Abstract
The Saccharomyces cerevisiae protein Hsl7 is a regulator of the Swe1 protein kinase in cell cycle checkpoint control. Hsl7 has been previously described as a type III protein arginine methyltransferase, catalyzing the formation of ω-monomethylarginine residues on non-physiological substrates. However, we show here that Hsl7 can also display type II activity, generating symmetric dimethylarginine residues on calf thymus histone H2A. Symmetric dimethylation is only observed when enzyme and the methyl-accepting substrate were incubated for extended times. We confirmed the Hsl7-dependent formation of symmetric dimethylarginine by amino acid analysis and thin layer chromatography with wild type and mutant recombinant enzymes expressed from both bacteria and yeast. This result is significant because no type II activity has been previously demonstrated in S. cerevisiae. We also show that Hsl7 has little or no activity on GST-GAR, a commonly used substrate for protein arginine methyltransferases, and only minimal activity on myelin basic protein. This enzyme thus may only recognize only a small subset of potential substrate proteins in yeast, in contrast to the situation with Rmt1, the major type I methyltransferase.
Methylation and demethylation of lysine and arginine residues of the N-terminal tails of histones play crucial roles in the regulation of gene expression [1]. In the budding yeast Saccharomyces cerevisiae, HSL7 was identified as an essential gene in mutants where the histone H3 tail was deleted [2]. The amino acid sequence of Hsl7 is similar to those of known yeast and mammalian protein arginine methyltransferases [3,4]. It is most similar (27.5% identity over 680 residues) to PRMT5, the predominant type II protein arginine methyltransferase of mammalian cells that catalyzes symmetric dimethylarginine formation in a variety of methyl-accepting substrates including histone H3 [5,6]. However, such an enzymatic function has not been shown to date for Hsl7. Indeed, evidence has been presented that Hsl7 is not involved in histone arginine methylation in S. cerevisiae, although it has a robust type III activity forming ω-monomethylarginine with mammalian histone H2A [7]. Hsl7 is present in a complex at the yeast bud neck with the Hsl1 and Swe1 proteins that acts to monitor septin assembly as a cell cycle checkpoint [8–11]. Disruption of the HSL7 gene in S. cerevisiae leads to an elongated bud phenotype and a delay in G2 of the cell cycle [2]. How this function of Hsl7 may be linked to chromatin modification in the nucleus is unclear, although it has been suggested that histone tail methylation and acetylation may act to bypass the morphogenesis checkpoint [11].
It was initially reported that Hsl7, expressed as a FLAG-tagged fusion protein in wild type yeast cells, had methyltransferase activity on substrates that included calf thymus histones H2A and H4 as well as myelin basic protein [12]. However, later studies were not able to detect methyltransferase activity in GST-Hsl7 or Hsl7-myc fusion proteins expressed in yeast or bacterial cells using a variety of methyl-accepting substrates [13]. A complication of these studies is that Hsl7 purified from wild type yeast cells can be contaminated with the major endogenous protein arginine methyltransferase Rmt1 [7]. When a FLAG-tagged fusion of Hsl7 was purified from yeast cells lacking the Rmt1 methyltransferase, a preparation was obtained that was unable to methylate histone H4 or myelin basic protein, but that was able to readily methylate calf thymus histone H2A [7]. Two mutant constructs of Hsl7 with either a deletion of the GAGRG sequence of its methyltransferase motif I or a double mutation of the motif to GAVRV were unable to catalyze any methylation reaction, demonstrating that the activity observed with the wild type enzyme on histone H2A was in fact due to Hsl7 methyltransferase activity [7]. Interestingly, Hsl7 is not essential for the major protein arginine methylation modification of endogenous S. cerevisiae histones [7]. Amino acid analysis indicated that the only product of the Hsl7 enzymatic reaction was ω-monomethylarginine, suggesting that this enzyme was not in fact the catalytic homolog of the mammalian PRMT5 enzyme, which is clearly a type II enzyme able to catalyze the addition of a second methyl group to the arginine residue to form symmetric dimethylarginine [5], but a potentially novel type III methyltransferase.
In this study we ask if Hsl7 has the ability to form dimethylated arginine residues if incubated for extensive times with methyl-accepting substrates. We show here that Hsl7 can in fact act as a type II protein arginine methyltransferase, forming symmetric dimethylarginine like the mammalian PRMT5 enzyme. However, Hsl7 is clearly not the functional homolog of PRMT5; the endogenous substrates of Hsl7 still remain to be identified.
Materials and methods
Purification of GST-GAR, GST-Hsl7 wild type and GST-Hsl7G387A
GST-GAR is a fusion protein containing the first 148 amino acids of human fibrillarin [19]. GST-Hsl7 and GST-Hsl7G387A are fusion proteins expressed from plasmids pDLB2211 and pDLB2212 respectively. These plasmids were obtained from Dr. Daniel Lew at the Duke University Medical Center and were constructed as described in Theesfeld et al. [14] to encode the wild type enzyme and a form with a glycine to alanine substitution in the second conserved glycine residue of motif I (ILVAGAGRG to ILVAGAARG). Plasmids encoding these proteins were used to transform Escherichia coli strains DH5α (GST-GAR) and BL21 (GST-Hsl7 and GSTHsl7G387A); in each case cells were grown in 1 L LB media in the presence of 100 μg/ml ampicillin at 37 °C until an optical density of OD600 of 0.6 was obtained. The cells were induced with 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 25 °C for 16 h, and the cells were harvested by centrifugation at 5,000 x g and washed twice with phosphate-buffered saline solution (PBS) containing 137 mM NaCl, 2.7 mM KCl, 1.4 mM KH2PO4, and 4.3 mM NaHPO4, pH 7.4. The cells were sonicated on ice in the presence of protease inhibitor (100 μM of phenylmethylsulfonyl fluoride from a 1000-fold concentrated stock in DMSO). Lysed cells were centrifuged at 23,300 x g for 50 min at 4 °C and the supernatant was mixed with 0.5 ml of glutathione-Sepharose 4B beads (GE Healthcare Biosciences AB, Uppsala, Sweden). After three washes with 10 ml of PBS, the GST-fusion proteins were eluted with 1 ml of 30 mM glutathione, 50 mM Tris-HCl, pH 7.5, and 120 mM NaCl.
Expression and purification of wild type and mutant FLAG-HSL7 in S. cerevisiae
A yeast-E. coli shuttle vector containing a gene engineered to express a FLAG-tagged fusion of Hsl7 (pTKB-FLAG-Hsl7; [12]) was a gift from Dr. Sidney Pestka at Rutgers University. A plasmid expressing a site-directed double mutant (FLAG-Hsl7 (GAGRG → GAVRV)) where valine residues replace the glycine residues at position 387and 389 of Hsl7 [7] was transformed into S. cerevisiae strain AFY5130 (MATa, ade2, rmt1::LEU2, hsl7::URA3) [7]. Expressed FLAG-tagged proteins were purified as described [7].
In vitro methylation of GST-GAR, H2A, and MBP
Methylation reactions were performed in duplicate using 2 μg of bacterially expressed GST-Hsl7 and GST-Hsl7G387A in the presence and absence of 10 μg of the substrates GST-GAR, H2A (purified from calf thymus, Roche Molecular Chemical, Indianapolis, IN), or MBP (purified from bovine brain, lyophilized powder, product M1891, Sigma Chemical, St. Louis, MO), and 0.61 μM S-Adenosyl-L-[methyl-3H]methionine ([3H]AdoMet, 81.0 Ci/mmol, dilute HCl: ethanol (9:1), pH 2.0 to 2.5, Amersham Pharmacia Biotech, Piscataway, NJ) in 100 mM sodium phosphate buffer (pH 7.5) in a final volume of 60 μl for 1 h, 5 h, or 20 h at 37 °C. Reactions were incubated at 37 °C for 1 h, 5 h, and 20 h, and frozen at −20 °C. Similar methylation reactions were performed with 2 μg of S. cerevisiae expressed wild type FLAG-Hsl7 or FLAG-Hsl7G387V, G389V enzyme and 10 μg of calf thymus H2A. These reactions were incubated for 20 h at 30 °C and stored at −20 °C.
Chemical analysis of 3H-methylated species by amino acid analysis
Proteins from in vitro methylation reactions were precipitated in 60 μl of 25 % (w/v) trichloroacetic acid with 20 μg of bovine serum albumin as a carrier, then hydrolyzed as previously described [15]. The hydrolyzed sample was added to 50 μl of water and 500 μl of citrate buffer (0.2 M Na+, pH 2.2) along with 1.0 μmol each of unlabeled standards of ω-NG – monomethylarginine (acetate salt, Sigma Chemical, St. Louis, MO)( ω-MMA) and ω-NG, NG-dimethylarginine (hydrochloride, Sigma Chemical, St. Louis, MO)(ADMA), and ω-NG, NG-dimethylarginine (di(p-hydroxyazobenzene-p’-sulfonate) salt, Sigma Chemical, St. Louis, MO)(SDMA). Amino acids were separated by high resolution cation-exchange chromatography as described [15].
Results and discussion
Yeast Hsl7 is a type II protein arginine methyltransferase forming symmetric dimethylarginine residues
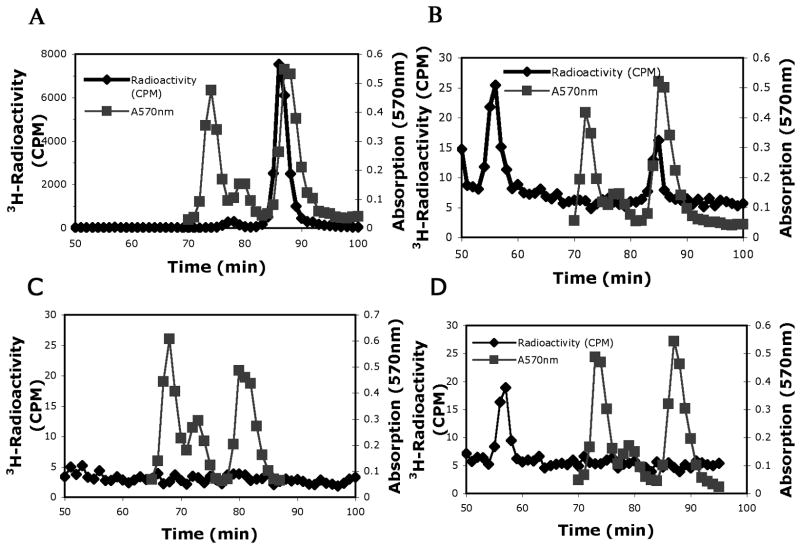
Amino acid analysis of short term incubation mixtures of wild type and mutant GST-Hsl7 with [3H]AdoMet and calf thymus histone H2A demonstrated that the GST-Hsl7 fusion protein is an active methyltransferase. In Fig. 1, we show the formation of ω-[3H]monomethylarginine (ω-MMA) eluting at 86 to 88 min from hydrolysates of reaction mixtures of wild type enzyme and histone H2A (panel A). No 3H-ω-MMA is detected in the absence of histone H2A (panel B). A control reaction where we incubated histone H2A with [3H]AdoMet and GST-Hs17 mutated in the methyltransferase active site (containing an alanine substitution at position 387 in motif I) also revealed no detectable formation of methylated products (panel C). These results demonstrate that GST fusions of Hsl7, as well as FLAG fusions [7], can function as type III protein arginine methyltransferases.
Fig. 1.
Predominant type III protein arginine methyltransferase activity of Hsl7 with calf thymus histone H2A. Amino acid analysis of 3H-methylated arginine residues when histone H2A was incubated for 1 h at 37 °C with wild type GST-Hsl7 (panel A) or mutant GST-Hsl7G387A (panel B) in the presence of 3H-AdoMet as described in the “Materials and methods” section. A control is also shown when the wild type enzyme is incubated in the absence of histone H2A (panel C). 3H-Radioactivity from mixing 200 μl of each fraction with 400 μl water and 5 mL scintillation fluor (Safety Solve, Research Products International, Mt. Prospect, IL) is seen in the black line with diamonds. Unlabeled standards of ω-MMA, ADMA, and SDMA were detected with a ninhydrin assay using 100 μl of each fraction [16] and are shown as absorption at 570 nm by gray lines with squares. As expected, the elution position of the tritiated methyl arginine derivatives is slightly ahead of the non-isotopically labeled standards [17].
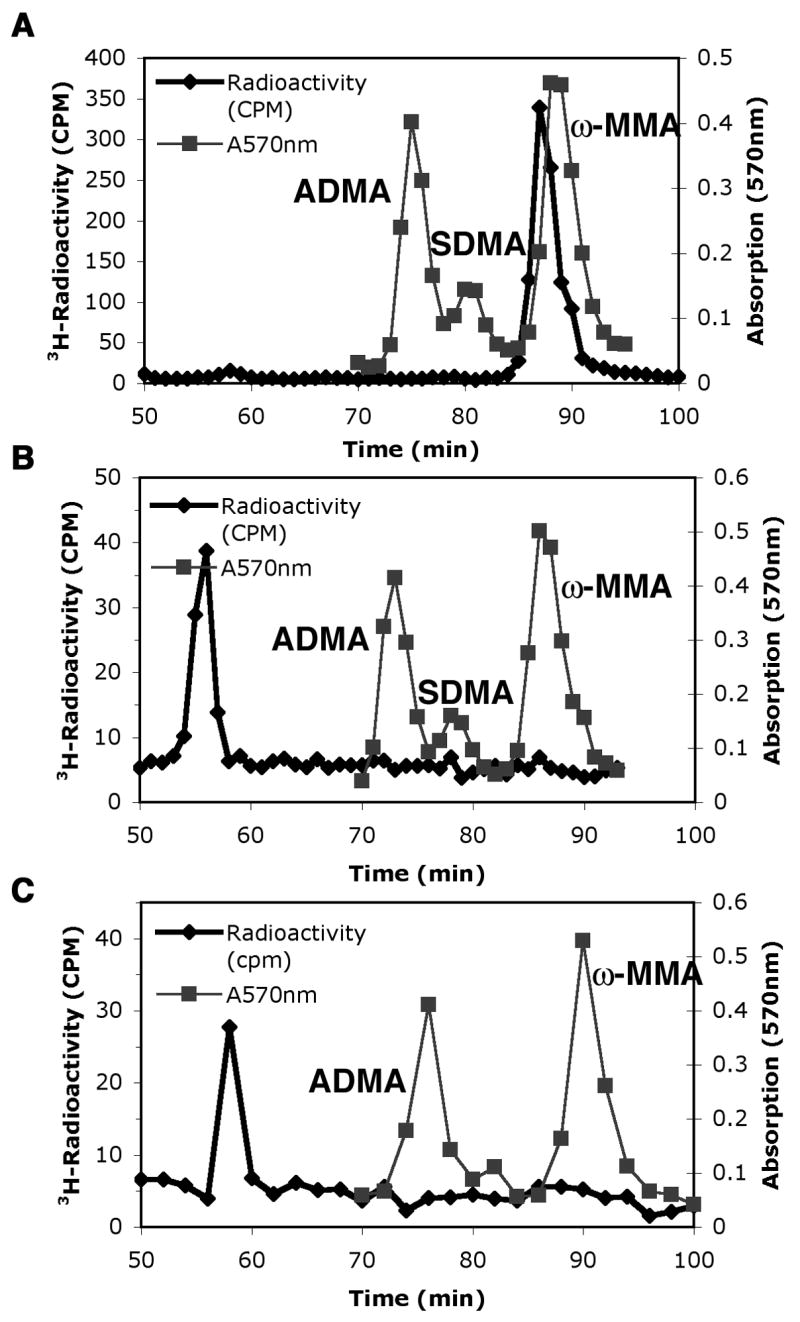
To ask if Hsl7 was capable of adding a second methyl group to arginine residues, these GST-fusion enzymes were incubated for 20 h rather than 1 h with [3H]AdoMet and histone H2A (Fig. 2). Under these conditions, there was approximately twenty-times more 3H-ω-MMA formed (panel A). However, we were now able to detect a small amount of radioactivity eluting at the position expected for symmetric dimethylarginine at 76 to 78 min. In the parallel control where the mutant GST-Hsl7G387A enzyme was used in place of GST-Hsl7, we could detect a small radioactive peak of ω-MMA that represented about 0.14% of that of the wild type enzyme (panel B). Further controls performed in the absence of histone H2A revealed no detectable formation of 3H-ω-MMA or other arginine derivatives for GST-Hsl7 (panel C) and for GST-Hsl7G387A (panel D). These results suggest that there is a small residual activity of GST-Hsl7G387A where the alanine substitution in motif I greatly slows, but does not completely inhibit the enzyme.
Fig. 2.
Detection of symmetric dimethylarginine (SDMA) as a minor product in histone H2A incubated with Hsl7 for extended times. Amino acid analysis of histone H2A methylation in wild type GST-Hsl7 (panel A) and mutant GST-Hsl7G387A (panel B) incubations for 20 h at 37 °C as described in “Materials and methods” and in the legend to Fig. 1. Controls are also shown where GST-Hsl7 (panel C) and GST-Hsl7G387A (panel D) were incubated under similar conditions in the absence of histone H2A. 3H-Radioactivity (cpm) is seen in the black line with diamonds corresponding to every fraction of the 1 ml/min elution of the cation exchange column. The ninhydrin peaks of unlabeled ω-MMA, SDMA, and ADMA are seen by gray lines with squares at 570 nm absorption corresponding to every fraction as well.
We confirmed the residual activity of GST-Hsl7G387A in an experiment where we measured radiolabeled polypeptides resulting from the incubation of this enzyme (as well as wild type GST-Hsl7) with [3H]AdoMet and calf thymus histone H2A for 1, 5, and 20 h (Fig. 3). For the wild type enzyme, we found robust incorporation of 3H-methyl groups into histone H2A at each of the time points. However, we also detected much lesser, but still significant, [3H]methylation of the histone H2A polypeptide at the 5 and 20 h time points for the mutated enzyme.
Fig. 3.
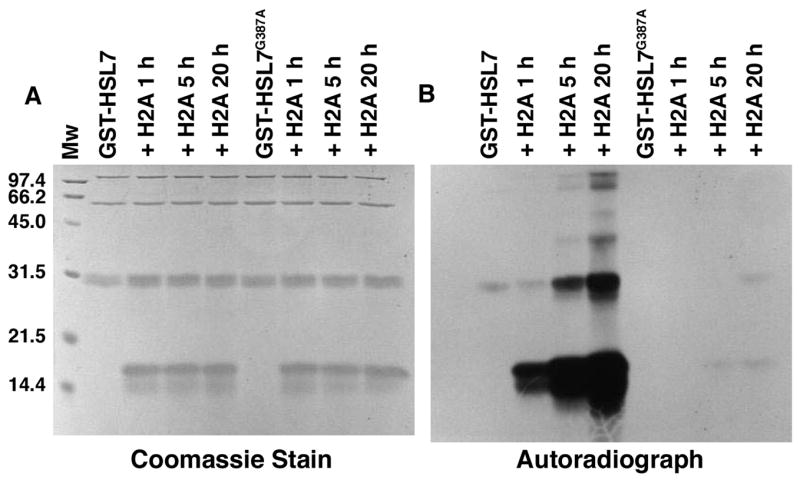
Methylation of calf thymus histone H2A by wild type and mutant GST-Hsl7 detected by SDS-PAGE. The indicated enzyme was incubated with [3H]AdoMet and calf thymus histone H2A for 1, 5, and 20 h at 37 °C as described in the “Materials and methods” section. Reactions were stopped with the addition of 60 μl of SDS sample buffer and polypeptides separated as previously described [15] except that electrophoresis was performed for 16 h at 4 mA. Molecular weight standards were electrophoresed in a parallel lane and included phosphorylase a, 97.4 kDa; serum albumin, 66.2 kDa; ovalbumin, 45.0 kDa; carbonic anhydrase, 31.0 kDa; soybean trypsin inhibitor, 21.5 kDa; and lysozyme, 14.4 kDa (BioRad Laboratories. Richmond, CA). The gel was stained with Coomassie Brilliant Blue R-250, destained, EN3HANCED, and fluorographed as described for 1 month [15].
We then wanted to confirm the ability of Hsl7 to act as a type II methyltransferase and catalyze the formation of symmetric dimethylarginine in the extended incubations of the GST-Hsl7 fusion enzyme prepared in E. coli as shown in Fig. 2. We thus prepared wild type and mutated FLAG-Hsl7 proteins that were expressed in S. cerevisiae in a strain lacking the major Rmt1 methyltransferase and endogenous Hsl7. When these enzymes were incubated with [3H]AdoMet and calf thymus histone H2A under conditions similar to those used in the experiment shown in Fig. 2, amino acid analysis also revealed the presence of a small peak of radioactivity that eluted in the position expected for 3H- SDMA (Fig. 4, panel A). In a control experiment, no 3H-radioactivity was seen when the mutant FLAG-Hsl7G387V, G389V protein was used. In this mutant, the replacement of two of the glycine residues in motif I with bulky valine residues completely abrogates all methyltransferase activity that can form either 3H-ω-MMA or 3H-SDMA (data not shown). To confirm the chemical identification of the 3H-SDMA formed with the wild type enzyme, the peak fractions were pooled, desalted, and analyzed by thin layer chromatography (Fig. 4, panel B). We found an exact coincidence of the position of the radioactivity and the unlabelled SDMA standard.
Fig. 4.
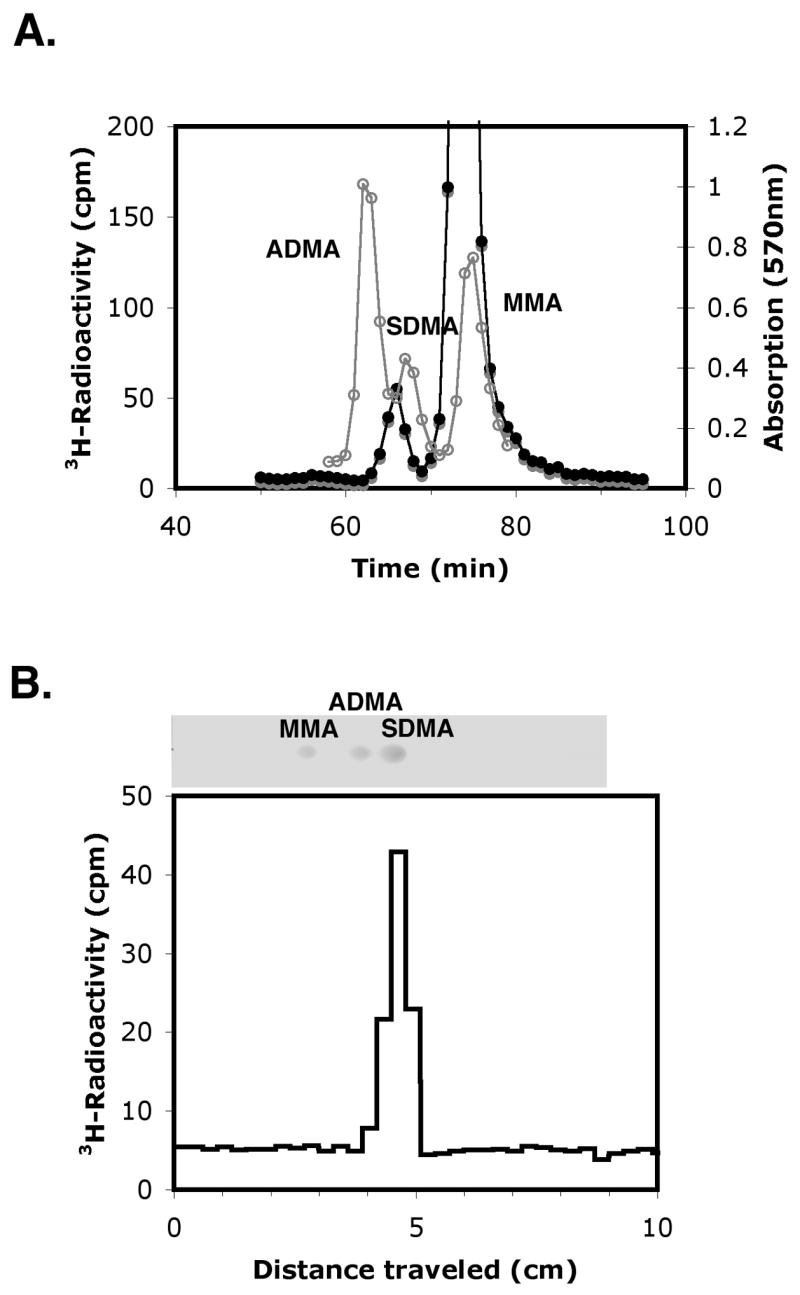
Amino acid analysis and thin layer chromatography of 3H-methylated histone H2A. (panel A) Purified FLAG-Hsl7 (2 μg) was incubated with calf thymus H2A (10 μg) and [3H]AdoMet (1 μM) for 20 hours at 30 °C. The methylation products were precipitated with trichloroacetic acid, acid hydrolyzed with 6 M HCl at 110 °C for 20 hours, and loaded on a cation exchange column with unlabeled ω-MMA, SDMA, ADMA standards as described in “Materials and methods”. Unlabeled standards were detected by absorbance at 570 nm with ninhydrin reagent (gray line) and 3H-radioactivity by scintillation counting (black line) as described in the Fig. 1 legend. (panel B) The 3H-radioactive fractions eluting with the SDMA standard at 62 min to 66 min from a total of four amino acid analyses (as shown in panel A above) were pooled and concentrated to 1 ml under vacuum centrifugation. This material was desalted at room temperature on a Sephadex G15 column (1.5 cm inner diameter x 77 cm column height) equilibrated with 0.1 M acetic acid. Radioactive fractions (1 ml) with the least salt content were pooled and concentrated to about 5 μl by vacuum centrifugation. After mixing with standards, 2 μl of a mixture containing 1.7 μl of the concentrated radioactive fractions and 0.3 μl of standards (containing 7 nmol ω-MMA, 3 nmol SDMA, and 7 nmol ADMA) was spotted onto a Whatman flexible silica thin layer chromatography (TLC) plate (20 cm x 20 cm, 250 μm layer). Chromatography was performed in a glass chamber pre-incubated for 1.5 h with 100 % ethanol:ammonium hydroxide (3:1) until the mobile phase reached to just below the top of the TLC plate. The plate was then air dried, sprayed with 0.2 % ninhydrin in ethanol, heated at 120 °C for approximately 1 min until colored spots developed (photograph). Spots were identified from the migration positions ω-MMA, ADMA, and SDMA chromatographed individually and in combination in parallel lanes. The silica was then removed from 0.3 cm slices, resuspended in 1 ml of water, and then the samples were counted for 60 min in 5 ml of fluor.
These results clearly demonstrate that Hsl7 can show type II methyltransferase activity. As such, this is the first documentation of the possibility of forming SDMA in S. cerevisiae. The total amount of SDMA in yeast is still very small compared to the levels of ω-MMA and ADMA, and no SDMA-containing proteins have yet been identified in S. cerevisiae [7]. These results now can allow us to view Hsl7 is a true catalytic homolog of the mammalian PRMT5 type II enzyme. These results also show that enzymes capable of catalyzing dimethylarginine formation may still only form monomethylarginine residues under some conditions. For example, the only methylated product observed with the mammalian type I PRMT1 and the peptide substrate GGFGGRGGFG-amide is ω-MMA, even though the major product is ADMA when protein substrates are used [18].
Substrate specificity of Hsl7
Our demonstration of SDMA formation upon extended incubation time prompted us to reconsider the specificity of GST-Hsl7 towards other methyl-accepting substrates. Previously, no detectable methylation was seen with two common substrates of mammalian PRMT5, GST-GAR and myelin basic protein [7]. The former species is a fusion protein of the N-terminal portion of fibrillarin that contains a number of arginine residues in sequences flanked by glycine residues [19]. In an experiment similar to that shown in panel A of Fig. 2 where we used GST-GAR as a methyl-accepting substrate in place of histone H2A, we could detect no formation of [3H] ω-MMA or other methylated arginine derivatives, even after incubation for 20 h under conditions where we would readily detect 0.1% of the methylation of histone H2A (data not shown). However, we were able to detect Hsl7-catalyzed [3H] ω-MMA formation in myelin basic protein under conditions similar to those in Fig. 1, panel A, but at only 10% of the extent of histone H2A (data not shown). GST-Hsl7G387A was not able to methylate MBP in a similar reaction (data not shown). We conclude that Hsl7 is distinct from the major type I yeast Rmt1 methyltransferase as well as the mammalian type II PRMT5 enzyme in that it does not catalyze the modification of arginine residues in a glycine rich context.
Physiological role of Hsl7 as a methyltransferase
It is clear that calf thymus histone H2A can mimic the endogenous substrate(s) of Hsl7; what these species are remains to be determined. These substrates appear to make up only a small fraction of the total methylated arginine species in yeast because there is no measurable reduction in methylation in hsl7−rmt1− strains compared to rmt1− strains [7]. Interestingly, Rmt1 and not Hsl7 is the major enzyme responsible for endogenous histone arginine methylation in S. cerevisiae [7,11]. These results do not exclude the possibility that Hsl7 may function by methylating histones associated with nucleosomes on a small number of genes. A recent study has identified interactions of several enzymes that modify chromatin with Hsl7 checkpoint control function, including the Set1 histone protein lysine methyltransferase [11]. However, it appears that these enzymes may function simply to bypass this checkpoint control [11]; thus Hsl7 may not function at all in the nucleus.
The question remains of how, or if, the methyltransferase activity of Hsl7 can be used to regulate the morphogenesis checkpoint when bud formation is disrupted. Based on the ability of the Hsl7G387A mutant protein to rescue of viability of yeast strains lacking the Mih1 tyrosine phosphatase involved in Cdc28 cell cycle control, Theesfeld et al., [14] suggested that the methyltransferase activity of Hsl7 was not required for its function. We do, however, detect a small residual methyltransferase activity of this mutant (0.14%); it is unclear whether this small amount of activity is sufficient for the observed rescue. Nevertheless, Ruault and Pillus [11] have also shown that a double mutant in motif I (GAGRG to GAAAG) can also complement a Hsl7 deletion change in gene silencing. It will be important to ask if the GAAAG enzyme has residual activity or not, and whether the double mutant described here (GAVRV) is also biologically active. Detection of the endogenous methylated products of Hsl7 will be an important next step in understanding its function.
Acknowledgments
This work was funded by the NIH Grant GM026020. We thank Dr. Daniel Lew (Duke University) for providing us with plasmids and helpful advice, and Drs. Michael Grunstein (UCLA) and Sidney Pestka (Rutgers University) for supplying strains and plasmids.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nature Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 2.Ma XJ, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–13807. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 3.Pollack BP, Kontenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274:31531–31542. doi: 10.1074/jbc.274.44.31531. [DOI] [PubMed] [Google Scholar]
- 4.Ma XJ. Cell-cycle regulatory proteins Hsl7p/skb1p belong to the protein methyltransferase superfamily. Trends Biochem Sci. 2000;25:11–12. doi: 10.1016/s0968-0004(99)01509-1. [DOI] [PubMed] [Google Scholar]
- 5.Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- 6.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 7.Miranda TB, Sayegh J, Frankel A, Katz JE, Miranda M, Clarke S. Yeast Hsl7 (histone synthetic lethal 7) catalyses the in vitro formation of ω-N(G)-monomethylarginine in calf thymus histone H2A. Biochem J. 2006;395:563–570. doi: 10.1042/BJ20051771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMillan JN, Longtine MS, Sia RAL, Theesfeld CL, Bardes ESG, Pringle JR, Lew DL. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. J Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruault M, Pillus L. Chromatin-modifying enzymes are essential when the Saccharomyces cerevisiae morphogenesis checkpoint is constitutively activated. Genetics. 2006;174:1135–1149. doi: 10.1534/genetics.106.059873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Cook JR, Pollack BP, Kinzy TG, Norris D, Pestka S. Hsl7p, the yeast homologue of human JBP1, is a protein methyltransferase. Biochem Biophys Res Commun. 2000;274:105–111. doi: 10.1006/bbrc.2000.3049. [DOI] [PubMed] [Google Scholar]
- 13.Cid VJ, Schulewitz MJ, McDonald KL, Thorner J. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. J Mol Biol Cell. 2001;12:1645–1669. doi: 10.1091/mbc.12.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theesfeld CL, Zyla TR, Bardes EGS, Lew DW. A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol Biol Cell. 2003;14:3280–3291. doi: 10.1091/mbc.E03-03-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayegh J, Webb K, Cheng D, Bedford MT, Clarke SG. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J Biol Chem. 2007;282:36444–36453. doi: 10.1074/jbc.M704650200. [DOI] [PubMed] [Google Scholar]
- 16.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 17.Gary JD, Lin WJ, Yang MC, Herschman HR, Clarke S. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- 18.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 19.Tang J, Gary JD, Clarke S, Herschman HR. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem. 1998;273:16935–16945. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]