Abstract
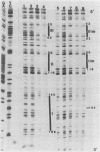
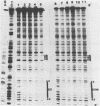
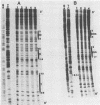
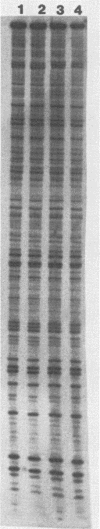
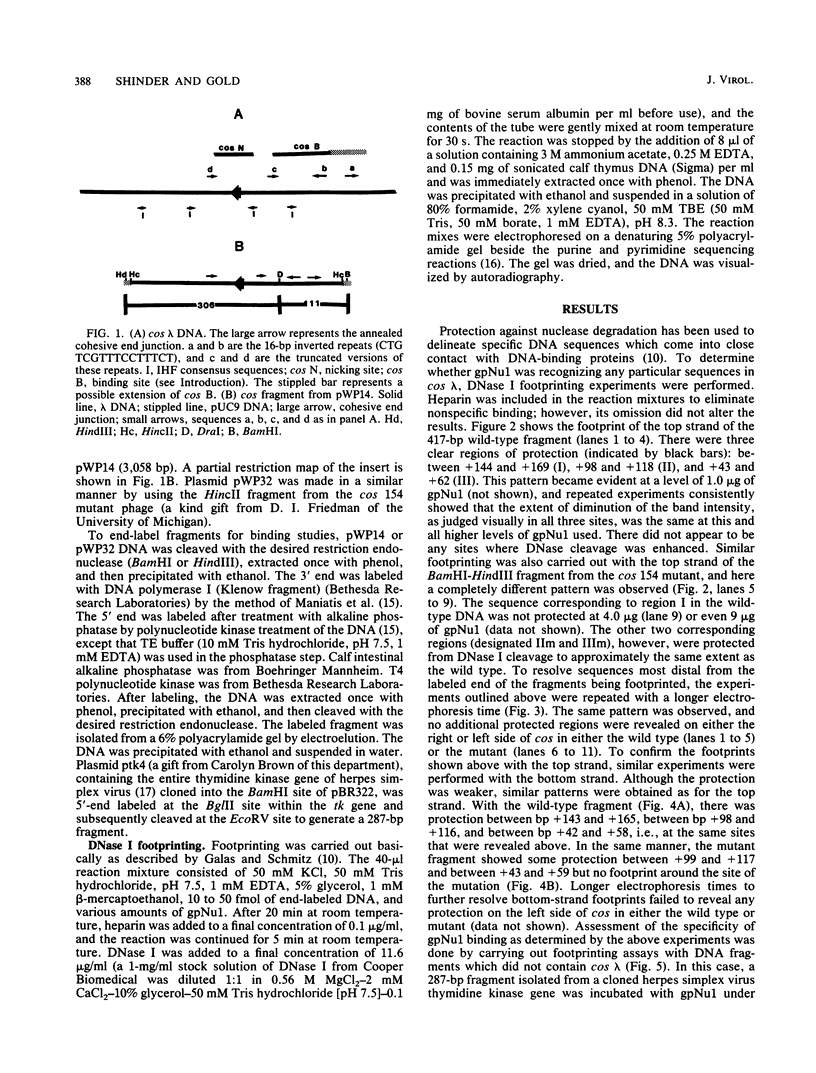
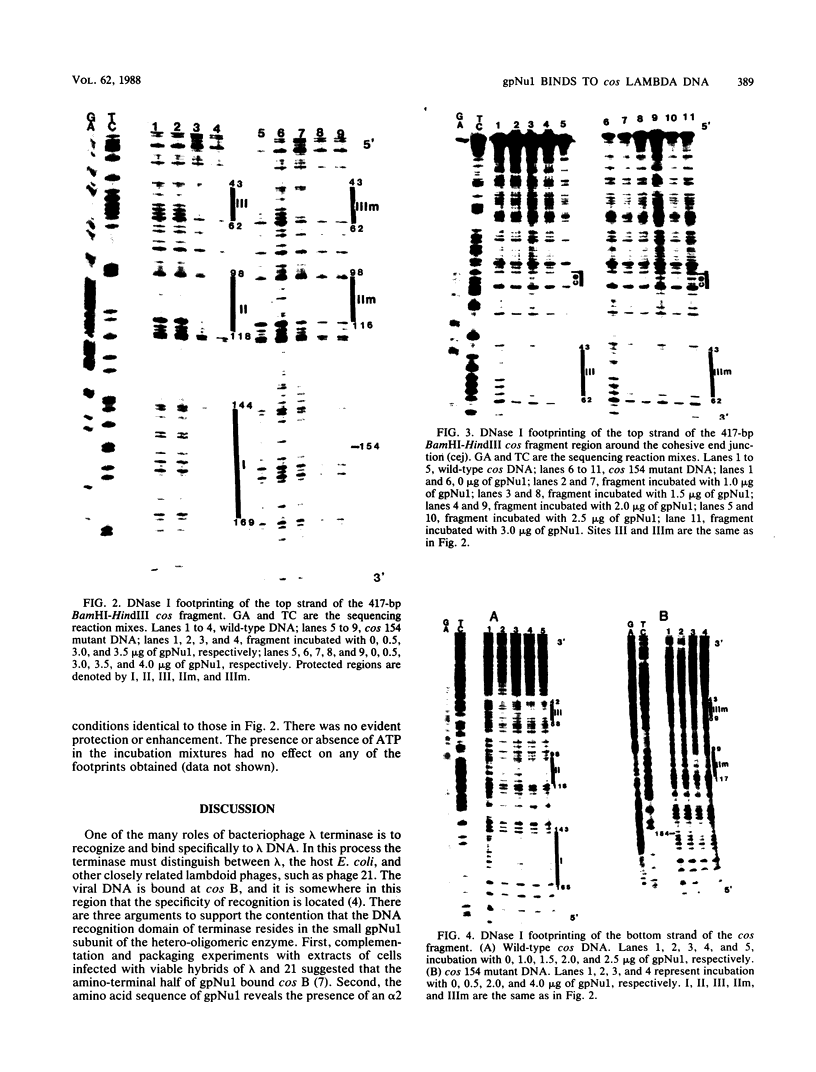
The maturation and packaging of bacteriophage lambda DNA are under the control of the multifunctional viral terminase enzyme, which is composed of the protein products of Nu1 and A, the two most leftward genes of the phage chromosome. Terminase binds selectively to the cohesive end site (cos) of multimeric replicating lambda DNA and introduces staggered nicks to regenerate the 12-base single-stranded cohesive ends of the mature phage genome. The purified gpNu1 subunit of terminase forms specific complexes with cos lambda DNA. DNase I footprinting experiments showed that gpNu1 bound to three distinct regions near the extreme left end of the lambda chromosome. These regions coincided with two 16-base-pair sequences (CTGTCGTTTCCTTTCT) that were in inverted orientation, as well as a truncated version of this sequence. Bear et al. (J. Virol. 52:966-972,1984) isolated a mutant phage which contained a CG to TA transition at the 10th position of the rightmost 16-base-pair sequence, and this phage (termed lambda cos 154) exhibits a defect in DNA maturation when it replicates in Escherichia coli which is deficient in integration host factor. Footprinting experiments with cos 154 DNA showed that gpNu1 could not bind to the site which contained the mutation but could protect the other two sites. Since the DNA-packaging specificity of terminase resides in the gpNu1 subunit, these studies suggest that terminase uses these three sites as recognition sequences for specific binding to cos lambda.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bear S. E., Court D. L., Friedman D. I. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984 Dec;52(3):966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- Feiss M., Widner W., Miller G., Johnson G., Christiansen S. Structure of the bacteriophage lambda cohesive end site: location of the sites of terminase binding (cosB) and nicking (cosN). Gene. 1983 Oct;24(2-3):207–218. doi: 10.1016/0378-1119(83)90081-1. [DOI] [PubMed] [Google Scholar]
- Frackman S., Siegele D. A., Feiss M. The terminase of bacteriophage lambda. Functional domains for cosB binding and multimer assembly. J Mol Biol. 1985 May 25;183(2):225–238. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold M., Becker A. The bacteriophage lambda terminase. Partial purification and preliminary characterization of properties. J Biol Chem. 1983 Dec 10;258(23):14619–14625. [PubMed] [Google Scholar]
- Gold M., Parris W. A bacterial protein requirement for the bacteriophage lambda terminase reaction. Nucleic Acids Res. 1986 Dec 22;14(24):9797–9809. doi: 10.1093/nar/14.24.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypr J., Mrázek J. Lambda phage protein Nu 1 contains the conserved DNA binding fold of repressors. J Mol Biol. 1986 Sep 5;191(1):139–140. doi: 10.1016/0022-2836(86)90430-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L. The nucleotide sequence and transcript map of the herpes simplex virus thymidine kinase gene. Nucleic Acids Res. 1980 Dec 20;8(24):5949–5964. doi: 10.1093/nar/8.24.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Feiss M. Sequence of the left end of phage 21 DNA. J Mol Biol. 1985 May 25;183(2):246–249. doi: 10.1016/0022-2836(85)90217-7. [DOI] [PubMed] [Google Scholar]
- Miwa T., Matsubara K. Lambda phage DNA sequences affecting the packaging process. Gene. 1983 Oct;24(2-3):199–206. doi: 10.1016/0378-1119(83)90080-x. [DOI] [PubMed] [Google Scholar]
- Murialdo H., Davidson A., Chow S., Gold M. The control of lambda DNA terminase synthesis. Nucleic Acids Res. 1987 Jan 12;15(1):119–140. doi: 10.1093/nar/15.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Hong G. F., Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982 Dec 25;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Sealey L., Chalkley R. At least two nuclear proteins bind specifically to the Rous sarcoma virus long terminal repeat enhancer. Mol Cell Biol. 1987 Feb;7(2):787–798. doi: 10.1128/mcb.7.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutman O., Cuttito M. J. Normal levels of natural cytotoxic cells against solid tumours in NK-deficient beige mice. Nature. 1981 Mar 19;290(5803):254–257. doi: 10.1038/290254a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wu R., Taylor E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J Mol Biol. 1971 May 14;57(3):491–511. doi: 10.1016/0022-2836(71)90105-7. [DOI] [PubMed] [Google Scholar]