Abstract
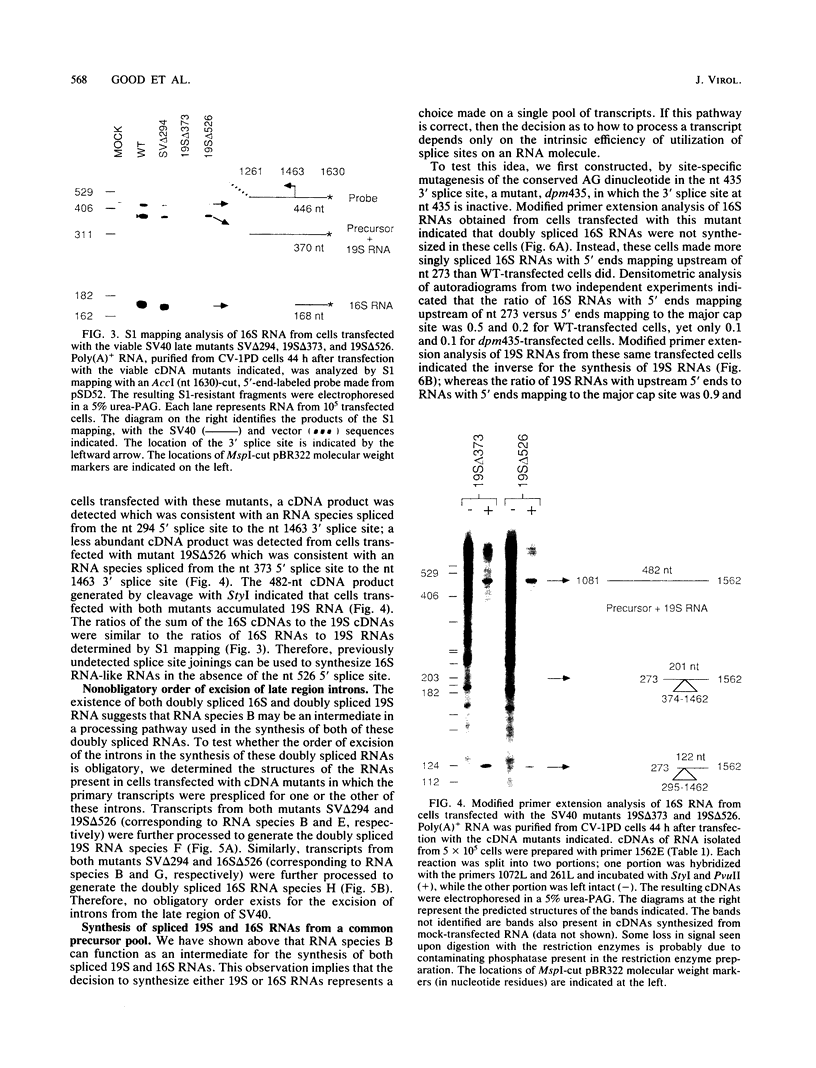
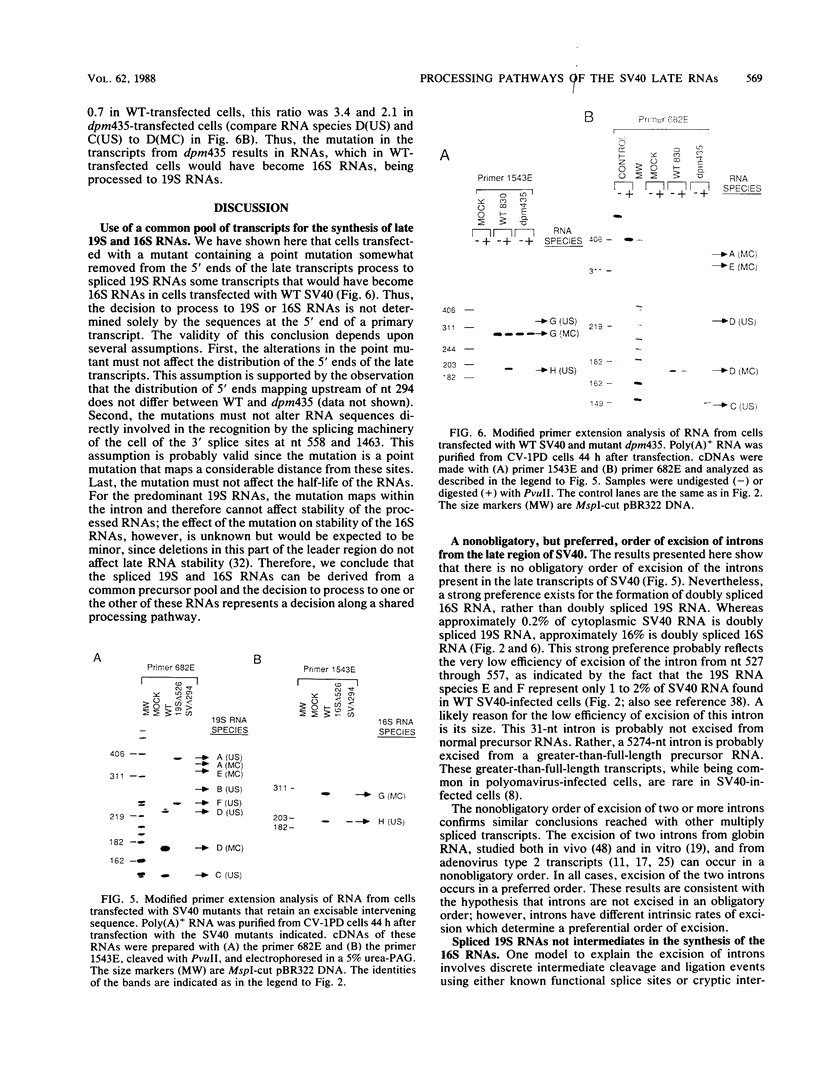
The late transcripts from the simian virus 40 (SV40) are alternatively spliced into two classes of spliced RNAs, 19S and 16S in size. We are interested in understanding the precursor-product relationships that result in the excision of different intervening sequences (introns) from the late transcripts. SV40 mutants containing precise deletions of the introns for each of the spliced 19S and 16S RNA species, including a previously undetected doubly spliced 19S RNA species, were isolated. Analysis by S1 mapping and a modified primer extension technique of the viral RNAs made in monkey cells transfected with each of these mutants led to the following conclusions. (i) Spliced late 19S RNA is not an intermediate in the synthesis of the late 16S RNAs. (ii) The 3' splice site used in the synthesis of the late 16S RNAs can join, albeit inefficiently, with alternative 5' splice sites in the absence of the 5' splice site normally used to synthesize 16S RNA. (iii) There is no obligatory order of excision of introns in the formation of the doubly spliced SV40 late 19S and 16S RNA species. A mutant was constructed by site-directed mutagenesis in which the 5'-proximal 3' splice site used in the synthesis of the doubly spliced RNAs is inactive. Cells transfected with this mutant processed transcripts into 19S RNA which, in wild-type-transfected cells, would have become doubly spliced 16S RNA. Therefore, we conclude that some of the spliced late 19S and 16S RNA can be synthesized from a common pool of transcripts.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y. Biogenesis and characterization of SV40 and polyoma RNAs in productively infected cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):165–178. doi: 10.1101/sqb.1974.039.01.023. [DOI] [PubMed] [Google Scholar]
- Aloni Y., Shani M., Reuveni Y. RNAs of simian virus 40 in productively infected monkey cells: kinetics of formation and decay in enucleate cells. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2587–2591. doi: 10.1073/pnas.72.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Mertz J. E. DNA sequence analysis of simian virus 40 mutants with deletions mapping in the leader region of the late viral mRNA's: mutants with deletions similar in size and position exhibit varied phenotypes. J Virol. 1981 Feb;37(2):730–737. doi: 10.1128/jvi.37.2.730-737.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Mertz J. E. The number of ribosomes on simian virus 40 late 16S mRNA is determined in part by the nucleotide sequence of its leader. Mol Cell Biol. 1984 Apr;4(4):813–816. doi: 10.1128/mcb.4.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chiu N. H., Radonovich M. F., Thoren M. M., Salzman N. P. Selective degradation of newly synthesized nonmessenger simian virus 40 transcripts. J Virol. 1978 Nov;28(2):590–599. doi: 10.1128/jvi.28.2.590-599.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. A gene chimaera of SV40 and mouse beta-globin is transcribed and properly spliced. Nature. 1981 Jan 29;289(5796):378–382. doi: 10.1038/289378a0. [DOI] [PubMed] [Google Scholar]
- Ford J. P., Cozzitorto J., Jr, Hsu M. T. Transcription pattern of in vivo-labeled late simian virus 40 RNA: evidence that 16S and 19S mRNA's are derived from distinct precursor RNA populations. J Virol. 1980 Sep;35(3):972–978. doi: 10.1128/jvi.35.3.972-978.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Fu X. Y., Manley J. L. Factors influencing alternative splice site utilization in vivo. Mol Cell Biol. 1987 Feb;7(2):738–748. doi: 10.1128/mcb.7.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattoni R., Keohavong P., Stévenin J. Splicing of the E2A premessenger RNA of adenovirus serotype 2. Multiple pathways in spite of excision of the entire large intron. J Mol Biol. 1986 Feb 5;187(3):379–397. doi: 10.1016/0022-2836(86)90440-7. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Piatak M., Mertz J. E., Weissman S. M., Lebowitz P. Altered utilization of splice sites and 5' termini in late RNAs produced by leader region mutants of simian virus 40. J Virol. 1982 Nov;44(2):610–624. doi: 10.1128/jvi.44.2.610-624.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Grosveld G. C., Koster A., Flavell R. A. A transcription map for the rabbit beta-globin gene. Cell. 1981 Feb;23(2):573–584. doi: 10.1016/0092-8674(81)90153-7. [DOI] [PubMed] [Google Scholar]
- Gruss P., Lai C. J., Dhar R., Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G. Z., Mertz J. E. Bidirectional promoter elements of simian virus 40 are required for efficient replication of the viral DNA. Mol Cell Biol. 1986 Oct;6(10):3513–3522. doi: 10.1128/mcb.6.10.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohavong P., Gattoni R., LeMoullec J. M., Jacob M., Stévenin J. The orderly splicing of the first three leaders of the adenovirus-2 major late transcript. Nucleic Acids Res. 1982 Feb 25;10(4):1215–1229. doi: 10.1093/nar/10.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinniburgh A. J., Ross J. Processing of the mouse beta-globin mRNA precursor: at least two cleavage-ligation reactions are necessary to excise the larger intervening sequence. Cell. 1979 Aug;17(4):915–921. doi: 10.1016/0092-8674(79)90331-3. [DOI] [PubMed] [Google Scholar]
- Lang K. M., van Santen V. L., Spritz R. A. The two intervening sequences of human beta- and gamma-globin pre-mRNAs are excised in a preferred temporal order in vitro. EMBO J. 1985 Aug;4(8):1991–1996. doi: 10.1002/j.1460-2075.1985.tb03882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebowitz P., Weissman S. M. Organization and transcription of the simian virus 40 genome. Curr Top Microbiol Immunol. 1979;87:43–172. doi: 10.1007/978-3-642-67344-3_3. [DOI] [PubMed] [Google Scholar]
- Lo K. M., Jones S. S., Hackett N. R., Khorana H. G. Specific amino acid substitutions in bacterioopsin: Replacement of a restriction fragment in the structural gene by synthetic DNA fragments containing altered codons. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2285–2289. doi: 10.1073/pnas.81.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan D. E., Danna K. J. S1 mapping of purified nascent transcripts of simian virus 40. Mol Cell Biol. 1984 Apr;4(4):625–633. doi: 10.1128/mcb.4.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariman E. C., van Beek-Reinders R. J., van Venrooij W. J. Alternative splicing pathways exist in the formation of adenoviral late messenger RNAs. J Mol Biol. 1983 Jan 15;163(2):239–256. doi: 10.1016/0022-2836(83)90005-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Norris K., Norris F., Christiansen L., Fiil N. Efficient site-directed mutagenesis by simultaneous use of two primers. Nucleic Acids Res. 1983 Aug 11;11(15):5103–5112. doi: 10.1093/nar/11.15.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A. C., Kirby E. M., Janda M. R., Drake N. L., Duke G. M., Potratz K. F., Collett M. S. The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 1984 Mar 26;12(6):2969–2985. doi: 10.1093/nar/12.6.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatak M., Ghosh P. K., Norkin L. C., Weissman S. M. Sequences locating the 5' ends of the major simian virus 40 late mRNA forms. J Virol. 1983 Nov;48(2):503–520. doi: 10.1128/jvi.48.2.503-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatak M., Subramanian K. N., Roy P., Weissman S. M. Late messenger RNA production by viable simian virus 40 mutants with deletions in the leader region. J Mol Biol. 1981 Dec 15;153(3):589–618. doi: 10.1016/0022-2836(81)90409-5. [DOI] [PubMed] [Google Scholar]
- Rautmann G., Matthes H. W., Gait M. J., Breathnach R. Synthetic donor and acceptor splice sites function in an RNA polymerase B (II) transcription unit. EMBO J. 1984 Sep;3(9):2021–2028. doi: 10.1002/j.1460-2075.1984.tb02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Ghosh P. K., Lebowitz P., Weissman S. M. Gaps and duplicated sequences in the leaders of SV40 16S RNA. Nucleic Acids Res. 1978 Nov;5(11):4195–4213. doi: 10.1093/nar/5.11.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Santangelo G. M., Cole C. N. Preparation of a "functional library" of African green monkey DNA fragments which substitute for the processing/polyadenylation signal in the herpes simplex virus type 1 thymidine kinase gene. Mol Cell Biol. 1983 Apr;3(4):643–653. doi: 10.1128/mcb.3.4.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Sequences involved in determining the locations of the 5' ends of the late RNAs of simian virus 40. J Virol. 1985 Dec;56(3):1002–1013. doi: 10.1128/jvi.56.3.1002-1013.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel L. F., Ward T. E., Jacobson A. Intron bypass: a rapid procedure for eliminating introns from cloned genomic DNA and its application to a cellulase gene. Nucleic Acids Res. 1984 Jul 25;12(14):5879–5895. doi: 10.1093/nar/12.14.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K. N. Segments of simian virus 40 DNA spanning most of the leader sequence of the major late viral messenger RNA are dispensable. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2556–2560. doi: 10.1073/pnas.76.6.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson C., Pettersson U., Akusjärvi G. Splicing of adenovirus 2 early region 1A mRNAs is non-sequential. J Mol Biol. 1983 Apr 15;165(3):475–495. doi: 10.1016/s0022-2836(83)80214-9. [DOI] [PubMed] [Google Scholar]
- Villarreal L. P., White R. T., Berg P. Mutational alterations within the simian virus 40 leader segment generate altered 16S and 19S mRNA's. J Virol. 1979 Jan;29(1):209–219. doi: 10.1128/jvi.29.1.209-219.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal L. A paranuclear extract contains a unique set of viral transcripts late in SV40 infection. Virology. 1981 Sep;113(2):663–671. doi: 10.1016/0042-6822(81)90195-1. [DOI] [PubMed] [Google Scholar]
- White R. T., Berg P., Villarreal L. P. Simian virus 40-rabbit beta-globin recombinants lacking late mRNA splice sites express cytoplasmic RNAs with altered structures. J Virol. 1982 Apr;42(1):262–274. doi: 10.1128/jvi.42.1.262-274.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M. P., Gurdon J. B. Post-transcriptional processing of simian virus 40 late transcripts in injected frog oocytes. J Mol Biol. 1983 Jan 5;163(1):1–26. doi: 10.1016/0022-2836(83)90027-x. [DOI] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]