Abstract
The term activator protein (AP)-1 describes homodimeric and heterodimeric transcription factors composed of members of the Jun, Fos, and cAMP response element-binding protein (CREB)/activating transcription factor (ATF) families of proteins. Distinct AP-1 dimers, for instance the prototypical c-Jun:c-Fos and c-Jun:ATF2 dimers, are differentially regulated by signaling pathways and bind related yet distinct response elements in the regulatory regions of AP-1 target genes. Little is known about the dimer-specific regulation of AP-1 activity at the promoter of its target genes. We have previously shown that nTrip6, the nuclear isoform of the LIM domain protein Trip6, acts as an AP-1 coactivator. Moreover, nTrip6 is an essential component of glucocorticoid receptor (GR)-mediated trans-repression of AP-1, in that it mediates the tethering of GR to the promoter-bound AP-1. We have now discovered a striking specificity of nTrip6 actions determined by the binding preference of its LIM domains. We show that nTrip6 interacts only with Fos family members. Consequently, nTrip6 is a selective coactivator for AP-1 dimers containing Fos. nTrip6 also assembles activated GR to c-Jun:c-Fos-driven promoters. Neither nTrip6 nor GR are recruited to a promoter occupied by c-Jun:ATF2. Thus, only Fos-containing dimers are trans-repressed by GR. Thus, the dimer composition of AP-1 determines the mechanism of both the positive and negative regulation of AP-1 transcriptional activity. Interestingly, on a second level of action, GR represses the increase in transcriptional activity of c-Jun:ATF2 induced by c-Jun N-terminal kinase (JNK)-dependent phosphorylation. This repression depends on GR-mediated induction of MAPK phosphatase 1 (MKP-1) expression, which results in c-Jun N-terminal kinase inactivation.
EUKARYOTIC GENE transcription is tightly regulated, such that a specific transcriptional program is turned on in an organ/tissue/cell-specific manner at any given moment. The set of genes that is turned on in response to an environmental cue is determined by the activation of a selective group of transcription factors, which can recognize DNA response elements in the regulatory region of the target genes.
Transcription factors very often belong to families of similar proteins, with related yet distinct DNA binding capacities. The term activator protein (AP)-1 defines such a family of mostly heterodimeric factors, composed of the Jun, Fos, and cAMP response element-binding protein (CREB)/activating transcription factor (ATF) subfamilies (1,2,3). AP-1 dimers bind the consensus 12-O-tetradecanoylphorbol-13-acetate (TPA) response element (TRE) or the related CREB response element (CRE). However, different AP-1 heterodimers have different affinities for the response elements. For instance, the c-Jun:c-Fos dimer binds with high affinity to the 7-bp consensus AP-1 binding site (TRE) TGAGTCA as it occurs e.g. in the collagenase I (MMP1) gene promoter, but only weakly to the 8-bp c-Jun:ATF2 motif (CRE) TTACCTCA, e.g. in the c-jun promoter. In contrast, c-Jun:ATF2 has a very low affinity for the c-Jun:c-Fos heptameric binding site and binds strongly to the octameric binding motif (4,5,6,7,8).
Thus, the transcription of an AP-1 target gene is primarily regulated by the relative expression level of each possible dimerization partner in the cell at a given time. In addition, not only the abundance but also the activation status of the different dimers determines the program of target genes expressed. Each AP-1 subunit is downstream of one or several signal transduction pathways. These activating pathways differ, however; for instance c-Jun:c-Fos is mainly activated by the Ras-Raf-Erk MAPK pathway, whereas both c-Jun and ATF2 are primarily activated by the stress-activated MAPKs c-Jun N-terminal kinase (JNK) and p38 (9,10).
Little is known about the specific regulation of the transcriptional activity of the different AP-1 dimers at the promoter of their target genes. In particular, although several transcriptional coactivators for AP-1 are known (11,12,13,14), the existence of AP-1 dimer-coactivator specificity has not yet been addressed.
Similarly, little is known about the mechanisms negatively regulating the response to specific AP-1 dimers. A physiologically and therapeutically relevant negative control of the AP-1 response is exerted by glucocorticoids. However, little is known about the dimer specificity of this repression. Glucocorticoids act through the GR, which regulates transcription by several mechanisms (15,16). GR is a ligand-activated transcription factor and induces the expression of numerous target genes (17). Liganded GR also interferes with several signal transduction pathways, including the Erk (18,19,20), JNK (21,22,23), and p38 pathways (24,25). Finally, GR directly represses the activity of several other transcription factors, including the AP-1 dimer c-Jun:c-Fos (26,27,28), at the promoter of their target genes. This process is referred to as trans-repression (15). Here, GR is recruited to the DNA-bound transcription factor through protein-protein interactions (tethering), resulting in transcriptional repression. Which of these different mechanisms are relevant for GR-mediated repression of the different AP-1 dimers remains unknown.
Both AP-1 transcriptional activity at promoters and its trans-repression by GR requires the presence of the platform protein nTrip6 (29). nTrip6 is an exclusive nuclear isoform of Trip6, a member of the zyxin family of LIM domain-containing protein expressed at sites of focal adhesion (30,31,32), regulating cell adhesion and migration (33,34,35). nTrip6 acts as a transcriptional coactivator for AP-1 at the promoter of the prototypical c-Jun:c-Fos target gene collagenase I (29). nTrip6 is also essential for the trans-repression of AP-1 by GR, by mediating the tethering of GR to the promoter-bound AP-1 (29).
We here report that nTrip6 specifically interacts with the Fos family members of AP-1, and not with the Jun family members or with ATF2. Consequently, nTrip6 acts as a coactivator for c-Jun:c-Fos, but not for c-Jun:ATF2. The ability of GR to trans-repress AP-1 is strictly dependent on the interaction between the AP-1 dimer and nTrip6; c-Jun:c-Fos is trans-repressed by GR but not c-Jun-ATF2. However, JNK-mediated activation of cJun:ATF2 is also inhibited by GR. This repression occurs in a transactivation-dependent manner, through the induction of MAPK phosphatase 1 (MKP-1) expression, a dual-specificity phosphatase, which dephosphorylates and inhibits activated JNK (36,37,38).
RESULTS
nTrip6 Is a Fos-Selective Coactivator
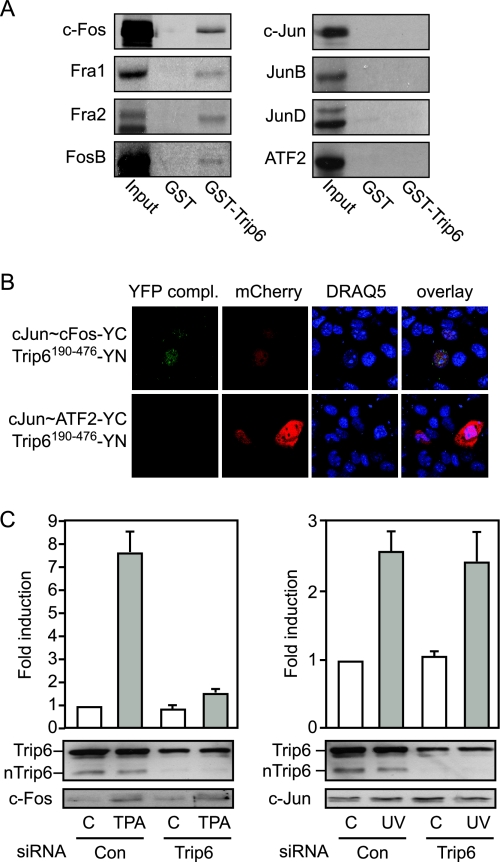
We studied the specificity of nTrip6 interaction with different AP-1 family members in GST pull-down experiments (Fig. 1A). GST-Trip6190–476, which we have previously reported to interact with c-Fos (29), interacted with all the Fos family members, c-Fos, Fra1, Fra2, and FosB, but with none of the Jun proteins, Jun JunB and JunD, or with ATF2 (Fig. 1A). For additional experiments, we focused on two prototypical AP-1 dimers, c-Jun:c-Fos and c-Jun:ATF2. We studied the interaction of nTrip6 with the two dimers in living cells using bimolecular fluorescence complementation (BiFC) (39). We used expression vectors for single-chain AP-1 constructs, in which the coding sequences of two AP-1 family members were fused in frame using a flexible linker (40). This strategy ensured that any detected interaction was specific for the investigated AP-1 dimer, and not due to dimerization of the studied exogenous AP-1 component with any of the endogenous AP-1 family members expressed in the cells. Yellow fluorescent protein (YFP) complementation was observed in the nucleus of 70–80% of the cells cotransfected with the single-chain c-Jun∼c-Fos fused to the C-terminal half of YFP (YC) and Trip6190–476 fused to the N-terminal half of YFP (YN). No complementation was detected between c-Jun∼ATF2-YC and Trip6190–476-YN, although the cells were efficiently transfected, as indicated by the expression of the red fluorescent protein mCherry used as a transfection control (Fig. 1B). The proper expression and subcellular localization of c-Jun∼c-Fos-YC, c-Jun∼ATF2-YC, and Trip6190–476-YN was confirmed by immunofluorescence (supplemental Fig. S1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Thus, nTrip6 interacts with c-Jun:c-Fos, but not with c-Jun:ATF2 in living cells, confirming the results of the in vitro interaction assays. The specificity of nTrip6 interaction suggests that although it exerts a coactivator function for c-Jun:c-Fos (29), it does not coactivate c-Jun:ATF2.
Figure 1.
nTrip6 Is a Fos Selective Coactivator
A, nTrip6 specifically interacts in vitro with Fos family members. 35S-labeled, in vitro-translated Fos family members (c-Fos, Fra1, and 2, FosB), Jun family members (c-Jun, JunB, and JunD), or ATF2 were subjected to GST pull-down assays using either GST or GST-Trip6190–476. Input represents 10% of the in vitro-translated material used in each assay. B, nTrip6 does not interact with c-Jun:ATF2 in living cells. Cos7 cells were transfected with either the single-chain AP-1 c-Jun∼c-Fos fused to YC and Trip6190–476 fused to YN or the single-chain AP-1 c-Jun∼ATF2 fused to YC and Trip6190–476 fused to YN, together with a mCherry expression vector as a transfection control. Nuclei were counterstained with DRAQ5. The reconstituted YFP fluorescence was allowed to mature for 1 h at 30 C before live cell imaging. The YFP complementation was observed in 80–90% of the cells transfected with c-Jun∼c-Fos-YC and Trip6190–476-YN. C, NIH-3T3 cells were transfected with either a control siRNA (Con) or a siRNA targeting Trip6, together with the −1977/−1858uPA-TATA-luciferase reporter gene and Ubi-Renilla. Cells were induced with either TPA or UV 48 h after transfection, and luciferase activities were determined 16 h later. Top, Luciferase activities normalized to Renilla activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate); bottom, cell lysates subjected to Western blotting using anti-Trip6, anti-c-Fos, and anti-c-Jun antibodies.
We addressed the suggested specific coactivator function by studying the effect of silencing nTrip6 on the activation of a reporter gene driven by the minimal AP-1-dependent enhancer of the urokinase-type plasminogen activator (uPA) gene (−1977/−1858uPA-TATA-Luc). uPA is a target gene for both cJun:c-Fos and c-Jun:ATF2, which specifically bind their respective response elements in the uPA gene enhancer (41,42,43). The uPA promoter is therefore a suitable model to study the regulation of c-Jun:c-Fos and c-Jun:ATF2 within the same promoter context. In NIH-3T3 cells transfected with the uPA reporter gene together with a control small interfering RNA (siRNA), treatment with the phorbol ester TPA, which activates cJun:c-Fos, induced the expression of the reporter gene. UV irradiation also induced the uPA reporter gene (Fig. 1C). This induction is mediated by the JNK pathway, which activates c-Jun:ATF2, and requires the octameric c-Jun:ATF2 binding site of the uPA enhancer (43). In cells transfected with the Trip6-specific siRNA, TPA-induced activation of the reporter gene was severely reduced, whereas UV-induced activation was not affected (Fig. 1C). Western blot analysis confirmed that the siRNA down-regulated nTrip6 without affecting c-Fos and c-Jun levels (Fig. 1C). These results show that nTrip6 is a coactivator for c-Jun:c-Fos but not for c-Jun:ATF2, in agreement with the protein-protein interaction results.
Only Fos-Containing AP-1 Dimers Are Trans-Repressed by GR
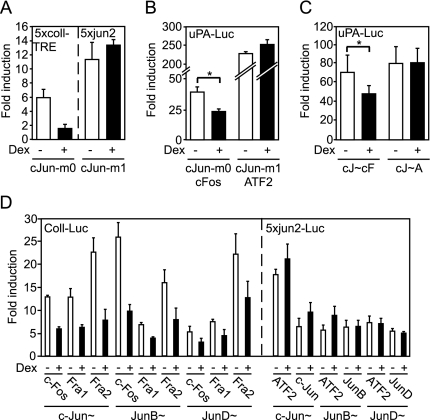
nTrip6 is not only a coactivator for c-Jun:c-Fos but is also required for the trans-repression of AP-1 by GR (29). Because the c-Jun:ATF2 dimer does not interact with nTrip6 and is not coactivated by nTrip6, we predicted that it would not be trans-repressed by GR. We first tested this hypothesis in reporter gene assays in chicken embryonic fibroblasts (CEFs) transfected with c-Jun mutants that preferentially dimerize and bind to DNA either as Jun:Fos or as Jun:ATF2. The c-Jun-m0 mutant preferentially dimerizes with c-Fos, whereas the c-Jun-m1 mutant preferentially dimerizes with ATF2 (8,44,45). The reporter genes used were 5xcoll-TRE-tata (26) and 5xjun2-tata (7), which have been shown to specifically respond to overexpressed c-Jun-m0 and c-Jun-m1 in CEFs, respectively (8,44,45). The induction of 5xcoll-TRE-tata by c-Jun-m0, the c-Fos-seeking mutant, was repressed by coexpressed GR upon treatment with the glucocorticoid dexamethasone, whereas the induction of 5xjun2-tata by c-Jun-m1, the ATF2-seeking mutant, was not repressed by GR (Fig. 2A). We next tested GR-mediated repression of the c-Jun mutants on the minimal uPA reporter gene. c-Jun-m0 and c-Jun-m1 both induced the reporter gene when cotransfected with their preferential dimerization partner c-Fos and ATF2, respectively. Only the activity of c-Jun-m0, and not that of c-Jun-m1, was repressed by GR (Fig. 2B). We confirmed these results using the single-chain AP-1 constructs. Overexpression of both c-Jun∼c-Fos and c-Jun∼ATF2 activated the −1977/−1858uPA reporter gene in the absence of c-Jun phosphorylation (supplemental Fig. S2A). c-Jun∼c-Fos-mediated activation was repressed by dexamethasone, whereas c-Jun∼ATF2-mediated activation was not repressed by dexamethasone (Fig. 2C). Similar results were obtained using a fragment of the ATF3 enhancer/promoter (−624/+35ATF3-Luc) containing both a TRE and a CRE (46,47,48). The activation mediated by overexpression of the single-chain c-Jun∼c-Fos was repressed by GR, whereas the activation by c-Jun∼ATF2 was not (supplemental Fig. S2B). Altogether, these results show that the AP-1 dimer c-Jun:c-Fos interacting with nTrip6, is trans-repressed by GR, whereas the dimer c-Jun:ATF2, not interacting with nTrip6, is not trans-repressed by GR. We extended these observations to other AP-1 single-chain constructs (Fig. 2D). All the single-chain AP-1 constructs containing a Fos family member (c-Fos, Fra1, or Fra2) were repressed by GR, irrespectively of the dimerization partner (c-Jun, JunB, or JunD). None of the Jun family homodimers and their heterodimers with ATF2 were repressed by GR (Fig. 2D).
Figure 2.
Only Fos-Containing AP-1 Dimers Are Trans-Repressed by GR
A, CEFs were cotransfected with pRSV-GR together with either the 5xcoll-TRE-TATA-luciferase reporter gene and an expression vector for the c-Fos seeking c-Jun mutant (c-Jun-m0) or the 5xjun2-TATA-luciferase reporter gene and an expression vector for the ATF2 seeking c-Jun mutant (c-Jun-m1). Luciferase activities were determined after treatment with dexamethasone (Dex) or solvent and are plotted as fold induction (mean ± sd of five independent experiments). B and C, HeLa cells were transiently cotransfected with the −1977/−1858uPA-TATA-luciferase reporter gene and Ubi-Renilla, together with expression vectors for either c-Jun-m0 and c-Fos or c-Jun-m1 and ATF2 (B) or expression vectors for either the single-chain AP-1 c-Jun∼c-Fos (cJ∼cF) or the single-chain AP-1 c-Jun∼ATF2 (cJ∼A) (C). Cells were treated with dexamethasone (Dex) as indicated. Luciferase activities normalized to Renilla activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate; *, P < 0.05). D, HeLa cells were transiently cotransfected with either the −517/+63-collagenaseI-luciferase reporter gene (Coll-Luc) or the 5xjun2-TATA-luciferase reporter gene, together with the indicated single-chain AP-1. Normalized luciferase activities were determined after treatment with dexamethasone (Dex) or solvent and are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate).
nTrip6 and GR Are Not Recruited to Promoter-Bound c-Jun:ATF2
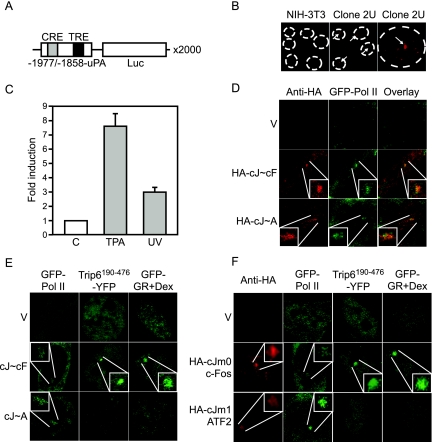
We have previously reported that the function of nTrip6 in GR-mediated repression of AP-1 is to provide a platform for the tethering of GR to the AP-1-occupied promoter. We thus hypothesized that the lack of trans-repression of c-Jun:ATF2 by GR might be due to the lack of recruitment of nTrip6 to the promoter, with a consequent lack of tethering of GR. To directly visualize nTrip6 and GR recruitment, we generated a reporter cell line containing an integrated array of multiple copies of the −1977/−1858uPA-Luc reporter gene. The array was generated by transfecting the reporter plasmid together with the pΔBN-AR1 plasmid (49). pΔBN-AR1 contains a mammalian replication initiation origin and a matrix attachment region from the Chinese hamster dhfr gene; when integrated into the genome, it initiates events similar to gene amplification in cancer cells, leading to tandem repeats of up to 10,000 copies (49,50). The high local concentration of binding sites on the promoter of the amplified gene unit enables the direct visualization of binding to the gene array of protein tagged with a fluorescent marker, as previously shown for an nuclear factor-κB-dependent array cell line (51). Using this amplification method, NIH-3T3 fibroblasts carrying several hundred integrated uPA-Luc plasmid copies in discrete loci were obtained (Fig. 3A). One of the cell clones, clone 2U, was estimated by real-time PCR to have an integrated array of about 2000 gene units (data not shown). The presence of the uPA-Luc array was confirmed by DNA fluorescent in situ hybridization using a fragment of the luciferase coding sequence as a probe. The staining was restricted to a single spot within the nucleus of the 2U cells and was seen in 100% of the cells, whereas only background staining was visible in the parental NIH-3T3 cells (Fig. 3B). Both TPA treatment and UV irradiation induced luciferase activity in clone 2U (Fig. 3C), suggesting that both c-Jun:c-Fos and c-Jun:ATF2 are able to bind to their cognate response elements on the array and to activate transcription from the uPA-Luc gene array. We then directly studied the recruitment of the two AP-1 dimers and their ability to mediate the recruitment of the RNA polymerase II (Pol II) to the array (Fig. 3D). Overexpression of an hemagglutinin (HA)-tagged version of the c-Jun∼c-Fos single-chain construct led to its recruitment to the array, as shown by the enrichment to a single bright spot in the nucleus after immunostaining with an anti-HA antibody. Moreover, c-Jun∼c-Fos mediated the recruitment of Pol II to the array, as shown by the colocalization of the HA staining with green fluorescent protein (GFP)-Pol II (Fig. 3D). Similarly, cotransfected HA-c-Jun∼ATF2 and GFP-Pol II were corecruited to the array. These specific enrichments to the array were observed in 70–80% of the transfected cells. GFP-Pol II was not enriched to the array when transfected alone, as shown by the homogenous nuclear fluorescence (Fig. 3D). These results confirm that the uPA reporter gene array is functional and can respond to c-Jun:c-Fos and c-Jun:ATF2.
Figure 3.
nTrip6 and GR Are Not Tethered to c-Jun:ATF2-Occupied Promoter
A, Schematic representation of the c-Jun:c-Fos- and c-Jun:ATF2-regulated gene unit amplified in the clone 2U. B, Clone 2U and parental NIH-3T3 fibroblasts were subjected to DNA in situ hybridization using a fluorescently labeled cDNA probe complementary to the luciferase coding sequence. A single gene array is visible in 100% of the 2U cells. C, The uPA gene array is functional. 2U cells were treated with either solvent alone (C) or TPA or UV irradiated. Luciferase activities were determined 6 h later and are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate). D, AP-1 is functionally recruited to the uPA gene array. 2U cells were transfected with GFP-Pol II, together with the empty vector (V), an expression vector for an HA-tagged c-Jun∼c-Fos single chain (HA-cJ∼cF), or an expression vector for an HA-tagged c-Jun∼ATF2 single chain (HA-cJ∼A). The localization of the AP-1 single-chain proteins was determined by immunofluorescence using an anti-HA antibody. The enrichment of the AP-1 proteins and of GFP-Pol II was observed in 70–80% of the cotransfected cells. E and F, nTrip6 and GR are not tethered to c-Jun:ATF2 occupied promoter. E, 2U cells were transfected with empty vector, an expression vector for c-Jun∼c-Fos single chain (cJ∼cF), or an expression vector for c-Jun∼ATF2 single chain (cJ∼A), together with an expression vector for GFP-Pol II, an expression vector for Trip6190–476 fused to YFP (YFP-Trip6190–476), or an expression vector for GR fused to GFP. Cells were treated with dexamethasone for 30 min and imaged by confocal microscopy. Nuclei of representative cells are shown. F, Clone 2U uPA gene array cells were cotransfected with either GFP-Pol II, Trip6190–476 fused to YFP (YFP-Trip6190–476), or GR fused to GFP, together with empty vector (V), HA-tagged c-Jun-m0 mutant (HA-cJm0, c-Fos seeking) and c-Fos, or HA-tagged c-Jun-m1 mutant (HA-cJm1, ATF2 seeking) and ATF2. Cells were treated with dexamethasone for 30 min (Dex). The localization of the c-Jun mutants was determined by immunofluorescence using an anti-HA antibody. Cells were imaged by confocal microscopy. Nuclei of representative cells are shown.
We then studied the recruitment of nTrip6 and GR to the array. When transfected alone, Trip6190–476-YFP showed a homogeneous distribution in the nucleus, without specific local enrichment. Cotransfection with c-Jun∼c-Fos promoted the recruitment of Trip6190–476-YFP to the promoter array, whereas cotransfection with c-Jun∼ATF2 did not (Fig. 3E). These results match our observation that nTrip6 is a coactivator for c-Jun:c-Fos but not for c-Jun:ATF2. GFP-GR was located in the cytosol and translocated to the nucleus upon dexamethasone treatment (not shown). GFP-GR was recruited to the uPA gene array upon dexamethasone treatment in cells cotransfected with c-Jun∼c-Fos, but not in cells cotransfected with c-Jun∼ATF2 or with vector only (Fig. 3E). Similar results were obtained using another clone of uPA gene array cells (supplemental Fig. S3).
We confirmed these specific recruitments using the c-Jun dimerization mutants. Cotransfection of either HA-c-Jun-m0 or HA-c-Jun-m1 with their dimerization partner c-Fos and ATF2, respectively, led to their enrichment to the array, as shown by immunostaining with an anti-HA antibody, and both c-Jun mutants mediated the recruitment of GFP-Pol II to the array (Fig. 3F). Transfection of c-Jun-m0 together with c-Fos promoted the recruitment of Trip6190–476-YFP to the array as well as the tethering of GFP-GR upon dexamethasone treatment. Conversely, transfection of c-Jun-m1 and ATF2 did not promote the recruitment of Trip6190–476-YFP and GFP-GR to the array (Fig. 3). Thus, GR is not tethered to the c-Jun:ATF2-bound promoter. Because the tethering of GR to the AP-1-bound promoter is mediated via an interaction with nTrip6 (29), the lack of interaction between c-Jun:ATF2 and nTrip6 is most probably responsible for the lack of GR tethering to the c-Jun:ATF2 at the promoter. These results provide a mechanistic explanation for the absence of GR-dependent trans-repression of the c-Jun:ATF2 AP-1 dimer.
JNK-Mediated Activation of c-Jun:ATF2 Is Repressed by GR
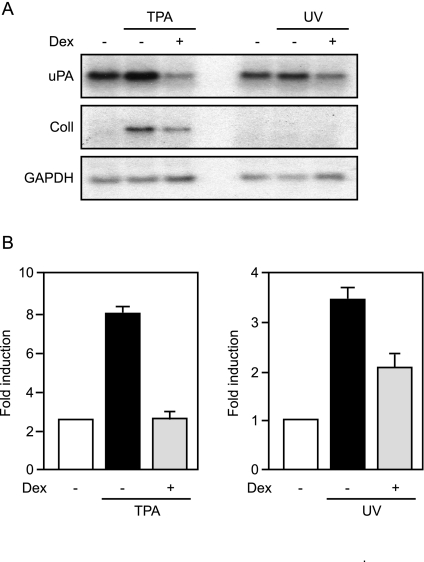
To confirm the data obtained with the reporter gene constructs, we studied dexamethasone-mediated repression of endogenous uPA, which is also targeted by both c-Jun:c-Fos and c-Jun:ATF2 (Fig. 4A). The Fos-dependent TPA-mediated induction of uPA mRNA, as well as of collagenase I mRNA, was repressed by dexamethasone, as expected. Surprisingly, UV-mediated induction of uPA mRNA was also repressed by dexamethasone (Fig. 4A). These results were confirmed using ATF3, another cJun:c-Fos and c-Jun:ATF2 target gene. As in the case of uPA, UV-induced ATF3 mRNA was repressed by the glucocorticoid (supplemental Fig. S4A). These results are in apparent contradiction with the observation that GR does not trans-repress c-Jun:ATF2 (see above). Both the uPA and ATF3 genes are transcriptionally controlled through complex enhancers, with binding sites for transcription factors other than c-Jun:c-Fos and c-Jun:ATF2 (47,52), which could be trans-repressed by GR. To test this option, we studied the effect of GR on TPA- and UV-mediated induction of the minimal uPA reporter gene containing only the c-Jun:c-Fos and c-Jun:ATF2 binding sites (Fig. 4B). As expected, the Fos-dependent TPA-induced expression of the reporter gene was repressed by GR. The c-Jun:ATF2-dependent UV-mediated induction was also repressed by GR (Fig. 4B). Similar results were obtained using the ATF3 minimal reporter gene containing only c-Jun:c-Fos and c-Jun:ATF2 binding sites (supplemental Fig. S4B).
Figure 4.
UV-Induced uPA Expression Is Repressed by GR
A, NIH-3T3 cells were either treated with TPA or UV irradiated in the presence or absence of dexamethasone (Dex). Total RNA was prepared and subjected to Northern blotting using probes specific for uPA, collagenase I (Coll), or GAPDH. B, HeLa cells were cotransfected with the −1977/−1858uPA-TATA-luciferase reporter gene and Ubi-Renilla and either treated with TPA or UV irradiated in the presence or absence of dexamethasone (Dex). Luciferase activities normalized to Renilla activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicates).
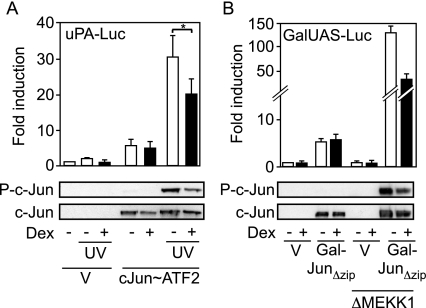
Thus, the activation of c-Jun:ATF2 target genes by UV irradiation treatment is repressed by GR, although the c-Jun:ATF2 dimer is not directly trans-repressed by GR. This can only be explained by an action of GR before promoter binding, thus on UV-mediated activation of c-Jun:ATF2 (Fig. 5A). To avoid the complication of several Jun family members in cells, we tested c-Jun-deficient mouse embryonic fibroblasts (MEFs) and found that UV irradiation only marginally activated the minimal uPA reporter gene, showing that the other Jun family members that might be expressed in these cells are at least not strong inducers of the reporter gene under the condition of our assay. The induction of the reporter gene by overexpression of the single-chain cJun∼ATF2 was not repressed by GR, as previously observed in other cell types (see above). UV irradiation strongly enhanced the activation by cJun∼ATF2, reflecting the activating phosphorylation of the dimer as seen on the Western blot using a phosphorylation-dependent anti-c-Jun antibody. Importantly, the UV-mediated phosphorylation and activation of cJun∼ATF2 was significantly repressed by dexamethasone (Fig. 5A). We confirmed this result using another approach. We used a GAL4 DNA binding domain fusion of a c-Jun mutant lacking the dimerization domain (Gal-JunΔzip) to directly measure the transcriptional activity of c-Jun in cells with an integrated GAL4-dependent reporter gene (53). The activating phosphorylation of Gal-JunΔzip was achieved by cotransfection of a constitutively active mutant of the MEK kinase 1 (ΔMEKK1), a direct activator of MAPK kinase 4, which in turn activates JNK (54). As expected, in the absence of ΔMEKK1, the induction of the reporter gene mediated by overexpression of Gal-JunΔzip was not repressed by GR (Fig. 5B). When transfected alone, ΔMEKK1 did not activate the reporter gene. ΔMEKK1 promoted the phosphorylation of Gal-JunΔzip and a strong increase in Gal-JunΔzip-mediated transcription. The ΔMEKK1-mediated phosphorylation and increase in c-Jun activity was partially repressed by dexamethasone (Fig. 5B).
Figure 5.
JNK-Stimulated c-Jun:ATF2 Activity Is Repressed by GR
A, c-Jun-deficient MEFs were transfected with the −1977/−1858uPA-TATA-luciferase reporter gene (uPA-Luc), together with Ubi-Renilla, and either empty vector (V) or an expression vector for c-Jun∼ATF2 single chain. Cells were UV irradiated in the presence or absence of dexamethasone (Dex). Top, Luciferase activities normalized to Renilla activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate, *, P < 0.05); bottom, the phosphorylation of the c-Jun moiety of the c-Jun∼ATF2 single chain was assessed by Western blotting using a phosphospecific antibody. The membrane was stripped and reprobed with a phosphorylation state-independent anti-c-Jun antibody. B, HEK-293 cells with an integrated GAL4-responsive reporter gene (GalUAS-Luc) were transfected with empty vector (V) or an expression vector for a GAL4 DNA binding domain fusion of c-Jun lacking its dimerization domain (Gal-JunΔzip), together with either empty vector or an expression vector for ΔMEKK1, and treated with dexamethasone (Dex), as indicated. Top, Luciferase activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate); bottom, the phosphorylation of the c-Jun moiety of Gal-JunΔzip was assessed by Western blotting using a phosphospecific antibody. The membrane was stripped and reprobed with a phosphorylation state-independent anti-c-Jun antibody.
GR Inhibits JNK-Mediated Activation of c-Jun:ATF2 through a Transactivation-Dependent Mechanism
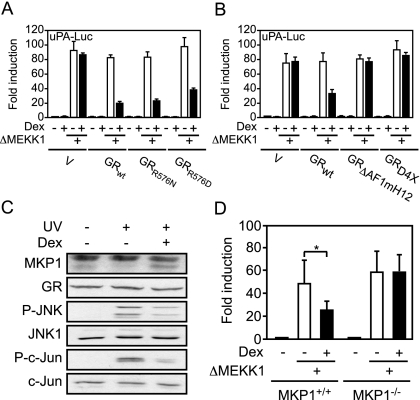
Altogether, c-Jun:ATF2 does not interact with nTrip6 and is consequently not trans-repressed by GR. However, the UV- and ΔMEKK1-mediated increase in c-Jun:ATF2 activity is inhibited by GR. UV- and MEKK1-mediated induction of uPA is dependent on JNK activation (43). One could therefore hypothesize that GR inhibits JNK-mediated phosphorylation and activation of the c-Jun:ATF2 dimer. GR has been shown to inhibit JNK activity through a nongenomic mechanism (21). This inhibition does not require the trans-activation function of GR (55) but strictly depends on a direct protein-protein interaction between GR and JNK (56). We thus studied the effect of two GR mutants, GRR576N and GRR576D, unable to interact with and inhibit JNK (56) on JNK-mediated activation of the minimal uPA reporter gene (Fig. 6A). In GR-deficient Cos7 cells, ΔMEKK1-mediated induction of the reporter gene was not inhibited by dexamethasone in the absence of cotransfected GR. GRwt, as well as GRR576N and GRR576D repressed the ΔMEKK1-mediated transcriptional induction (Fig. 6A), speaking against the proposed mechanism of direct interaction. The two GR mutants were as potent as GRwt to transactivate a reporter gene driven by the glucocorticoid response element-containing mouse mammary tumor virus long terminal repeat (MMTV-Luc, supplemental Fig. S5A), and to trans-repress the TPA-induced reporter gene driven by the c-Jun:c-Fos-dependent collagenase I promoter (Coll-Luc, supplemental Fig. S5B). Thus, GR does not inhibit the JNK-mediated phosphorylation and activation of c-Jun:ATF2 through a direct interaction with JNK.
Figure 6.
The Induction of MKP-1 by GR Mediates the Repression of JNK-Activated c-Jun:ATF2
A, GR does not inhibit the JNK-mediated activation of c-Jun:ATF2 through a direct interaction with JNK. Cos7 cells were transfected with the −1977/−1858uPA-TATA-luciferase reporter gene (uPA-Luc), together with Ubi-Renilla, and with either empty vector or expression vectors for wild-type GR (GRwt) or two GR mutants unable to interact with and inhibit JNK (GRR576N and GRR576D), in the presence or absence of ΔMEKK1. Cells were treated with dexamethasone (Dex) as indicated. Luciferase activities normalized to Renilla activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate). B, The repression of JNK-mediated activation of c-Jun:ATF2 requires the transactivation function of GR. Cos7 cells were transfected with the −1977/−1858uPA-TATA-luciferase reporter gene (uPA-Luc), together with Ubi-Renilla, and either empty vector or expression vectors for wild-type GR (GRwt) or two GR mutants unable to transactivate (GRΔAF1mH12 and GRRD4X), in the presence or absence of ΔMEKK1. Cells were treated with dexamethasone (Dex) as indicated. Luciferase activities normalized to Renilla activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate). C, GR-mediated induction of MKP-1 is associated with an inhibition of JNK and c-Jun phosphorylation. NIH-3T3 fibroblasts were treated with dexamethasone (Dex) for 3 h and then UV irradiated as indicated. MKP-1 expression was assessed 20 min later by Western blotting (*, nonspecific band). GR was used as a loading control. JNK and c-Jun phosphorylation (p-JNK and p-c-Jun) was assessed by Western blotting using phosphospecific antibodies. The membranes were stripped and reprobed with phosphorylation state-independent anti-JNK1 and anti-c-Jun antibodies. D, The induction of MKP-1 by GR mediates the repression of JNK-activated c-Jun:ATF2. Primary MEFs from MKP-1 knockout mice (MKP-1−/−) or littermate wild-type controls (MKP-1+/+) were transiently transfected with the −1977/−1858uPA-TATA-luciferase reporter gene (uPA-Luc), together with Ubi-Renilla, and with either empty vector or an expression vector for ΔMEKK1. Cells were treated with dexamethasone (Dex) as indicated. Luciferase activities normalized to Renilla activities are plotted as fold induction (mean ± sd of one representative experiment performed in triplicate; *, P < 0.05).
Because the repression of phosphorylated cJun:ATF2 involves neither trans-repression nor a direct inhibition of JNK, we studied whether it might require the trans-activation function of GR. In Cos7 cells, GRwt repressed the induction of the minimal uPA reporter gene by ΔMEKK1. However, two trans-activation-deficient GR mutants, GRΔAF1mH12 (29) and GRD4X (57), failed to repress ΔMEKK1-mediated activation of the reporter gene (Fig. 6B). GRΔAF1mH12 was unable to transactivate MMTV-Luc, and GRD4X had a severely reduced ability to induce MMTV-Luc (supplemental Fig. S5C). Both mutants still trans-repressed TPA-induced Coll-Luc (supplemental Fig. S5D). Thus, the repression by GR of JNK-activated c-Jun:ATF2 does not involve trans-repression nor the direct inhibition of JNK but requires trans-activation by GR.
MKP-1 Induction Mediates the Inhibition of JNK-Activated c-Jun:ATF2 by GR
A GR target gene that might be responsible for the inhibition of JNK-activated c-Jun:ATF2 is MKP-1, also known as dual-specificity phosphatase 1 (DUSP1). Indeed, this dual-specificity phosphatase, which dephosphorylates and inhibits activated JNK (36,37,38), is induced by GR through a classical trans-activation mechanism (19). We studied MKP-1 expression in conditions of c-Jun:ATF2 repression by GR (Fig. 6C). MKP-1 was barely detectable in untreated cells or in UV-irradiated cells. Cotreatment of the cells with dexamethasone induced MKP-1 expression (Fig. 6C). Dexamethasone treatment resulted in a partial inhibition of UV-induced JNK phosphorylation, associated with a partial inhibition of c-Jun phosphorylation (Fig. 6C). Thus, the glucocorticoid-induced expression of MKP-1 correlates with an inhibition of JNK and c-Jun phosphorylation. We then studied the contribution of MKP-1 in the inhibition of JNK-stimulated transcriptional activity of c-Jun:ATF2. In transient transfection experiments in primary MEFs derived from MKP-1-deficient mice, dexamethasone did not repress the ΔMEKK1-mediated induction of the minimal uPA reporter gene. The repression was observed in primary MEFs derived from wild-type littermates (Fig. 6D). These results demonstrate that the induction of MKP-1 expression by GR is responsible for the repression of JNK-stimulated transcriptional activity of c-Jun:ATF2.
DISCUSSION
The AP-1 family of heterodimeric transcription factors has been extensively studied regarding the physiological and pathological functions of the different dimers (2,3). However, little is known about the specific regulation of different dimers when bound to DNA. In particular, an open question is whether their function requires dimer-specific coactivators. Some known AP-1 coactivators exert their function independently of the dimer composition. For instance, CREB-binding protein/p300 and Src1 interact with and coactivate c-Jun and c-Fos as well as ATF2 (11,13,58,59,60). For other AP-1 coactivators, the specificity has not been addressed. We report here that nTrip6, the nuclear isoform of the LIM domain-containing protein Trip6, is a Fos family-specific coactivator. nTrip6 interacted in vitro with c-Fos, FosB, Fra1, and Fra2 but not with any of the Jun family members or with ATF2. In living cells, nTrip6 interacted with the c-Jun:c-Fos dimer but not with the c-Jun:ATF2 dimer. nTrip6 was required for c-Jun:c-Fos transcriptional activity, as already reported (29), but not for c-Jun:ATF2 activity, as shown by RNA interference experiments. Furthermore, nTrip6 was recruited together with c-Jun:c-Fos, but not with c-Jun:ATF2, to the uPA enhancer, which is responsive to both dimers (41,42,43). Thus, nTrip6 is a specific coactivator for Fos-containing AP-1 dimers.
nTrip6 interacts with c-Fos through its C-terminal LIM domains (29). Other LIM domain proteins are known for their coactivator-like functions (61,62,63). For instance, lipoma-preferred partner (LPP), another zyxin-family LIM domain protein closely related to Trip6, acts as a coactivator for the ETS domain transcription factor polyomavirus enhancer activator 3 (PEA3) (64). Hic-5, a member of the paxillin family, is a coactivator for androgen receptor (65) and for GR (66). Because nTrip6 does not carry any known coactivator domain or function, it presumably exerts its action as a coactivator platform, enabling the recruitment of other coactivators. The AP-1 dimer specificity of nTrip6 interaction might therefore confer specificity in the assembly of other coactivators, depending on the dimer composition. Alternatively, one cannot exclude the possibility that another LIM domain protein plays a similar role for the coactivation of c-Jun:ATF2.
The second question we addressed in this report is whether there is a dimer specificity for GR-mediated trans-repression of AP-1. Several pieces of evidence have suggested that c-Fos and not c-Jun is the target of GR in the trans-repression of AP-1. 1) GR was co-immunoprecipitated with c-Jun after cross-linking (26,28). However, down-regulation of c-Fos expression by an antisense construct reduced the amount of c-Jun co immunoprecipitated with GR (67), suggesting that c-Fos, and not c-Jun, directly or indirectly interacted with GR. 2) In vitro transcription experiments showed that c-Fos is essential for GR-mediated repression of AP-1 (68). 3) At the composite response element of the proliferin gene, GR repressed c-Jun:c-Fos dimers, but not c-Jun homodimers (69,70). 4) The c-Jun:c-Fos-mediated activation of a reporter gene driven by the AP-1-dependent collagenase I enhancer is repressed by GR, but not that mediated by c-Jun homodimers (71). All these observations point toward a requirement for c-Fos, or another Fos family member, in GR-mediated repression of AP-1. By using single-chain AP-1 dimers (40) and c-Jun mutants preferentially dimerizing with either c-Fos or ATF2 (8,44,45), we confirmed here that c-Fos is the target for GR-mediated trans-repression of AP-1. We have reported that nTrip6 is essential for the repression of AP-1 by GR (29). nTrip6 serves as a tethering platform for GR at the promoter of AP-1 target genes. We show here that AP-1 dimers not containing c-Fos, as exemplified by c-Jun:ATF2, do not interact with nTrip6 and cannot recruit GR to a c-Jun:ATF2 target promoter. We propose that the lack of interaction with nTrip6 is causal to the inability of GR to tether to and to trans-repress chromatin-bound c-Jun:ATF2 and other AP-1 dimers not containing c-Fos or other Fos family members.
Although GR did not trans-repress c-Jun:ATF2, dexamethasone reduced the enhancement of c-Jun:ATF2 activity upon phosphorylation by JNK. One hypothesis could be that the JNK-mediated phosphorylation of c-Jun:ATF2 promotes its interaction with nTrip6, thus enabling GR tethering and trans-repression. We were, however, unable to detect any interaction between phosphorylated c-Jun:ATF2 and nTrip6 (data not shown). Thus GR cannot directly trans-repress phosphorylated c-Jun:ATF2 transcriptional activity but rather targets the activation of c-Jun:ATF2 by JNK. It has been reported that GR inhibits JNK activity through a rapid mechanism (21), which does not require the trans-activation function of GR (55) but relies on the hormone-induced interaction between GR and JNK (56). However, two GR constructs mutated in the JNK docking site, and therefore unable to interact with JNK and to inhibit its activity (56), still repressed the JNK-mediated activation of c-Jun:ATF2. Furthermore, both GRD4X, a dimerization-deficient GR mutant that fails to bind DNA and to trans-activate (57), and GRΔAF1mH12, devoid of its trans-activation functions (29), failed to repress JNK-induced c-Jun:ATF2 activity. Thus, the putative nongenomic inhibition of JNK by direct interaction with GR is irrelevant for the inhibition of phosphorylated c-Jun:ATF2.
MKP-1 can dephosphorylate the three classes of MAPKs, Erk1/2, p38, and JNK (36,37,38). We have previously shown that MKP-1 is a GR target gene, whose induction by glucocorticoids is responsible for the dephosphorylation of Erk1/2 (19). Similarly, MKP-1 induction by GR mediates the dephosphorylation of p38 and JNK (72,73,74,75). We here provide evidence that the GR-induced, MKP-1-mediated dephosphorylation of JNK is necessary for the repression of c-Jun:ATF2 by glucocorticoids.
Thus, GR employs different mechanisms to repress AP-1, depending on the AP-1 dimer composition. c-Jun:c-Fos and other Fos-containing dimers are trans-repressed by GR by direct protein-protein interaction, whereas c-Jun:ATF2 phosphorylation and activation is inhibited by GR in a trans-activation-dependent manner. In trans-repression, GR has been suggested to inhibit a relatively late step, after preinitiation has occurred (76,77). The repression of c-Jun:ATF2 requires the de novo expression of MKP-1 and might thus be considered as a late effect in the kinetics of glucocorticoid action. However, the GR-mediated inhibition of c-Jun phosphorylation might be considered as a very early step, when considering the kinetics of c-Jun-ATF2 activation. In resting conditions, c-Jun is constitutively bound to the promoter of AP-1 target genes, associated with the corepressors nuclear receptor corepressor (NCoR) and histone deacetylase 3 (HDAC3). JNK-mediated phosphorylation of c-Jun promotes a de-repression through the release of NCoR and HDAC3 from the promoter (53,78). GR-mediated inhibition of c-Jun phosphorylation thus most probably inhibits this de-repression step and prevents preinitiation.
At the level of the organism, in response to a stress condition, glucocorticoids are released in the circulation to prevent an excessive response or to terminate the response. Because the AP-1 response to stress relies on the activation of different dimers, GR probably employs both mechanisms, trans-repression and induction of MKP-1 expression, to terminate the AP-1 response. Trans-repression is indeed known to be relevant in vivo for the termination of a stress response (79,80,81). In parallel, the increase in circulating glucocorticoids may also lead to an induction of MKP-1 expression, which would then contribute to the termination of the c-Jun:ATF2 AP-1 response.
In conclusion, this work uncovers specificity in the coactivation and repression of different AP-1 dimers at the promoter of their target genes. The nature of the dimer determines the use of nTrip6 as a coactivator platform. The presence of nTrip6 in turn determines the mechanism by which GR represses AP-1.
MATERIALS AND METHODS
Plasmids
For in vitro translation, pCS2-Fra1, pCS2-Fra2, pBAT-c-Jun, and pBAT-JunB were from Peter Angel (Deutsches Krebsforschungszentrum, Heidelberg, Germany). pGEM-c-Fos, and pBAT-ATF2 were described previously (8). pEP-FosB was from Marc Timmers (University Medical Center Utrecht, Utrecht, The Netherlands). GST-Trip6190–476 has been described (29). pBiFC-Trip6190–476-YN was constructed by PCR amplification of Trip6190–476 and cloning into pBiFC-YN155 (39). Trip6190–476-YFP was generated as follows. The YFP coding sequence was PCR amplified and cloned between the NotI and XbaI sites of pcDNA3.1 (Invitrogen, Karlsruhe, Germany) to generate pcDNA3.1-YFP, and Trip6190–476 sequence was PCR amplified to delete the stop codon and cloned between the KpnI and NotI sites of pcDNA3.1-YFP. The pCG expression vectors for the single-chain AP-1 constructs (Bakiri) were from Latifa Bakiri (IMP, Vienna, Austria). c-Jun∼ATF2 and c-Jun∼c-Fos coding sequences were PCR amplified and coned into pBiFC-YC155 (39) to obtain pBiFC-c-Jun∼ATF2-YC and pBiFC-c-Jun∼c-Fos-YC. pRSV-c-Jun-mo and pRSV-c-Jun-m1 were previously described (8). pUbi-cJunΔzip and pCMV5-ΔMEKK1 (53) were from Carsten Weiss (Forschungszentrum Karlsruhe, Karlsruhe, Germany). mCherry coding sequence was PCR-amplified from pRSET-B-mCherry (82), a gift from Roger Y. Tsien (University of California, San Diego, La Jolla CA), and recloned into pcDNA3.1 (Invitrogen) to obtain pcDNA3.1-mCherry. pEGFP-Pol II, encoding an EGFP fusion of the largest subunit of Pol II (83), was from Kimihiko Sugaya (National Institute of Radiological Sciences, Chiba, Japan). The GFP-GR construct, pRSV-GR, and pRSV-GRD4X were from Andrew C. B. Cato (Forschungszentrum Karlsruhe). pRSV-GRmH12ΔAF-1 has been described (29). pMTG-myc-GR, pMTG-myc-GRR546N, and pMTG-myc-GRR546D (56) were from Carme Caelles (Institut de Recerca Biomèdica de Barcelona, Barcelona, Spain). The luciferase reporter constructs were as described: MMTV-Luc (84), Coll-Luc (85), 5Xcoll-TRE-TATA-Luc, and 5Xjun2-TATA-Luc (8) and GAL-UAS-Luc (53). −624/+35ATF3-Luc was generated by PCR amplification and subcloning in pGL3 (Promega, Mannheim, Germany). −1977/−1858uPA-TATA-Luc was generated as follows: the XhoI/SmaI- fragment of pBL-tata-CAT5 (26) was replaced by the XhoI/(blunted) BamHI fragment of pGL3 basic (Promega), resulting in pBL-TATA-Luc. −1977/−1858uPA-TATA-Luc was subsequently constructed by replacing the BamHI-KpnI fragments of −1977/−1858-uPA-TK-CAT4 (42) by the BamHI-KpnI fragment of pBL-TATA-Luc. The Ubi-Renilla construct has been described (29). pΔBN-AR1 (49) for the generation of the uPA array cell lines was from Noriaki Shimizu (Hiroshima University, Hiroshima, Japan).
Cell Culture and Transfections
CEFs chronically and stably infected by the R-c-Jun-m1 or R-c-Jun-m0 retroviruses (8) were transfected secondarily using Fugene 6 (Roche, Grenzach-Wyhlen, Germany). NIH-3T3 fibroblasts, HeLa and Cos7 cells, and immortalized c-Jun−/− and wild-type MEFs (obtained from Carsten Weiss, Forschungszentrum Karlsruhe) were cultured in DMEM supplemented with 10% fetal calf serum and transfected using Lipofectamine 2000 (Invitrogen). To obtain MKP-1−/− primary MEFs and wild-type littermate controls, C57BL/6 MKP-1+/− mice generated by the laboratory of Bravo and co-workers at Bristol-Myers Squibb Pharmaceutical Research Institute (86) were mated, and embryos were isolated at d 12.5 postcoitum. After removal of the head and internal organs as well as tail biopsies for genotyping, the remaining tissue was minced and incubated four times for 5 min at 37 C in 0.25% trypsin/1 mm EDTA. Cells were then harvested by centrifugation at 300 × g for 10 min and seeded in DMEM, 10% fetal calf serum, 2 mm l-glutamine in a T25 flask per embryo. Identification of littermates homozygous for the erp/mkp-1 wild type or mutant allele was carried out by PCR analysis of genomic DNA as described previously (74). MKP-1−/− primary MEFs and wild-type littermate control MEFs were transfected using Fugene HD (Roche).
A synthetic siRNA duplex targeting the mouse Trip6 mRNA sequence GUCUGGAUGCUGAGAUAGA and a control siRNA duplex targeting dsRed (87) were purchased from MWG Biotech (Ebersberg, Germany). NIH-3T3 fibroblasts were transfected with the siRNAs using Lipofectamine 2000 (Invitrogen).
Stable NIH-3T3 clones bearing an array of amplified uPA-Luc gene unit were obtained by blasticidin selection after cotransfection of equimolar amounts of −1977/−1858uPA-TATA-Luc and pΔBN-AR1, which promotes an amplification of cotransfected plasmids (49). Copy number was estimated by real-time PCR analysis, using primers amplifying both the Chinese hamster DHFR genomic region contained in pΔBN-AR1 and the endogenous mouse counterpart, as described by Bosisio et al. (51). The presence of the array was confirmed by fluorescence in situ hybridization, as described by Müller et al. (88). Briefly, cells grown on coverslips were fixed for 30 min at room temperature in 3.5% paraformaldehyde, permeabilized for 10 min in 0.5% Triton X-100 in PBS, and treated for 1 h with ribonuclease (50μg/ml). DNA was denatured in 50% formamide/2× standard saline citrate (SSC) for 5 min at 95 C and then 5 min on ice. Cells were then dehydrated on ice in 70, 90, and then 100% ethanol for 5 min each. A fluorescent probe targeting the luciferase coding sequence was generated by PCR in the presence of chromatide Alexa 546 deoxyuridine triphosphate (Molecular Probes, Invitrogen). The purified probe was denatured for 5 min in 25% formamide at 95 C and diluted at 5–10 μg/ml in hybridization solution (50% formamide, 2× SSC, 10% dextran sulfate, 1 mg/ml Escherichia coli tRNA). Hybridization was performed for 16 h at 37 C, followed by washes for 10 min in 2× SSC/0.05% Triton X-100, for 15 min in 2× SSC, and then for 5 min in 4× SSC.
All experiments were performed in serum-starved (24 h) cells. Unless otherwise stated, cells were treated 24 h after transfection with TPA (50 ng/ml) or UV-C irradiated (30 J/m2) in the presence or absence of dexamethasone (10 nm) and harvested either 4 h after treatment for Northern blotting or 16 h after treatment for reporter gene assays. Firefly luciferase activities were normalized to Renilla luciferase activity (Ubi-Renilla). Where indicated, significant differences were assessed by t test analysis, with values of P < 0.05 sufficient to reject the null hypothesis. In siRNA experiments, cells were treated 48 h after transfection.
BiFC, Immunofluorescence, and Laser Scanning Microscopy
For BiFC (39), cells were grown and transfected in eight-well chamber slides (NUNC, Roskilde, Denmark). Twenty-four hours after transfection with the BiFC constructs and pcDNA3.1-mCherry as a transfection control, cells were incubated for 2 h at 30 C to allow the maturation of the complemented YFP fluorescence.
For immunofluorescence, cells grown and transfected on coverslips were fixed for 20 min in 8% paraformaldehyde in PBS and permeabilized for 10 min in 0.5% Triton X-100 in PBS. Primary antibodies were a rat anti-HA monoclonal antibody (Roche) and an anti-Flag antibody (Sigma-Aldrich, Munich, Germany). When indicated, nuclei were counterstained with DRAQ5 (Biostatus Ltd., Shepshed, UK).
Microscopy (fluorescence in situ hybridization, immunofluorescence, and live cell imaging of BiFC and of uPA-Luc array cells) was performed using a Zeiss LSM 510 Meta in confocal multitracking mode, with a ×63/1.3-oil C-Apochromat objective (Zeiss) to generate 0.5-μm optical sections.
GST Pull-Down Assays
GST pull-down assays were performed as described (29). Briefly, pGEM-c-Fos, pCS2-Fra1, pCS2-Fra2, pEP-FosB, pBAT-c-Jun, pBAT-JunB, and pBAT-ATF2 were transcribed and translated in vitro, using 1 μg DNA, in the presence of [35S]methionine using the TNTCoupled Reticulocyte Lysate System (Promega) in a 50-μl reaction. Ten micrograms of purified GST-Trip6190–476 protein or GST alone were coupled to glutathione-Sepharose beads (GE Healthcare, Braunschweig, Germany) and incubated at 4 C for 2 h under agitation with 5μl in vitro translated proteins in pull-down buffer [40 mm HEPES-KOH (pH 7.9), 100 mm NaCl, 0,1 mm EDTA, 0.2% Triton X-100, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride]. Proteins bound were washed three times with pull-down buffer, resolved by SDS-PAGE, and analyzed by autoradiography.
Northern Blotting
Total RNA extraction and Northern blotting were performed as described (19), using as probes cDNA fragments for collagenase I (26), uPA (XbaI/SmaI), ATF3 (XbaI/BamHI), Trip6 (ScaI/SacI), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, PstI).
Western Blotting
Western blot analysis was performed as described (19) using the following antibodies: anti-MKP-1 (M18; Santa Cruz Biotechnology, Heidelberg, Germany), anti-GR (P20; Santa Cruz), anti-phospho-SAPK/JNK (Thr183/Tyr185; Cell Signaling Technology, Frankfurt a.M., Germany), anti-phospho-c-Jun (Ser63; Cell Signaling), anti-JNK1 (C17; Santa Cruz), anti-c-Jun (BD Biosciences, Heidelberg, Germany), anti-c-Fos (Upstate, Schwalbach, Germany), anti-ATF2 (N96; Santa Cruz), and anti-Trip6 (BD Transduction Laboratories).
Supplementary Material
Acknowledgments
We are grateful to Carme Caelles, Andrew Cato, Latifa Bakiri, Pascuale Verde, Gioacchino Natoli, Kimihiko Sugaya, Noriaki Shimizu, Peter Angel, Tom Kerppola, Carsten Weiss, and Roger Y. Tsien for providing plasmids and cells. We thank Carsten Weiss and Andrew Cato for stimulating discussion and Gioacchino Natoli for help and suggestions in the establishment of the reporter array cells.
Footnotes
This work was supported by the European Commission (HPRN-CT-2002-00256) and by the German Science Foundation (DFG Grant HE 551/13-1).
Disclosure Statement: The authors have nothing to disclose.
First Published Online June 5, 2008
Abbreviations: AP, Activator protein; ATF, activating transcription factor; BiFC, bimolecular fluorescence complementation; CEF, chicken embryonic fibroblast; CRE, CREB response element; CREB, cAMP response element-binding protein; GR, glucocorticoid receptor; HA, hemagglutinin; JNK, c-Jun N-terminal kinase; MEF, mouse embryonic fibroblast; ΔMEKK1, constitutively active mutant of the MEK kinase 1; MKP-1, MAPK phosphatase 1; Pol II, RNA polymerase II; siRNA, small interfering RNA; SSC, standard saline citrate; TPA, 12-O-tetradecanoylphorbol-13-acetate; TRE, TPA response element; uPA, urokinase-type plasminogen activator; YC, C-terminal half of YFP; YFP, yellow fluorescent protein; YN, N-terminal half of YFP.
References
- Chinenov Y, Kerppola TK 2001 Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438–2452 [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF 2003 AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3:859–868 [DOI] [PubMed] [Google Scholar]
- Hess J, Angel P, Schorpp-Kistner M 2004 AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117:5965–5973 [DOI] [PubMed] [Google Scholar]
- Benbrook DM, Jones NC 1990 Heterodimer formation between CREB and JUN proteins. Oncogene 5:295–302 [PubMed] [Google Scholar]
- Hai T, Curran T 1991 Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA 88:3720–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Liou HC, Kara CJ, Lamph WW, Verma IM, Glimcher LH 1990 mXBP/CRE-BP2 and c-Jun form a complex which binds to the cyclic AMP, but not to the 12-O-tetradecanoylphorbol-13-acetate, response element. Mol Cell Biol 10:1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H, Duyndam M, Rottier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb AJ 1993 Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J 12:479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H, Huguier S, Kooistra K, Baguet J, Vial E, van der Eb AJ, Herrlich P, Angel P, Castellazzi M 1998 Autocrine growth and anchorage independence: two complementing Jun-controlled genetic programs of cellular transformation. Genes Dev 12:1227–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M 1996 The regulation of AP-1 activity by mitogen-activated protein kinases. Phil Trans R Soc London 351:127–134 [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ 1996 Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med 74:589–607 [DOI] [PubMed] [Google Scholar]
- Arias J, Alberts AS, Brindle P, Claret FX, Smeal T, Karin M, Feramisco J, Montminy M 1994 Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226–229 [DOI] [PubMed] [Google Scholar]
- Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H 2001 Identification of SWI.SNF complex subunit BAF60a as a determinant of the transactivation potential of Fos/Jun dimers. J Biol Chem 276:2852–2857 [DOI] [PubMed] [Google Scholar]
- Lee SK, Kim HJ, Na SY, Kim TS, Choi HS, Im SY, Lee JW 1998 Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through interaction with the c-Jun and c-Fos subunits. J Biol Chem 273:16651–16654 [DOI] [PubMed] [Google Scholar]
- Martens JH, Verlaan M, Kalkhoven E, Zantema A 2003 Cascade of distinct histone modifications during collagenase gene activation. Mol Cell Biol 23:1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassel O, Herrlich P 2007 Crosstalk between the glucocorticoid receptor and other transcription factors: molecular aspects. Mol Cell Endocrinol 275:13–29 [DOI] [PubMed] [Google Scholar]
- Necela BM, Cidlowski JA 2004 Mechanisms of glucocorticoid receptor action in noninflammatory and inflammatory cells. Proc Am Thorac Soc 1:239–246 [DOI] [PubMed] [Google Scholar]
- Schoneveld OJ, Gaemers IC, Lamers WH 2004 Mechanisms of glucocorticoid signalling. Biochim Biophys Acta 1680:114–128 [DOI] [PubMed] [Google Scholar]
- Hulley PA, Gordon F, Hough FS 1998 Inhibition of mitogen-activated protein kinase activity and proliferation of an early osteoblast cell line (MBA 15.4) by dexamethasone: role of protein phosphatases. Endocrinology 139:2423–2431 [DOI] [PubMed] [Google Scholar]
- Kassel O, Sancono A, Kratzschmar J, Kreft B, Stassen M, Cato AC 2001 Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J 20:7108–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider LG, Hirasawa N, Santini F, Beaven MA 1996 Activation of the mitogen-activated protein kinase cascade is suppressed by low concentrations of dexamethasone in mast cells. J Immunol 157:2374–2380 [PubMed] [Google Scholar]
- Caelles C, Gonzalez-Sancho JM, Munoz A 1997 Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes Dev 11:3351–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa N, Sato Y, Fujita Y, Mue S, Ohuchi K 1998 Inhibition by dexamethasone of antigen-induced c-Jun N-terminal kinase activation in rat basophilic leukemia cells. J Immunol 161:4939–4943 [PubMed] [Google Scholar]
- Swantek JL, Cobb MH, Geppert TD 1997 Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) is required for lipopolysaccharide stimulation of tumor necrosis factor α (TNF-α) translation: glucocorticoids inhibit TNF-α translation by blocking JNK/SAPK. Mol Cell Biol 17:6274–6282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imasato A, Desbois-Mouthon C, Han J, Kai H, Cato AC, Akira S, Li JD 2002 Inhibition of p38 MAPK by glucocorticoids via induction of MAPK phosphatase-1 enhances nontypeable Haemophilus influenzae-induced expression of toll-like receptor 2. J Biol Chem 277:47444–47450 [DOI] [PubMed] [Google Scholar]
- Lasa M, Brook M, Saklatvala J, Clark AR 2001 Dexamethasone destabilizes cyclooxygenase 2 mRNA by inhibiting mitogen-activated protein kinase p38. Mol Cell Biol 21:771–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, Herrlich P 1990 Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell 62:1189–1204 [DOI] [PubMed] [Google Scholar]
- Schüle R, Rangarajan P, Kliewer S, Ransone LJ, Bolado J, Yang N, Verma IM, Evans RM 1990 Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell 62:1217–1226 [DOI] [PubMed] [Google Scholar]
- Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, Karin M 1990 Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell 62:1205–1215 [DOI] [PubMed] [Google Scholar]
- Kassel O, Schneider S, Heilbock C, Litfin M, Göttlicher M, Herrlich P 2004 A nuclear isoform of the focal adhesion LIM-domain protein Trip6 integrates activating and repressing signals at AP-1- and NF-κB-regulated promoters. Genes Dev 18:2518–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD 1995 Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol 9:243–254 [DOI] [PubMed] [Google Scholar]
- Wang Y, Dooher JE, Koedood Zhao M, Gilmore TD 1999 Characterization of mouse Trip6: a putative intracellular signaling protein. Gene 234:403–409 [DOI] [PubMed] [Google Scholar]
- Yi J, Beckerle MC 1998 The human TRIP6 gene encodes a LIM domain protein and maps to chromosome 7q22, a region associated with tumorigenesis. Genomics 49:314–316 [DOI] [PubMed] [Google Scholar]
- Guryanova OA, Sablina AA, Chumakov PM, Frolova EI 2005 Downregulation of TRIP6 gene expression induces actin cytoskeleton rearrangements in human carcinoma cell lines. Mol Biol 39:792–795 [PMC free article] [PubMed] [Google Scholar]
- Worley MJ, Nieman GS, Geddes K, Heffron F 2006 Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc Natl Acad Sci USA 103:17915–17920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lai YJ, Lin WC, Lin FT 2004 TRIP6 enhances lysophosphatidic acid-induced cell migration by interacting with the lysophosphatidic acid 2 receptor. J Biol Chem 279:10459–10468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi DR, Smythe C, Keyse SM 1993 The human CL100 gene encodes a Tyr/Thr-protein phosphatase which potently and specifically inactivates MAP kinase and suppresses its activation by oncogenic ras in Xenopus oocyte extracts. Oncogene 8:2015–2020 [PubMed] [Google Scholar]
- Groom LA, Sneddon AA, Alessi DR, Dowd S, Keyse SM 1996 Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J 15:3621–3632 [PMC free article] [PubMed] [Google Scholar]
- Sun H, Charles CH, Lau LF, Tonks NK 1993 MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell 75:487–493 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK 2002 Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9:789–798 [DOI] [PubMed] [Google Scholar]
- Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M 2002 Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol 22:4952–4964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo G, Casalino L, Vallone D, Caracciolo A, De Cesare D, Verde P 1999 Role of distinct mitogen-activated protein kinase pathways and cooperation between Ets-2, ATF-2, and Jun family members in human urokinase-type plasminogen activator gene induction by interleukin-1 and tetradecanoyl phorbol acetate. Mol Cell Biol 19:6240–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cesare D, Vallone D, Caracciolo A, Sassone-Corsi P, Nerlov C, Verde P 1995 Heterodimerization of c-Jun with ATF-2 and c-Fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene 11:365–376 [PubMed] [Google Scholar]
- Miralles F, Parra M, Caelles C, Nagamine Y, Felez J, Munoz-Canoves P 1998 UV irradiation induces the murine urokinase-type plasminogen activator gene via the c-Jun N-terminal kinase signaling pathway: requirement of an AP1 enhancer element. Mol Cell Biol 18:4537–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguier S, Baguet J, Perez S, van Dam H, Castellazzi M 1998 Transcription factor ATF2 cooperates with v-Jun to promote growth factor-independent proliferation in vitro and tumor formation in vivo. Mol Cell Biol 18:7020–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H, Castellazzi M 2001 Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene 20:2453–2464 [DOI] [PubMed] [Google Scholar]
- Cai Y, Zhang C, Nawa T, Aso T, Tanaka M, Oshiro S, Ichijo H, Kitajima S 2000 Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH2-terminal kinase and promoter response element. Blood 96:2140–2148 [PubMed] [Google Scholar]
- Liang G, Wolfgang CD, Chen BP, Chen TH, Hai T 1996 ATF3 gene. Genomic organization, promoter, and regulation. J Biol Chem 271:1695–1701 [DOI] [PubMed] [Google Scholar]
- Tamura K, Hua B, Adachi S, Guney I, Kawauchi J, Morioka M, Tamamori-Adachi M, Tanaka Y, Nakabeppu Y, Sunamori M, Sedivy JM, Kitajima S 2005 Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. EMBO J 24:2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Hashizume T, Shingaki K, Kawamoto JK 2003 Amplification of plasmids containing a mammalian replication initiation region is mediated by controllable conflict between replication and transcription. Cancer Res 63:5281–5290 [PubMed] [Google Scholar]
- Shimizu N, Miura Y, Sakamoto Y, Tsutsui K 2001 Plasmids with a mammalian replication origin and a matrix attachment region initiate the event similar to gene amplification. Cancer Res 61:6987–6990 [PubMed] [Google Scholar]
- Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G 2006 A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-κB-dependent gene activity. EMBO J 25:798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde P, Boast S, Franze A, Robbiati F, Blasi F 1988 An upstream enhancer and a negative element in the 5′ flanking region of the human urokinase plasminogen activator gene. Nucleic Acids Res 16:10699–10716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss C, Schneider S, Wagner EF, Zhang X, Seto E, Bohmann D 2003 JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-Jun. EMBO J 22:3686–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Dai T, Deak JC, Kyriakis JM, Zon LI, Woodgett JR, Templeton DJ 1994 Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature 372:798–800 [DOI] [PubMed] [Google Scholar]
- Gonzalez MV, Jimenez B, Berciano MT, Gonzalez-Sancho JM, Caelles C, Lafarga M, Munoz A 2000 Glucocorticoids antagonize AP-1 by inhibiting the activation/phosphorylation of JNK without affecting its subcellular distribution. J Cell Biol 150:1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna A, Nicolas M, Munoz A, Kyriakis JM, Caelles C 2003 Glucocorticoid receptor-JNK interaction mediates inhibition of the JNK pathway by glucocorticoids. EMBO J 22:6035–6044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf HJ, Herrlich P, Cato AC 1994 A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J 13:4087–4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Oehler T, Wilhelm D, Angel P, Kouzarides T 1995 Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene 11:2509–2514 [PubMed] [Google Scholar]
- Duyndam MC, van Dam H, Smits PH, Verlaan M, van der Eb AJ, Zantema A 1999 The N-terminal transactivation domain of ATF2 is a target for the co-operative activation of the c-jun promoter by p300 and 12S E1A. Oncogene 18:2311–2321 [DOI] [PubMed] [Google Scholar]
- Sano Y, Tokitou F, Dai P, Maekawa T, Yamamoto T, Ishii S 1998 CBP alleviates the intramolecular inhibition of ATF-2 function. J Biol Chem 273:29098–29105 [DOI] [PubMed] [Google Scholar]
- Bach I 2000 The LIM domain: regulation by association. Mech Dev 91:5–17 [DOI] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R 1998 LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet 14:156–162 [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC 2004 The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 5:920–931 [DOI] [PubMed] [Google Scholar]
- Guo B, Sallis RE, Greenall A, Petit MM, Jansen E, Young L, Van de Ven WJ, Sharrocks AD 2006 The LIM domain protein LPP is a coactivator for the ETS domain transcription factor PEA3. Mol Cell Biol 26:4529–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N, Yeh S, Kang HY, Inui S, Chang HC, Mizokami A, Chang C 1999 Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J Biol Chem 274:8316–8321 [DOI] [PubMed] [Google Scholar]
- Yang L, Guerrero J, Hong H, DeFranco DB, Stallcup MR 2000 Interaction of the τ transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol Biol Cell 11:2007–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touray M, Ryan F, Jaggi R, Martin F 1991 Characterisation of functional inhibition of the glucocorticoid receptor by Fos/Jun. Oncogene 6:1227–1234 [PubMed] [Google Scholar]
- Kerppola TK, Luk D, Curran T 1993 Fos is a preferential target of glucocorticoid receptor inhibition of AP-1 activity in vitro. Mol Cell Biol 13:3782–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR 1990 Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science 249:1266–1272 [DOI] [PubMed] [Google Scholar]
- Miner JN, Yamamoto KR 1992 The basic region of AP-1 specifies glucocorticoid receptor activity at a composite response element. Genes Dev 6:2491–2501 [DOI] [PubMed] [Google Scholar]
- Teurich S, Angel P 1995 The glucocorticoid receptor synergizes with Jun homodimers to activate AP-1-regulated promoters lacking GR binding sites. Chem Senses 20:251–255 [DOI] [PubMed] [Google Scholar]
- Abraham SM, Lawrence T, Kleiman A, Warden P, Medghalchi M, Tuckermann J, Saklatvala J, Clark AR 2006 Antiinflammatory effects of dexamethasone are partly dependent on induction of dual specificity phosphatase 1. J Exp Med 203:1883–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch K, de Wet H, Schuurmans MM, Allie-Reid F, Cato AC, Cunningham J, Burrin JM, Hough FS, Hulley PA 2007 Mitogen-activated protein kinase phosphatase 1/dual specificity phosphatase 1 mediates glucocorticoid inhibition of osteoblast proliferation. Mol Endocrinol 21:2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier JV, Brema S, Tuckermann J, Herzer U, Klein M, Stassen M, Moorthy A, Cato AC 2007 DUSP1 knockout mice show enhanced susceptibility to anaphylaxis but are sensitive to glucocorticoids. Mol Endocrinol 21:2663–2671 [DOI] [PubMed] [Google Scholar]
- Wu W, Pew T, Zou M, Pang D, Conzen SD 2005 Glucocorticoid receptor-induced MAPK phosphatase-1 (MPK-1) expression inhibits paclitaxel-associated MAPK activation and contributes to breast cancer cell survival. J Biol Chem 280:4117–4124 [DOI] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR 2005 The glucocorticoid receptor blocks P-TEFb recruitment by NFκB to effect promoter-specific transcriptional repression. Genes Dev 19:1116–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yamamoto KR 2000 The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 14:2314–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK 2004 A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci USA 101:14461–14466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G 1998 DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93:531–541 [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Umland T, Bauer A, Kretz O, Schutz G 2000 Mice with an increased glucocorticoid receptor gene dosage show enhanced resistance to stress and endotoxic shock. Mol Cell Biol 20:9009–9017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckermann JP, Reichardt HM, Arribas R, Richter KH, Schutz G, Angel P 1999 The DNA binding-independent function of the glucocorticoid receptor mediates repression of AP-1-dependent genes in skin. J Cell Biol 147:1365–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY 2004 Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572 [DOI] [PubMed] [Google Scholar]
- Sugaya K, Vigneron M, Cook PR 2000 Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J Cell Sci 113:2679–2683 [DOI] [PubMed] [Google Scholar]
- Heck S, Bender K, Kullmann M, Göttlicher M, Herrlich P, Cato AC 1997 IκBα-independent downregulation of NF-κB activity by glucocorticoid receptor. EMBO J 16:4698–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneikert J, Peterziel H, Defossez PA, Klocker H, Launoit Y, Cato AC 1996 Androgen receptor-Ets protein interaction is a novel mechanism for steroid hormone-mediated down-modulation of matrix metalloproteinase expression. J Biol Chem 271:23907–23913 [DOI] [PubMed] [Google Scholar]
- Dorfman K, Carrasco D, Gruda M, Ryan C, Lira SA, BravoR 1996 Disruption of the erp/mkp-1 gene does not affect mouse development: normal MAP kinase activity in ERP/MKP-1-deficient fibroblasts. Oncogene 13:925–931 [PubMed] [Google Scholar]
- Weiss C, Faust D, Durk H, Kolluri SK, Pelzer A, Schneider S, Dietrich C, Oesch F, Gottlicher M 2005 TCDD induces c-jun expression via a novel Ah (dioxin) receptor-mediated p38-MAPK-dependent pathway. Oncogene 24:4975–4983 [DOI] [PubMed] [Google Scholar]
- Müller WG, Walker D, Hager GL, McNally JG 2001 Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J Cell Biol 154:33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.