Abstract
Polymerase chain reaction (PCR) assays have been used to detect bacteria adherent to failed orthopaedic implants, but some PCR assays have had problems with probable false-positive results. We used a combination of a Staphylococcus species-specific PCR and a universal PCR followed by DNA sequencing to identify bacteria on implants retrieved from 52 patients (92 implants) at revision arthroplasty. We addressed two questions in this study: (1) Is this method able to show the existence of bacterial DNA on presumed aseptic loosed implants?; and (2) What proportion of presumed aseptic or culture-negative implants was positive for bacterial DNA by PCR? Fourteen implants (15%) were believed infected, whereas 74 implants (85%) were believed aseptic. Each implant was sonicated and the resulting solution was submitted for dual real-time PCR assay and culture. All implants believed aseptically loose were culture-negative, but nine of the 74 (12%) had bacterial DNA by PCR; two (2.7%) were PCR-positive and also showed histologic findings suggestive of infection. Uniquely developed PCR and bacterial sequencing assays showed bacterial DNA on 12% of implants removed for presumed aseptic loosening. Additional studies are needed to determine the clinical importance of bacterial DNA detected by PCR but not by conventional culture.
Level of Evidence: Level III, diagnostic study. See the Guidelines for Authors for a complete description of levels of evidence.
Introduction
Total joint arthroplasty provides a high percentage of patients with excellent clinical results, but aseptic loosening and infection remain important complications [5]. Some believe the common mechanism of aseptic loosening is particle-induced bone resorption [9, 23]. However, several reports provide evidence of bacteria associated with implants removed for clinically aseptic loosening [4, 21, 26–28]. Several reports suggest a reduced incidence of aseptic loosening for implants fixed with antibiotic-containing bone cement compared with those fixed with conventional cement [7, 11, 16, 29]. These observations suggest the incidence of occult infections involving loose orthopaedic implants may be underestimated [19].
The most commonly detected bacteria from infected total joint arthroplasties are Staphylococcus aureus and Staphylococcus epidermidis [8], which tend to exist in a biofilm on implant surfaces [1, 20]. If implants have a low concentration of adherent organisms, then it may be necessary to disrupt the biofilm using, for example, an ultrasonication technique to detect them [14, 21, 27, 28]. Furthermore, some type of molecular detection technique may be useful for identifying small quantities of organisms or slow-growing bacteria that may not be detected with conventional methods [24–26].
The most popular molecular method for detecting a broad range of bacteria is a PCR based on the 16S rRNA gene [30], a gene present in almost all bacteria. However, so-called universal assays of this type may have problems with false-positive results either from contaminating or commensal organisms [3] or from residual Escherichia coli DNA in one of the reagents [6].
We have developed a real-time PCR assay that can detect S. aureus and coagulase-negative Staphylococcus (CNS) with a high degree of specificity and sensitivity [22]. (Real-time means the quantification of amplified DNA in real time without delay at each amplification cycle.) This assay is desirable in cases of orthopaedic infection given the high frequency of the Staphylococcus species as a causative agent of infection [12]. In addition, we developed a broad-range universal PCR combined with pyrosequencing technology that can identify bacterial subgroups [13, 15]. This dual real-time PCR approach has made it possible to improve specificity, which was difficult to achieve using only conventional universal PCR [4, 10, 27].
We addressed two questions in this study: (1) Are two different real-time PCR assays with pyrosequencing after ultrasonication able to detect adherent bacteria on implants removed at revision arthroplasty for presumed aseptic loosening?; and (2) What percent of presumed aseptic or culture-negative implants was positive for bacterial DNA by PCR?
Materials and Methods
To clarify the positive rate of PCR assay for presumed aseptic loosening, we applied two different real-time PCR assays prospectively to retrieved failed joint implants. After brief ultrasonication processing, each sonicate solution was submitted for microbiologic culture and DNA extraction after PCR assays. For discordant samples, histologic findings also were reviewed.
We retrieved failed implants in 52 consecutive patients (92 implants) undergoing revision arthroplasty for failed hip (n = 24) or knee (n = 28) arthroplasty between April 2003 and December 2005. Fourteen of the 92 implants (15%) were believed infected based on preoperative diagnosis made by a surgeon (VK) as described below; 74 implants (85%) were believed aseptic. Twenty-nine of these patients were reported in a previous study in which the molecular Gram-stain characteristics of the PCR were validated [15]. For the purposes of this study, a Staphylococcus genus-specific PCR [22] was used in addition to the molecular Gram-stain characteristics, and PCR was defined as positive if at least one assay showed a positive result. Institutional Review Board approval was obtained for the study.
Before implant retrieval, unimplanted, company-packaged sterile acetabular components (SECUR-FIT-HA PSL; Stryker Orthopaedics, Mahwah, NJ) were submitted to the same sonication processing and DNA extraction and served as a negative control. After sterilization, the negative control test was repeated twice. Three negative control tests revealed all negative results for both PCR assays.
Each retrieved implant was packaged in the bioclean operating room with a laminar air-flow system in an individual sterile container and then immediately transported to our microbiology department. Preoperative diagnoses were made by the surgeon (VK) based on clinical symptoms, local clinical findings, physical findings, and serologic testing. Briefly, we believed patients had aseptic loosening if symptoms of pain or instability were accompanied by radiographic features suggestive of loosening but other serologic tests that usually suggest infection were negative. Patients were believed to have infection if they had local pain, swelling, or redness with evidence of positive serologic testing such as C-reactive protein and sedimentation rate and/or a positive culture of aspirated joint fluid. Preoperative diagnoses for all patients are summarized in Fig. 1. All patients believed not to have infection were considered to be aseptic and were culture-negative, although intraoperative smears in two samples were interpreted as showing Gram-positive cocci. Because of the intraoperative Gram-stain results, the two patients were treated as having infection and underwent two-stage revisions.
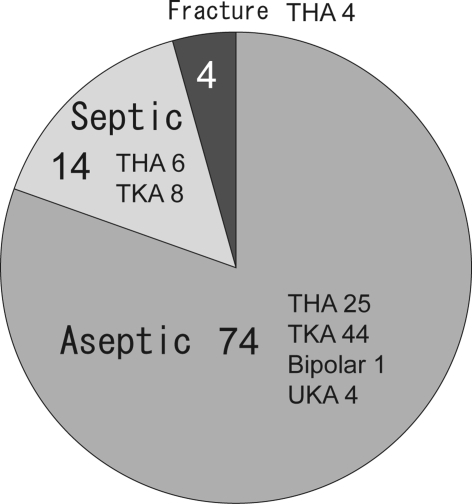
Fig. 1.
A diagram shows the preoperative diagnoses. Of the 92 tested implants, 74 (80.4%) were clinically identified as having aseptic loosening, 14 (15%) were clinically infected, and four (4%) were associated with periprosthetic fractures and believed aseptic. UKA = unilateral knee arthroplasty.
In all cases, antibiotics were withheld until fluid and tissue cultures had been taken, then the patients were given 1 mg cephazolin intravenous drip infusion, or if truly allergic to PCN/cephalosporins, they received 1 mg vancomycin hydrochloride.
We placed the retrieved implants in a water-bath sonicator (Branson Ultrasonic Cleaner; Branson Ultrasonics, Danbury, CT) and sonicated for 5 minutes. Twenty-five milliliters of sonicate solution was centrifuged (10,000 × g 20 minutes), the supernatant was discarded, and DNA was extracted using a Qiagen DNA mini kit (Qiagen Inc, Valencia, CA) according to the manufacturer’s instructions.
For each implant, we submitted 1 mL of sonicates to standard microbiologic culture to detect aerobic and anaerobic bacteria without subculturing. The microbiologic culture was performed and the bacteria were allowed to grow for up to 2 weeks.
We obtained all oligonucleotide primers and probes from BioChem (Salt Lake City, UT). The entire tuf gene sequences of Staphylococcus species available from public databases were analyzed with the ClustalW multiple sequence alignment program provided by the European Bioinformatics Institute (www.ebi.ac.uk/clustalw/). Based on multiple sequence alignment, we chose regions of the tuf gene, highly conserved among Staphylococcus species. The selection of PCR primers and probes from these regions was enhanced using LightCycler Probe Design software (version 1.0; Idaho Technology Inc, Salt Lake City, UT). The primers and probes used for this test have been described [22]. Briefly, the sequence of the forward primer was 5′-CAATGCCACAAACTCG-3′ (position 33–48), whereas the sequence of the reverse primer was 5′-GCTTCAGCGTAGTCTA-3′ (position 510–494). The broad-range Staphylococcus genus-specific FRET hybridization probes were 5′-ACGGCCTGTAGCAACAGTAC-FITC-3′ (position 372–391) and 5′-LCRed640-CGACCAGTGATTGAGAATACGTCC-phosphate-3′ (position 369–346), whereas the S. aureus species-specific FRET hybridization probes were 5′-GGCGATGCTCAATACGAAGAAAAAATC-FITC-3′ (position 239–265) and 5′-LCRed705-AGAATCAATGGAAGCTGTAGATAC-phosphate-3′ (position 268–291).
The primers and probes used for the universal PCR and pyrosequencing have been described [13]. Briefly, the selection of these primers was enhanced with the use of primer design software (LC Probe Design Software, Roche, IN). The forward primer was biotinylated for amplicon capture, which is necessary for pyrosequencing. The forward and reverse primer sequences were 5′-biotin-GGATTAGATACCCTGGTAGT-3′ and 5′-GGTAAGGTTCTTCGCG-3′, respectively. The selection of sequencing primer was enhanced using the SNP Primer Design software (Pyrosequencing AB version 1.0.1, Biotage, MA). The sequencing primer was 5′-CGTACTCCCCAGGC-3′.
Polymerase chain reaction mixtures consisted of 3.0 mmol/L MgCl2, 1.0 μmol/L concentration of each Staphylococcus generic primer, 0.2 μmol/L concentration of each Staphylococcus generic probe, and 2 μL of 10× LightCycler FastStart DNA Master Hybridization Probes mixture (Roche, IN). We added 2 μL of template DNA extract to obtain a reaction volume of 20 μL for each capillary tube. The cycling parameters consisted of one 95°C-incubation for 10 minutes for enzyme activation and DNA denaturation followed by 45 PCR amplification cycles. Each cycle consisted of three different incubation temperatures and times: 95°C for 10 seconds, 56°C for 10 seconds, and 72°C for 23 seconds. We performed fluorescence readings after annealing at 56°C for 1 second. Polymerase chain reaction cycling was followed by melting curve analysis of 40°C to 75°C (temperature transition rate of 0.5°C/second) with continuous fluorescence readings.
The presence of an amplification or quantification curve for the LC640 signal captured in the F2 channel of the LightCycler in conjunction with a melt curve with a temperature greater than 58°C was considered a positive result for CNS. The absence of a quantification curve or the presence of a quantification curve in the absence of a corresponding melting curve greater than 58°C was considered a negative result for a member of the CNS. We considered the presence of a melt curve for the LC705 signal captured in the F3 channel of the LightCycler a positive result for S. aureus. The specimen was considered negative for S. aureus if this melt curve was absent.
The extracted DNA was submitted for real-time PCR using the RotorGene 3000 (Corbett Research, Sydney, Australia). The PCR mixture consisted of 2.0 μmol/L MgCl2, 0.2 μmol/L of each primer, and 2 μL of 10× LightCycler FastStart DNA Master SYBR green I (Roche Diagnostics, Indianapolis, IN) for a volume of 15 μL master mix per reaction. We added 5 μL of template DNA extract to the reaction mixture for a final reaction volume of 20 μL for each tube. After incubation of the reaction mixture at 95°C for 10 minutes, 45 cycles of PCR amplification were performed using the following parameters: 95°C for 5 seconds, 57°C for 5 seconds, and 72°C for 15 seconds.
Samples were interpreted as positive for bacterial DNA if amplification occurred before the cycle in which the nuclease-free water (ie, the negative control) showed amplification.
We submitted the samples identified as positive by universal PCR for pyrosequencing assay (Pyrosequencing, Biotage, MA). The details of this procedure have been described [13]. Briefly, 15 μL of biotinylated PCR products was mixed in streptavidin sepharose beads and binding buffer and incubated at 40°C for 20 minutes. The immobilized products were transferred to a 96-well filter plate and processed according to the manufacturer’s protocol.
As described [13], if the first three nucleotides obtained during pyrosequencing were GGA or GGG, then the bacteria was most likely Gram-positive, whereas if GGT was obtained, then the bacteria was most likely Gram-negative. The reliability of this definition has been validated using a large number of bacterial strains [13].
The PCR result was defined as positive when either the Staphylococcus PCR or universal PCR was positive.
For culture-positive or for patients in whom there was preoperative suspicion of infection, we also reviewed microscope slides of tissue that had been submitted routinely during surgery, and histologic findings were used to help establish a diagnosis. The microscope slides were considered suggestive of infection if a minimum of five high-power fields (x400) contained five or more neutrophils [18].
Results
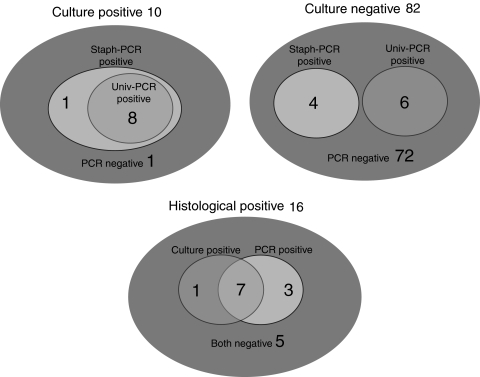
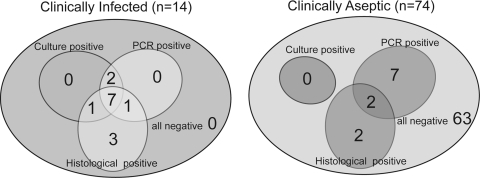
Of 82 culture-negative implants, 10 were PCR-positive (12%) (Fig. 2). Of the 74 implants retrieved from a patient believed to have aseptic failure, nine were PCR-positive (12%) (Fig. 3) and two of these (3%) also showed histologic features of infection. Sixty-three of the 82 (77%) culture-negative implants were negative using all test methods (Fig. 3).
Fig. 2.
A diagram shows PCR and culture results for the culture-positive, culture-negative, and histology-positive implants. In 82 culture-negative implants, 10 were PCR-positive.
Fig. 3.
A diagram shows PCR, culture, and histologic results for each preoperative diagnosis. Of the 74 implants believed to have undergone aseptic failure, nine were PCR-positive.
The dual PCR assay (Staphylococcus PCR and universal PCR) showed 90% sensitivity and 87.8% specificity compared with conventional microbiologic culture. The positive predictive value was 0.47, accuracy was 0.88, and likelihood ratio for a positive result was 7.38. Interpreted from the opposite perspective, microbiologic cultures showed 47.4% sensitivity and 98.6% specificity compared with the dual PCR assay results.
For nine of the 10 culture-positive implants, melting curve analysis differentiated CNS from S. aureus correctly (compared with culture results). Our molecular Gram-stain using PCR and pyrosequencing matched the conventional Gram-stain results in six implants. Four patients with misidentified molecular Gram stains include two with Gram-negative bacilli by tissue culture (the sonicate cultures grew CNS) and two with CNS for which the universal PCR was negative (therefore pyrosequencing was not performed). One patient with a positive culture result but negative by both PCR assays had acute inflammation in the tissue and we believed this was a false-negative PCR result (Table 1).
Table 1.
Clinical data
| Implant | Patient number | Implant | Time in vivo (months) | Preoperative diagnosis | Culture | PCR | Histopathology |
|---|---|---|---|---|---|---|---|
| 1 | 1 | TKA femoral | 24 | I | – | − | Infection |
| 2 | TKA tibial | 24 | I | – | − | Infection | |
| 3 | 2 | THA femoral | 20 | I | S. aureus | + | Infection |
| 4 | THA acetabular | 20 | I | S. aureus | + | Infection | |
| 5 | 3 | THA stem | 324 | F | – | − | N/A |
| 6 | THA acetabular | 324 | F | – | − | N/A | |
| 7 | 4 | THA femoral | N/A | A | – | − | N/A |
| 8 | 5 | THA femoral | 48 | I | CNS | + | N/A |
| 9 | THA acetabular | 48 | I | CNS | + | N/A | |
| 10 | 6 | THA femoral | 173 | A | – | + | Negative |
| 11 | THA acetabular | 173 | A | – | − | N/A | |
| 12 | 7 | TKA femoral | 120 | A | – | − | N/A |
| 13 | TKA tibial | 120 | A | – | − | N/A | |
| 14 | 8 | TKA femoral | 11 | I | – | − | Infection |
| 15 | TKA tibial | 11 | I | – | + | Infection | |
| 16 | 9 | Bipolar femoral | 108 | A | – | + | N/A |
| 17 | 10 | THA femoral | 216 | F | – | − | N/A |
| 18 | THA acetabular | 216 | F | – | − | N/A | |
| 19 | 11 | TKA femoral | 88 | I | CNS | + | Infection |
| 20 | TKA tibial | 88 | I | CNS | + | Infection | |
| 21 | 12 | TKA femoral | 10 | A | – | − | N/A |
| 22 | TKA tibial | 10 | A | – | − | N/A | |
| 23 | 13 | THA acetabular | 252 | A | – | − | N/A |
| 24 | 14 | THA acetabular | 210 | A | – | − | N/A |
| 25 | 15 | TKA femoral | 22 | A | – | − | N/A |
| 26 | TKA tibial | 22 | A | – | − | N/A | |
| 27 | 16 | TKA femoral | 204 | A | - but Smear Gram-positive cocci | + | Negative |
| 28 | TKA tibial | 204 | A | - but Smear Gram-positive cocci | − | Negative | |
| 29 | 17 | TKA femoral | 180 | A | – | + | Negative |
| 30 | TKA tibial | 180 | A | – | − | N/A | |
| 31 | 18 | TKA femoral | 204 | A | – | − | N/A |
| 32 | TKA tibial | 204 | A | – | − | N/A | |
| 33 | 19 | THA cup | 156 | A | – | + | Negative |
| 34 | 20 | THA femoral | 276 | A | – | − | N/A |
| 35 | THA acetabular | 276 | A | – | − | N/A | |
| 36 | 21 | TKA femoral | 52 | A | – | − | N/A |
| 37 | TKA tibial | 52 | A | – | − | N/A | |
| 38 | 22 | THA femoral | N/A | A | – | − | N/A |
| 39 | THA acetabular | N/A | A | – | − | N/A | |
| 40 | 23 | THA femoral | 252 | A | – | − | N/A |
| 41 | THA acetabular | 252 | A | – | − | N/A | |
| 42 | 24 | TKA femoral | 60 | A | – | − | N/A |
| 43 | TKA tibial | 60 | A | – | − | N/A | |
| 44 | 25 | TKA femoral | 180 | A | – | + | Negative |
| 45 | TKA tibial | 180 | A | – | + | Negative | |
| 46 | 26 | THR femoral | 168 | A | – | − | N/A |
| 47 | THR acetabular | 168 | A | – | − | N/A | |
| 48 | 27 | THR femoral | 12 | I | CNS | − | Infection |
| 49 | THR acetabular | 12 | I | CNS | + | Infection | |
| 50 | 28 | THR acetabular | 72 | A | – | − | N/A |
| 51 | 29 | TKR femoral | N/A | A | – | − | N/A |
| 52 | TKR tibial | N/A | A | – | − | N/A | |
| 53 | 30 | THA acetabular | 240 | A | – | − | N/A |
| 54 | 31 | TKA femoral | 72 | A | – | − | N/A |
| 55 | TKA tibial | 72 | A | – | − | N/A | |
| 56 | 32 | TKA femoral | 36 | A | – | − | N/A |
| 57 | TKA tibial | 36 | A | – | − | N/A | |
| 58 | 33 | THA femoral | 16 | A | – | + | Infection |
| 59 | THA acetabular | 120 | A | – | + | Infection | |
| 60 | 34 | TKA femoral | 11 | A | – | − | N/A |
| 61 | TKA tibial | 11 | A | – | − | N/A | |
| 62 | 35 | TKA femoral | 3 | A | – | − | N/A |
| 63 | TKA tibial | 3 | A | – | − | N/A | |
| 64 | 36 | THA femoral | 30 | A | – | − | N/A |
| 65 | 37 | THA femoral | N/A | A | – | − | N/A |
| 66 | 38 | THA femoral | 13 | A | – | − | N/A |
| 67 | 39 | THA femoral | 276 | A | – | − | N/A |
| 68 | 40 | TKA femoral | 48 | A | – | − | N/A |
| 69 | TKA tibial | 48 | A | – | − | N/A | |
| 70 | 41 | TKA femoral | 24 | A | – | − | N/A |
| 71 | TKA tibial | 24 | A | – | − | N/A | |
| 72 | 42 | UKA femoral | 36 | A | – | − | N/A |
| 73 | UKA tibial | 36 | A | – | − | N/A | |
| 74 | 43 | TKA femoral | 144 | A | – | − | N/A |
| 75 | TKA tibial | 144 | A | – | − | N/A | |
| 76 | 44 | TKA femoral | N/A | I | CNS | + | Infection |
| 77 | TKA tibial | N/A | I | CNS | + | Infection | |
| 78 | 45 | UKA femoral | 24 | A | – | − | N/A |
| 79 | UKA tibial | 24 | A | – | − | N/A | |
| 80 | 46 | THA femoral | 216 | A | – | − | N/A |
| 81 | 47 | TKA femoral | N/A | A | – | − | N/A |
| 82 | TKA tibial | N/A | A | – | − | N/A | |
| 83 | 48 | TKA femoral | 30 | A | – | − | N/A |
| 84 | TKA tibial | 30 | A | – | − | N/A | |
| 85 | 49 | TKA femoral | N/A | A | – | − | N/A |
| 86 | TKA tibial | N/A | A | – | − | N/A | |
| 87 | 50 | TKA femoral | 18 | A | – | − | Infection |
| 88 | TKA tibial | 18 | A | – | − | Infection | |
| 89 | 51 | TKA femoral | N/A | A | – | − | N/A |
| 90 | TKA tibial | N/A | A | – | − | N/A | |
| 91 | 52 | THA femoral | 72 | A | – | − | N/A |
| 92 | THA acetabular | 72 | A | – | − | N/A |
PCR = polymerase chain reaction; N/A = not applicable; I = infected; F = fracture; A = aseptic failure; PCR+; PCR-positive with at least one assay; PCR− = negative for both PCR tests; CNS = coagulase-negative staphylococcus; negative results for histology means no significant acute inflammation.
Among the eight patients with culture-negative, PCR-positive results, three were treated as having infection and underwent two-stage revision surgery; two (Patients 8, 33) had histologically positive results by frozen section, and one (Patient 16) was believed to have infection based on the intraoperative fast smear Gram-stain findings (Table 2). These three patients (Patients 8, 16, 33) who underwent two-stage revisions currently are without clinical evidence of recurrent infection.
Table 2.
Culture-negative, PCR-positive data
| Patient number/PCR | Preoperative diagnosis | Operative findings/procedure | Molecular subtyping by PCR or sequencing | Histologic findings | Clinical outcome |
|---|---|---|---|---|---|
| 6/univ PCR | Aseptic loosening THA | Grossly loose femoral component/revision | Gram-negative species by sequencing | No acute inflammation | Stable up to 31 months |
| 8/univ PCR | Infected TKA | Granulation tissue/débridement, insertion of antibiotic-impregnated spacer | Gram-negative species by sequencing | Acute inflammation | After two-stage revision, stable up to 8 months |
| 9/staph PCR | Aseptic loosening of bipolar hip | Grossly loosed femoral component/revision | CNS by PCR | N/A | Stable up to 5 months |
| 16/univ PCR | Aseptic loosening of TKA | No gross purulence, but fast smear of joint fluid was positive for Gram-positive cocci/débridement, insertion of antibiotic-impregnated spacer | Gram-positive species by sequencing | No acute inflammation | After two- stage revision, stable up to 12 months |
| 17/staph PCR | Aseptic loosening of TKA | Grossly loose patella component/revision | CNS by PCR | No acute inflammation | Knee pain after 14 months |
| 19/staph PCR | Aseptic loosening THA | Grossly loose acetabular component | CNS by PCR | No acute inflammation | Stable up to 13 months |
| 25*/univ PCR | Unstable TKA, severe femoral osteolysis | Large cavity at distal femur, tissue culture grew S. aureus after surgery | Gram-negative species by sequencing | No acute inflammation | After antibiotic therapy for 6 weeks, stable up to 5 months |
| 33/univ PCR staph PCR | Aseptic loosening of THA | Severe pelvic bone loss/débridement, insertion of antibiotic-impregnated spacer | Gram-negative species by sequencing/CNS by PCR | Acute inflammation | After two-stage revision, stable up to 9 months |
*Specimen was culture-negative from sonicate solutions, but tissue cultures were positive several days later; frozen sections of periimplant tissue for cases showed acute inflammation and histologic features suggestive of ongoing infection; PCR = polymerase chain reaction; univ = universal; staph = staphylococci; CNS = coagulase-negative staphylococcus; N/A = not available.
Discussion
Numerous observations suggest the incidence of occult infections involving loose orthopaedic implants may be underestimated. The research questions of this study were (1) Are two different real-time PCR assays after ultrasonication able to detect the adherent bacteria on implants removed at revision arthroplasty for presumed aseptic loosening?; and (2) What percent of presumed aseptic or culture-negative implants was positive for bacterial DNA by PCR?
The major limitation of this study is the lack of a gold standard for diagnosis of infection, especially with respect to the culture-negative/PCR-positive results. We are not able to determine the viability of bacteria in culture-negative/PCR-positive cases. In this situation, other information can help define infection status, including histologic analysis of periimplant tissues. Several, but not all, of our patients with culture-negative, PCR-positive results showed histologic features of infection. Polymerase chain reaction does not determine bacterial viability, therefore the clinical importance of organisms detected with this method needs to be clarified with long-term prospective studies and with molecular techniques intended to distinguish viable from necrotic organisms.
Conventional microbiologic culture has long been the gold standard for diagnosis of periprosthetic infection although several other tests are available, and the generally good clinical results based on those cultures suggest we should apply PCR to routine clinical use only with caution. Some studies report differences in sensitivity and specificity between PCR and culture results such that it is difficult to know the clinical importance of discrepancies (culture-negative/PCR-positive or culture-positive/PCR-negative). For example, Mariani et al. [17] were the first investigators who applied PCR assay for detection of bacterial DNA in knee fluid aspirates and reported its high sensitivity. Tunney et al. [27] used a universal PCR based on the 16S rRNA gene and reported positive results for 72% of implants revised for aseptic loosening. Although universal PCR assays of that type have very high sensitivity, specificity sometimes is compromised because of contamination of some commercially available Taq polymerase reagents with Escherichia coli DNA, because the polymerase is derived from a recombinant E. coli source [6, 8]. The possibility of Propionibacterium acnes (P. acnes) was described in the study by Tunney et al. [27], whereas no specimens were P. acnes-positive by culture in our study. Our molecular Gram stain suggested Gram-negative infection in five patients despite negative culture results possibly related to P. acnes infection. Clarke et al. [4] also investigated the use of a universal PCR to identify evidence of bacteria in patients with presumed aseptic loosening. They reported a 46% positive rate in specimens from revision surgery and a 21.4% positive rate in specimens from primary surgery, again suggesting high sensitivity but possibly low specificity with respect to clinically important organisms. However, Ince et al. [10] reported universal PCR was not superior to routine bacteriologic culture techniques for detecting low-grade infections associated with failed hip arthroplasty. The differences of PCR methods and results among these prior studies and current study are summarized in Table 3, although it is difficult to compare the positive PCR rates in each study. The discrepancy between prior studies and ours might be attributable to several factors such as the primer and probe sequences, other technical factors related to PCR methods, patient criteria, and tissue sampling.
Table 3.
Comparison of PCR studies
| Study | PCR method/Primer design | DNA sequencing | Specimen |
|---|---|---|---|
| Mariani et al. [17] | Conventional/broad-range | None | Joint fluid |
| Tunney et al. [27] | Conventional/broad-range | None | Implant |
| Clarke et al. [4] | Conventional/broad-range | None | Tissue around implant/joint fluid |
| Ince et al. [10] | Conventional/broad-range | None | Tissue around implant |
| Current study | Real-time/species-specific and broad-range | Done after broad-range PCR | Implant |
PCR = polymerase chain reaction.
The selection of the PCR assay (universal versus species-specific) is an important and difficult issue for every study based on PCR. The most important concept of the current study is the use of two different real-time PCR assays: a genus-specific PCR that detects the most frequent bacteria associated with orthopaedic infections (ie, Staphylococcus species) and a broad-range universal PCR followed by a pyrosequencing assay to address the issue of false-positive reactions secondary to residual E. coli DNA in the DNA polymerase. We also intentionally reduced the sensitivity of our universal PCR because of our stringent definition of a positive PCR result. Our PCR-positive rate in patients believed to have aseptic loosening was 12.2% (nine of 74 patients) at the maximum, considerably lower than the results reported by Tunney et al. [27] and Clarke et al. [4].
Instead of only comparing PCR and culture results, other possible outcome measures include the histology of periimplant membranes, preoperative clinical findings, serologic tests, and the ultimate clinical followup [2]. Unfortunately, it still is not possible to define with certainty the existence of bacteria or the clinical importance of a positive test result using only one assay. Interpreting all outcome measures together in our study, two of 74 patients with culture-negative results (2.7%) who were believed to have aseptic loosening now are believed to have had infection based on the combination of positive PCR results and histologic findings. Seven of 74 patients with culture-negative results believed to have experienced aseptic loosening (9.5%) had PCR-positive (only) results. It is unclear if these represent false-positive PCR results or very low-grade infections, but the patients were treated as not having infection and no evidence of infection was seen at least with a short followup.
Detection of bacteria using dual real-time PCR assays provides evidence of bacterial DNA in some patients believed to have experienced aseptic loosening. If all of our PCR results are interpreted as true-positives, then 12% of the patients with aseptic loosening actually were colonized with bacteria, which is lower than the rates reported in many other PCR studies.
Footnotes
One or more of the authors (TW) has received funding from Stryker Orthopedics (Mahwah, NJ) and Nippon Stryker KK (Tokyo, Japan).
References
- 1.Arciola CR, Campoccia D, Montanaro L. Detection of biofilm-forming strains of Staphylococcus epidermidis and S. aureus. Expert Rev Mol Diagn. 2002;2:478–484. [DOI] [PubMed]
- 2.Bauer TW, Parvizi J, Kobayashi N, Krebs V. Diagnosis of periprosthetic infection. J Bone Joint Surg Am. 2006;88:869–882. [DOI] [PubMed]
- 3.Borst A, Box AT, Fluit AC. False-positive results and contamination in nucleic acid amplification assays: suggestions for a prevent and destroy strategy. Eur J Clin Microbiol Infect Dis. 2004;23:289–299. [DOI] [PubMed]
- 4.Clarke MT, Roberts CP, Lee PT, Gray J, Keene GS, Rushton N. Polymerase chain reaction can detect bacterial DNA in aseptically loose total hip arthroplasties. Clin Orthop Relat Res. 2004;427:132–137. [DOI] [PubMed]
- 5.Clohisy JC, Calvert G, Tull F, McDonald D, Maloney WJ. Reasons for revision hip surgery: a retrospective review. Clin Orthop Relat Res. 2004;429:188–192. [DOI] [PubMed]
- 6.Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol. 2000;38:1747–1752. [DOI] [PMC free article] [PubMed]
- 7.Espehaug B, Engesaeter LB, Vollset SE, Havelin LI, Langeland N. Antibiotic prophylaxis in total hip arthroplasty: review of 10,905 primary cemented total hip replacements reported to the Norwegian arthroplasty register, 1987 to 1995. J Bone Joint Surg Br. 1997;79:590–595. [DOI] [PubMed]
- 8.Grahn N, Olofsson M, Ellnebo-Svedlund K, Monstein HJ, Jonasson J. Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol Lett. 2003;219:87–91. [DOI] [PubMed]
- 9.Hirakawa K, Jacobs JJ, Urban R, Saito T. Mechanisms of failure of total hip replacements: lessons learned from retrieval studies. Clin Orthop Relat Res. 2004;420:10–17. [DOI] [PubMed]
- 10.Ince A, Rupp J, Frommelt L, Katzer A, Gille J, Lohr JF. Is “aseptic” loosening of the prosthetic cup after total hip replacement due to nonculturable bacterial pathogens in patients with low-grade infection? Clin Infect Dis. 2004;39:1599–1603. [DOI] [PubMed]
- 11.Josefsson G, Lindberg L, Wiklander B. Systemic antibiotics and gentamicin-containing bone cement in the prophylaxis of postoperative infections in total hip arthroplasty. Clin Orthop Relat Res. 1981;159:194–200. [PubMed]
- 12.Kobayashi N, Bauer TW, Sakai H, Togawa D, Lieberman IH, Fujishiro T, Procop GW. The use of newly developed real-time PCR for the rapid identification of bacteria in culture-negative osteomyelitis: a case report. Joint Bone Spine. 2006;73:745–747. [DOI] [PubMed]
- 13.Kobayashi N, Bauer TW, Togawa D, Lieberman IH, Sakai H, Fujishiro T, Tuohy MJ, Procop GW. A molecular Gram stain using broad range PCR and pyrosequencing technology: a potentially useful tool for diagnosing orthopaedic infections. Diagn Mol Pathol. 2005;14:83–89. [DOI] [PubMed]
- 14.Kobayashi N, Bauer TW, Tuohy MJ, Fujishiro T, Procop GW. Brief ultrasonication improves detection of biofilm-formative bacteria around a metal implant. Clin Orthop Relat Res. 2007;457:210–213. [DOI] [PubMed]
- 15.Kobayashi N, Bauer TW, Tuohy MJ, Lieberman IH, Krebs V, Togawa D, Procop GW. The comparison of pyrosequencing molecular Gram stain, culture, and conventional Gram stain for diagnosing orthopaedic infections. J Orthop Res. 2006;24:1641–1649. [DOI] [PubMed]
- 16.Malchau H, Herberts P, Eisler T, Garellick G, Soderman P. The Swedish Total Hip Replacement Register. J Bone Joint Surg Am. 2002;84(suppl 2):2–20. [DOI] [PubMed]
- 17.Mariani BD, Martin DS, Levine MJ, Booth RE Jr, Tuan RS. The Coventry Award. Polymerase chain reaction detection of bacterial infection in total knee arthroplasty. Clin Orthop Relat Res. 1996;331:11–22. [DOI] [PubMed]
- 18.Mirra JM, Amstutz HC, Matos M, Gold R. The pathology of the joint tissues and its clinical relevance in prosthesis failure. Clin Orthop Relat Res. 1976;117:221–240. [PubMed]
- 19.Nelson CL, McLaren AC, McLaren SG, Johnson JW, Smeltzer MS. Is aseptic loosening truly aseptic? Clin Orthop Relat Res. 2005;437:25–30. [DOI] [PubMed]
- 20.Neut D, van Horn JR, van Kooten TG, van der Mei HC, Busscher HJ. Detection of biomaterial-associated infections in orthopaedic joint implants. Clin Orthop Relat Res. 2003;413:261–268. [DOI] [PubMed]
- 21.Nguyen LL, Nelson CL, Saccente M, Smeltzer MS, Wassell DL, McLaren SG. Detecting bacterial colonization of implanted orthopaedic devices by ultrasonication. Clin Orthop Relat Res. 2002;403:29–37. [DOI] [PubMed]
- 22.Sakai H, Procop GW, Kobayashi N, Togawa D, Wilson DA, Borden L, Krebs V, Bauer TW. Simultaneous detection of Staphylococcus aureus and coagulase-negative staphylococci in positive blood cultures by real-time PCR with two fluorescence resonance energy transfer probe sets. J Clin Microbiol. 2004;42:5739–5744. [DOI] [PMC free article] [PubMed]
- 23.Schmalzried TP, Kwong LM, Jasty M, Sedlacek RC, Haire TC, O’Connor DO, Bragdon CR, Kabo JM, Malcolm AJ, Harris WH. The mechanism of loosening of cemented acetabular components in total hip arthroplasty: analysis of specimens retrieved at autopsy. Clin Orthop Relat Res. 1992;274:60–78. [PubMed]
- 24.Stoodley P, Kathju S, Hu FZ, Erdos G, Levenson JE, Mehta N, Dice B, Johnson S, Hall-Stoodley L, Nistico L, Sotereanos N, Sewecke J, Post JC, Ehrlich GD. Molecular and imaging techniques for bacterial biofilms in joint arthroplasty infections. Clin Orthop Relat Res. 2005;437:31–40. [DOI] [PubMed]
- 25.Trampuz A, Osmon DR, Hanssen AD, Steckelberg JM, Patel R. Molecular and antibiofilm approaches to prosthetic joint infection. Clin Orthop Relat Res. 2003;414:69–88. [DOI] [PubMed]
- 26.Tunney MM, Patrick S, Curran MD, Ramage G, Anderson N, Davis RI, Gorman SP, Nixon JR. Detection of prosthetic joint biofilm infection using immunological and molecular techniques. Methods Enzymol. 1999;310:566–576. [DOI] [PubMed]
- 27.Tunney MM, Patrick S, Curran MD, Ramage G, Hanna D, Nixon JR, Gorman SP, Davis RI, Anderson N. Detection of prosthetic hip infection at revision arthroplasty by immunofluorescence microscopy and PCR amplification of the bacterial 16 rRNA gene. J Clin Microbiol. 1999;37:3281–3290. [DOI] [PMC free article] [PubMed]
- 28.Tunney MM, Patrick S, Gorman SP, Nixon JR, Anderson N, Davis RI, Hanna D, Ramage G. Improved detection of infection in hip replacements: a currently underestimated problem. J Bone Joint Surg Br. 1998;80:568–572. [DOI] [PubMed]
- 29.van de Belt H, Neut D, Schenk W, van Horn JR, van der Mei HC, Busscher HJ. Infection of orthopedic implants and the use of antibiotic-loaded bone cements: a review. Acta Orthop Scand. 2001;72:557–571. [DOI] [PubMed]
- 30.Yang S, Rothman RE. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis. 2004;4:337–348. [DOI] [PMC free article] [PubMed]