were conducted by Real Time PCR. Cells from three placentas were separated into highly purified, matching villous CTB cells and villous MC.
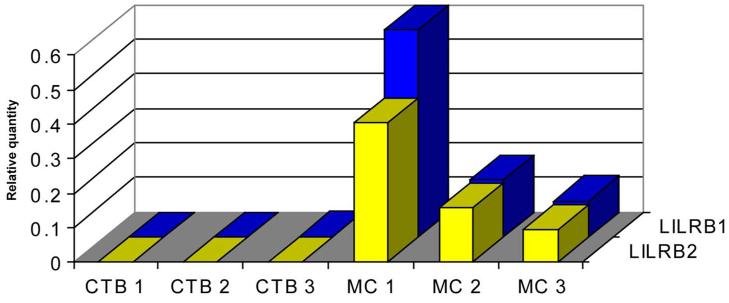
Figure 3 shows that, consistent with the immunohistochemical results, LILRB1 and LILRB2 mRNAs were absent from villous CTB cells but were abundant in all preparations of villous MC. In these experiments, PBMC cDNA served as the standard against which CTB and MC LILR mRNA were quantified.
Fig. 3. Use of Real Time PCR to identify LILRB1 and LILRB2 mRNA in highly purified term placental villous CTB cells and villous MC.
Three preparations of purified CTB cells from three different term placentas failed to demonstrate LILRB1 (blue bars) or LILRB2 (yellow bars) mRNA. The matching villous MC from the same three placentas all demonstrated LILRB1 and LILRB2 mRNA. Levels of LILRB mRNA in placental cells were quantified against levels in PBMC.
These data supplied convincing evidence that both LILRB1 and LILRB2 are synthesized in non-trophoblast in villous placenta.
3.4 LILRB2 mRNA is present in umbilical cord vascular smooth muscle
To investigate expression of LILRB2 in umbilical cord, we performed a preliminary Real Time PCR experiment. Samples of villous placenta (positive control), umbilical cord and chorion membrane as well as JEG-3 human trophoblastic tumor cells and HUVEC human venous endothelial cells were tested (one sample of each). Relative quantities of message were obtained by comparison with LILRB2 cDNA in PBMC.
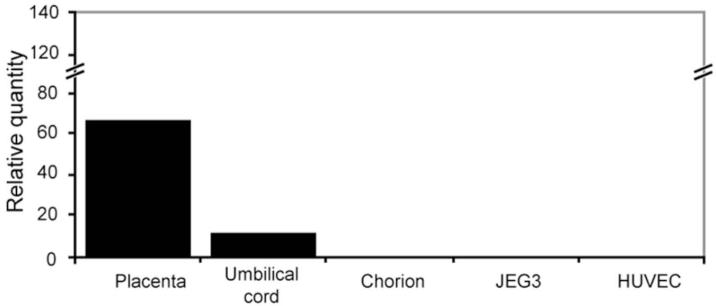
As shown in Figure 4, LILRB2 mRNA was present in term placenta (lane 1) and term umbilical cord (lane 2). By contrast, LILRB2 transcripts were not detected in chorionic CTB cells dissected from term amniochorion (lane 3), JEG-3 cells (lane 4) or HUVEC cells (lane 5). These results indicated that the LILRB2 gene is transcribed in umbilical cord as well as in fetal villous placenta and, consistent with other results given above, did not support a postulate for expression in either CTB or endothelial cells.
Fig. 4. Identification of LILRB2 mRNA in villous placenta, umbilical cord and cell lines by Real Time PCR.
Levels of LILRB2 mRNA were assessed against levels of the mRNA in PBMC. (Lane 1) term villous placenta; (Lane 2) term umbilical cord; (Lane 3) chorion membrane CTB cells; (Lane 4) JEG-3 trophoblast tumor cell line; (Lane 5) human umbilical vein endothelial cells (HUVEC). Placenta and umbilical cord contained detectable levels of LILRB2 mRNA whereas chorion, JEG-3 and HUVEC cells did not contain the specific messages.
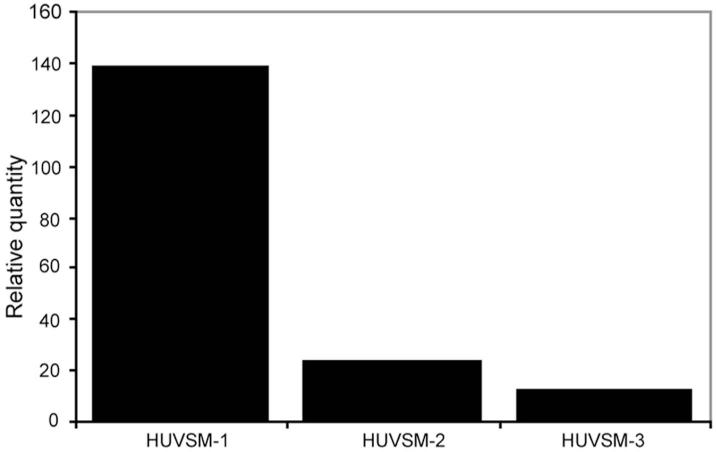
In a second set of Real Time PCR experiments, venous smooth muscle cells from three different term umbilical cords were dissected by laser microscopy and their RNA was tested for LILRB2 mRNA. Figure 5 shows that all three smooth muscle samples contained LILRB2 mRNA as determined by quantifying against LILRB2 cDNA in PBMC.
Fig. 5. Use of Real Time PCR to identify LILRB2 mRNA in 3 different samples of umbilical cord vascular smooth muscle obtained by laser microdissection.
Levels of LILRB2 mRNA in human umbilical vein smooth muscle (HUVSM) were compared with levels in PBMC to obtain relative quantity.
3.5 Double labeling immunofluorescence experiments identify LILRB2 in umbilical cord smooth muscle
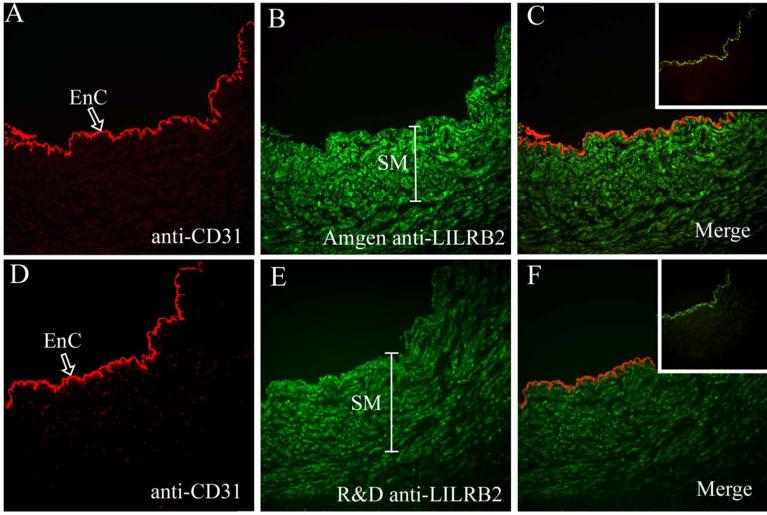
Double labeling immunofluorescence was again performed to determine with certainty that the LILRB2 mRNA identified in umbilical cords (n, 3 different cords) was translated into protein and to establish expression patterns in vascular smooth muscle and endothelia.
Figure 6A shows that endothelial cells in the umbilical cord were positive for CD31. In Figure 6B, vascular smooth muscle exhibited LILRB2 (Amgen). When the images were merged in Figure 6C, endothelial cells were clearly CD31+/LILRB2neg whereas vascular smooth muscle cells were CD31neg/LILRB2+. The same pattern was observed when additional sections of the same umbilical cord were immunostained with anti-CD31 and anti-LILRB2 (R & D) (Fig. 6D, 6E) and were merged to detect double expression (Fig. 6F). Inserts show that only background staining (green) on the inner elastic membrane (between the endothelium and smooth muscle layers) was observed when the specific mAbs were replaced with isotype-matched mouse Ig.
Fig. 6. Double label immunofluorescence identification of CD31 and LILRB2 in umbilical cord vein.
(A) Anti-CD31 recognizes venous endothelial cells (EnC, arrow). (B) Anti-LILRB2 (Amgen) identifies venous smooth muscle (SM, bracket), (C) Merging of the anti-CD31 and anti-LILRB2 (Amgen) demonstrates no bright yellow staining and therefore no overlap in expression of the two cellular markers. (D) Anti-CD31 recognizes venous endothelial cells, (EnC, arrow), (E) Anti-LILRB2 (R & D) identifies venous smooth muscle (SM, bracket), (F) Merging of the anti-CD31 and anti-LILRB2 (R & D) demonstrates no overlap in staining. Inserts to (C) and (F) are merged images of staining controls. Normal sheep Ig followed by the fluorochrome-labeled secondary antibody yielded no red signal but isotype-matched normal mouse Ig followed by the fluorochrome-labeled secondary antibody yielded a weak green signal on the inner elastic membrane located between the endothelium and smooth muscle layers. Original magnifications, ×200.
4.0 Discussion
The results of this study indicate potential binding of LILR by HLA-G, with major cellular recipients of targeting being villous stromal cells via LILRB1 and vascular smooth muscle cells via LILRB2. The experiments indicated that neither endothelial cells nor trophoblast in these fetal tissues will be signaled through LILRB1 or LILRB2.
The identification of LILRB1 in stromal cells was not unexpected as ∼15% of these cells are macrophages [20], a cell type that participates importantly in the innate immune response and is known to express LILRB1 [10]. Furthermore, testing of total RNA from purified villous placental MC showed that LILRB1-specific mRNA was present, indicating that the LILRB1 gene is transcribed and translated in villous placental mesenchyme. A previous study demonstrated that in the adjacent maternal decidua, macrophages contain LILRB1 mRNA and to express the receptor [9]. Thus, the data we have collected suggests that macrophages in both fetal extraembryonic tissue and in maternal decidua may be governed by binding of HLA class I molecules via this inhibitory receptor. However, more experiments on the placenta are required to eliminate the possibility that fibroblasts and/or stem-like cells that populate the placenta are not LILRB1 expressors as no mesenchymal cell separation experiments have as yet been done.
Unlike our finding of LILRB1 mRNA and protein in stromal cells, which were predictable, the three observations reported here on LILRB2 expression in human term placentas and umbilical cords, were surprising. The findings included observations of (1) diffuse, punctuate expression of LILRB2 in term villous placental stroma, (2) strong immunostaining of placental vascular smooth muscle as well as umbilical cord smooth muscle by anti-LILRB2 mAb, and (3) no detection of LILRB2 expression in endothelial cells. We anticipated that as with other mononuclear phagocytes [10], our immunostains would reveal placental stromal cell staining not unlike that we observed with anti-LILRB1. This was not the case. The punctuate staining we here report suggests the possibility that receptor is synthesized in these immature cells but is retained within granules rather than moved into position for receptivity. Alternatively, placental stromal cells may synthesize soluble LILRB2 for export. Both possibilities are amenable to experimental evaluation. Regarding the second observation, we observed that vascular smooth muscle, a cell type that has never been reported as exhibiting LILRB2, was uniformly stained with anti-LILRB2. Furthermore, Real Time PCR experiments showed that the receptor mRNA is transcribed within the cells. For these experiments we used laser-dissected smooth muscle from umbilical cord because of the convenience of working with the large vessels and because immunohistochemical stains revealed LILRB2 protein in the cord vascular smooth muscle as well as in placental vessel walls. Possibly, the smooth muscle cells respond to HLA-G via LILRB2 with alterations in their functions, but this remains entirely speculative at this time.
As for endothelial cells, our negative results are inconsistent with the report of LILRB2 on endothelial cells in transplanted hearts [12] as endothelial cells in placentas and umbilical cords were clearly negative for LILRB2. Further, we cannot confirm the results of the early immunohistochemical study by Blaschitz et al. [21], who reported HLA-G on endothelial cells in chorionic fetal vessels of first trimester placentas. Since it is unlikely that endothelial cells produce HLA-G, another explanation is needed. Possibly, endothelial cells in first trimester placentas, unlike those in term placental villi, express one or another of the HLA-G receptors, thus trapping HLA-G. CD160 [14] might serve this function as its expression in placentas is not known. Other potential explanations might include variability in levels of LILRB2 expression in the samples tested here and by Baschitz et al. as LILRB2 are subject to modification by interleukin-10 [22], which is reportedly a product of trophoblast cells [23].
The question arises as to whether either the LILRB1 or the LILRB2 exhibited in placentas do, in fact, bind HLA-G. Although definitive experiments remain to be done, several observations suggest that this may take place. Signaling stromal cells via LILRB1 appears particularly feasible as LILRB1 in placental villous stromal cells appears to be distributed similiarly to receptor display by normal blood monocytes and macrophages. As for signaling vascular smooth muscle via targeting LILRB2 on vascular smooth muscle, lack of precedent for this suggests that a novel pathway might be in place, as follows. LILRB2 demonstrates a preference for HLA-G over other HLA class I proteins, and receptor binding of HLA-G is increased by the absence of the HLA class I light chain, β2-microglobulin (β2m) [24]. HLA-G5, a soluble isoform of HLA-G, is synthesized by placental CTB cells as heavy chain homodimers free of β2m [4]. Thus, placental β2m-free HLA-G5 may be specifically designed to interact with placental LILRB2. Finally, in rhesus monkeys (Macaca mulatta), passive administration of antibodies to the macaque equivalent of HLA-G, Mamu-AG, alters both the growth and vascularization of the macaque placenta [25], which markedly resembles that of humans. Seval and coworkers [26] have recently reported that placental macrophages (Hofbauer cells) in early human placentas are statistically closely associated with cells undergoing vasculogenesis and angiogenesis. Collectively these experiments offer the intriguing possibility that HLA-G5 produced in placental CTB cells may target in a paracrine fashion to fetal LILRB2-expressing placental and umbilical cord cells and, secondarily, modify vascular development and function.
Although studies performed by ourselves and others indicate that stage of development (i.e., fetal vs. adult) may be one of the conditions influencing LILRB expression, it remains to be seen whether fetal extraembryonic tissue cell receptors have a specific preference for HLA-G over other HLA. Nor have we eliminated the possibility that cells in the placenta and cord which do not express LILRB display other HLA-G receptors, as mentioned above. Specific binding properties, signal transduction and functional implications remain to be investigated that may well require establishment of primary cultures of placental or umbilical cord mononuclear phagocytes and smooth muscle cells. These studies, which could assist investigations of HLA-G signal transduction via LILR, remain an area of active investigation.
Acknowledgements
This study was supported by National Institutes of Health grants HD39878 Project III and HD049480 to J.S.H. and the University of Kansas School of Medicine Biomedical Training Grant to R.H.M. The authors appreciate the provision of the mouse mAbs to LILRB by Amgen Inc., and thank S. Fernald of the Kansas University School of Medicine Imaging Core for technical assistance with figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ellis SA, Sargent IL, Redman CW, McMichael AJ. Evidence for a novel HLA antigen found on human extravillous trophoblast and a choriocarcinoma cell line. Immunol. 1986;59:595–601. [PMC free article] [PubMed] [Google Scholar]
- [2].Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–23. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- [3].Morales PJ, Pace JL, Platt JS, Phillips TA, Morgan K, Fazleabas AT, Hunt JS. Placental cell expression of HLA-G2 isoforms is limited to the invasive trophoblast phenotype. J Immunol. 2003;171:6215–24. doi: 10.4049/jimmunol.171.11.6215. [DOI] [PubMed] [Google Scholar]
- [4].Morales PJ, Pace JL, Platt JS, Langat DL, Hunt JS. Synthesis of β2-microglobulin-free, disulfide-linked HLA-G5 homodimers in human placental villous cytotrophoblast cells. Immunol. 2007;122:179–88. doi: 10.1111/j.1365-2567.2007.02623.x. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hunt JS, Petroff MG, McIntire RH, Ober C. HLA-G and immune tolerance in pregnancy. FASEB J. 2005;19:681–93. doi: 10.1096/fj.04-2078rev. [DOI] [PubMed] [Google Scholar]
- [6].Hunt JS. Stranger in a strange land. Immunol Rev. 2006;213:36–47. doi: 10.1111/j.1600-065X.2006.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pistoia V, Morandi F, Wang X, Ferrone S. Soluble HLA-G: are they clinically relevant? Semin Cancer Biol. 2007;17:469–79. doi: 10.1016/j.semcancer.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Favier B, LeMaoult J, Carosella ED. Functions of HLA-G in the immune system. Tissue Antigens. 2007;69(Suppl 1):150–52. doi: 10.1111/j.1399-0039.2006.763_6.x. [DOI] [PubMed] [Google Scholar]
- [9].Petroff MG, Sedlmayr P, Azzola D, Hunt JS. Decidual macrophages are potentially susceptible to inhibition by class Ia and class Ib HLA molecules. J Reprod Immunol. 2002;56:3–17. doi: 10.1016/s0165-0378(02)00024-4. [DOI] [PubMed] [Google Scholar]
- [10].Allan DS, McMichael AJ, Braud VM. The ILT family of leukocyte receptors. Immunobiol. 2000;202:34–41. doi: 10.1016/S0171-2985(00)80050-9. [DOI] [PubMed] [Google Scholar]
- [11].Shiroishi M, Tsumoto K, Amano K, Shirakihara Y, Colonna M, Braud VM, Allan DS, Makadzange A, Rowland-Jones S, Willcox B, Jones EY, van der Merwe PA, Kumagai I, Maenaka K. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc Natl Acad Sci USA. 2003;100:8856–61. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Manavalan JS, Kim-Schulze S, Scotto L, Naiyer AJ, Vlad G, Colombo PC, Marboe C, Mancini D, Cortesini R, Suciu-Foca N. Alloantigen specific CD8+CD28- FOXP3+ T suppressor cells induce ILT3+ ILT4+ tolerogenic endothelial cells, inhibiting alloreactivity. Int Immunol. 2004;16:1055–68. doi: 10.1093/intimm/dxh107. [DOI] [PubMed] [Google Scholar]
- [13].Le Bouteiller P, Pizzato N, Barakonyi A, Solier C. HLA-G, pre-eclampsia, immunity and vascular events. J Reprod Immunol. 2003;59:219–34. doi: 10.1016/s0165-0378(03)00049-4. [DOI] [PubMed] [Google Scholar]
- [14].Fons P, Chabot S, Cartwright JE, Lenfant F, L’Faqihi F, Giustiniani J, Herault JP, Gueguen G, Bono F, Savi P, Aguerre-Girr M, Fournel S, Malecaze F, Bensussan A, Plouet J, Le Bouteiller P. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood. 2006;108:2608–15. doi: 10.1182/blood-2005-12-019919. [DOI] [PubMed] [Google Scholar]
- [15].Le Bouteiller P, Fons P, Herault J-P, Bono F, Chabot S, Cartwright JE, Bensussan A. Soluble HLA-G and control of angiogenesis. J Reprod Immunol. 2007;76:17–22. doi: 10.1016/j.jri.2007.03.007. [DOI] [PubMed] [Google Scholar]
- [16].Langat DL, Wheaton DA, Platt JS, Sifers T, Hunt JS. BAFF (B cell activating factor) and APRIL (a proliferation-inducing ligand): potential signaling pathways in human placental villi. Amer J Pathol. doi: 10.2353/ajpath.2008.071139. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens: new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- [18].Pace JL, Morales PJ, Phillips TA, Hunt JS. Soares MJ, Hunt JS, editors. Analysis of the soluble isoforms of HLA-G. Placenta and Trophoblast Methods and Protocols. 2006;122:181–203. doi: 10.1385/1-59259-989-3:181. [DOI] [PubMed] [Google Scholar]
- [19].Lien E, Liabakk NB, Austgulen R. Detection of soluble receptors for tumor necrosis factor, interleukin-2 and interleukin-6 in retroplacental serum from normal pregnant women. Gynecol Obstet Invest. 1996;41:1–4. doi: 10.1159/000292024. [DOI] [PubMed] [Google Scholar]
- [20].Vince GS, Johnson PM. Immunobiology of human uteroplacental macrophages - friend and foe? Placenta. 1996:191–99. doi: 10.1016/s0143-4004(96)90038-7. [DOI] [PubMed] [Google Scholar]
- [21].Blaschitz A, Lenfant F, Mallet V, Hartmann M, Bensussan A, Geraghty DE, Le Bouteiller P, Dohr G. Endothelial cells in chorionic fetal vessels of first trimester placenta express HLA-G. Eur J Immunol. 1997;27:3380–88. doi: 10.1002/eji.1830271237. [DOI] [PubMed] [Google Scholar]
- [22].Gleissner CA, Zastrow A, Klingenberg R, Kluger MS, Konstandin M, Celik S, Haemmerling S, Shankar V, Giese T, Katus HA, Dengler TJ. IL-10 inhibits endothelium-dependent T cell costimulation by upregulation of ILT3/4 in human vascular endothelial cells. Eur J Immunol. 2007;37:177–92. doi: 10.1002/eji.200636498. [DOI] [PubMed] [Google Scholar]
- [23].Roth I, Corry DB, Locksley RM, Abrams JS, Litton MJ, Fisher SJ. Human placental cytotrophoblasts produce the immunosuppressive cytokine interleukin-10. J Exp Med. 1996;184:536–48. doi: 10.1084/jem.184.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, Kato K, Kohda D, Maenaka K. Structural basis for recognition of the non-classical MHC molecule HLA-G by the leukocyte Ig-like receptor B2. Proc Natl Acad Sci USA. 2006;103:16412–7. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bondarenko GI, Burleigh DW, Durning M, Breburda EE, Grendell RL, Golos TG. Passive immunization against the MHC class I molecule Mamu-AG disrupts rhesus placental development and endometrial responses. J Immunol. 2007;179:8042–50. doi: 10.4049/jimmunol.179.12.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Seval Y, Korgun ET, Demir R. Hofbauer cells in early human placenta: possible implications in vasculogenesis and angiogenesis. Placenta. 2007;28:841–45. doi: 10.1016/j.placenta.2007.01.010. [DOI] [PubMed] [Google Scholar]