Abstract
The Epstein–Barr virus (EBV) encoded nuclear antigen (EBNA) 1 is expressed in latently infected B lymphocytes that persist for life in healthy virus carriers and is the only viral protein regularly detected in all EBV associated malignancies. The Gly-Ala repeat domain of EBNA1 was shown to inhibit in cis the presentation of major histocompatibility complex (MHC) class I restricted cytotoxic T cell epitopes from EBNA4. It appears that the majority of antigens presented via the MHC I pathway are subject to ATP-dependent ubiquitination and degradation by the proteasome. We have investigated the influence of the repeat on this process by comparing the degradation of EBNA1, EBNA4, and Gly-Ala containing EBNA4 chimeras in a cell-free system. EBNA4 was efficiently degraded in an ATP/ubiquitin/proteasome-dependent fashion whereas EBNA1 was resistant to degradation. Processing of EBNA1 was restored by deletion of the Gly-Ala domain whereas insertion of Gly-Ala repeats of various lengths and in different positions prevented the degradation of EBNA4 without appreciable effect on ubiquitination. Inhibition was also achieved by insertion of a Pro-Ala coding sequence. The results suggest that the repeat may affect MHC I restricted responses by inhibiting antigen processing via the ubiquitin/proteasome pathway. The presence of regularly interspersed Ala residues appears to be important for the effect.
Keywords: antigen processing
The Epstein–Barr virus (EBV) nuclear antigen (EBNA) 1 is the only viral protein regularly detected in all EBV-associated malignancies and is likely to be expressed also in latently infected B lymphocytes that serve as the viral reservoir in healthy virus carriers (reviewed in ref. 1). Several lines of evidence suggest that EBNA1 may be exempted from the efficient major histocompatibility complex (MHC) class I-restricted cytotoxic T lymphocyte (CTL)-mediated surveillance that prevents the uncontrolled proliferation of EBV-transformed blasts. Thus, EBNA1-expressing cells are not recognized by EBV-specific CTLs isolated from blood during acute primary infections (2) or by effectors reactivated by stimulation of lymphocytes from healthy EBV carriers with autologous virus infected cells (3, 4). Furthermore, expression of EBNA1 did not confer immunogenicity to transfected murine mammary carcinoma cells whereas immunogenicity was induced by expression of other EBV proteins that are highly immunogenic in humans, suggesting that CTL evasion may occur across species (5).
Previous evidence has implicated the internal Gly-Ala repeat domain of EBNA1 as a cis-acting inhibitor of MHC class I-restricted presentation (6). Insertion of the domain downstream of the immunodominant human leukocyte antigen (HLA) A11 restricted epitope in EBNA4 was shown to prevent recognition of the chimeric protein by EBV-specific CTLs. Conversely, the epitope was presented in the context of an EBNA1 deletion mutant lacking the Gly-Ala repeat but not in the context of the native, Gly-Ala containing protein. Two possible mechanisms may explain this effect. Presence of the repeat may interfere with the proteolytic machinery that leads to the production of antigenic peptides. Alternatively, Gly-Ala containing proteins may be sequestered in cellular compartments that are not accessible to the appropriate type of processing.
An increasing body of evidence implicates the ubiquitin/proteasome-dependent protein degradation system in the production of peptides for MHC class I-restricted presentation (7, 8). Treatment with peptide aldehydes that inhibit the major peptidase activity of the 20S subunit was shown to prevent the degradation of ubiquitinated proteins by the 26S proteasome (9). Cells treated with the proteasome specific inhibitor, lactacystin, failed to present influenza nucleoprotein to specific CTLs but retained the capacity to present defined epitopes expressed as cytosolic peptides by recombinant vaccinia viruses (10). Targeting of this protein degradation pathway by the Gly-Ala repeat could provide an efficient means to selectively hamper MHC class I-restricted responses. We have tested this possibility by comparing the capacity of native and Gly-Ala containing EBNA4 to act as substrate for ubiquitin/proteasome-dependent degradation in in vitro processing assays.
MATERIALS AND METHODS
Reagents.
Phenylhydrazine, ubiquitin, ATP, phosphokreatine, and creatine phosphokinase were purchased from Sigma. Hexokinase was purchased from Boehringer Mannheim. The proteasome inhibitor carbobenzoxy-l-leucil-l-leucil-l-leucinal (MG 132) was obtained from Peptides International. Ubiquitin aldehyde was prepared as described (11).
Plasmids.
Plasmids containing the EBNA1 gene from the B95.8 virus (pBS-E1) and an EBNA1 deletion mutant lacking the Gly-Ala repeat (pBS-E1ΔGA) were described (6). An EBNA4 cDNA was amplified by reverse transcription–PCR using the primers 5′-GCGGATCCATAATGAAGAAAGCGTGGCTC-3′ and 5′-ATGAATTCTCACTCATCGTTCGATGTT-3′. The PCR product was cloned in the pBS-KS(+)II vector (Stratagene) and sequenced using terminal and internal primers. The full-length Gly-Ala domain of the B95.8 EBNA1 (flGA, coordinates 108,201–108,930) was subcloned into the SmaI site of the pBS-KS(+)II vector, and the E4-flGA chimera was obtained by inserting the cassette in the MscI site of EBNA4. Insertion in reverse orientation yielded a chimera encoding a Pro-Ala repeat, E4-PPA. A short Gly-Ala fragment with unique EBNA1 flanking sequences was obtained by low stringency PCR amplification using the primers 5′-ATGGATCCACGTCCATTGCATTGGCTGCA-3′ and 5′-GCGAATTCCTGGCTCTTTCACGTCCTCTAC-3′. The PCR products were cloned in the BamHI/EcoRI sites of the pGEX2TK plasmid (Pharmacia), and recombinants with in-frame insertions were identified by Western blot analysis using Gly-Ala-specific reagents (12). The GA39 cassette was constructed by recloning the insert in the BamHI/EcoRI sites of pBS-KS(+)II, and the coding capacity was confirmed by sequencing. Insertion of the cassette in the SspI and MscI sites of EBNA4 yielded the E4-GA39(S) and E4-GA39(M) chimeras that contain a 17-amino acid-long Gly-Ala repeat. Wild-type and chimeric genes were subcloned into the pSP64poly(A) plasmid (Promega), and the constructs were used as templates for transcription/translation in a coupled reticulocyte system (TnT; Promega). In vitro-translated proteins were resolved by SDS/PAGE, transferred onto nitrocellulose, and visualized by enhanced chemiluminescence (Amersham Life Science) using the EBNA1-specific mouse mAb OT1x (13), affinity purified human antibodies specific for the EBNA1 Gly-Ala repeat (12), and a previously characterized polyclonal human serum containing high titers of antibodies to all EBNAs (6).
In Vitro Processing Assays.
Crude rabbit reticulocyte lysates were prepared as described (14). Reticulocyte rich blood was obtained from 3-week-old phenylhydrazine-treated female rabbits. Endogenous ubiquitin/protein conjugates were disrupted by incubation for 2 hr at 37°C in PBS containing 0.2 mM 2,4-dinitrophenol and 20 mM 2-deoxyglucose, and the reticulocytes were lysed in 1.5 volumes of 1 mM DTT. Particulate material was removed by centrifugation at 80,000 × g for 90 min, and aliquots of the supernatant were snap frozen and stored at −80°C. For reconstitution assays, the crude lysate were fractionated by ion exchange chromatography (DE-52, Whatman) (15). The nonadsorbed material (fraction I) contains haemoglobin, ubiquitin, and several E2 conjugating enzymes. The adsorbed material (fraction II) contains the remaining enzymes required for ubiquitin-dependent degradation.
The 35S-labeled proteins were obtained by in vitro transcription/translation in the presence of [35S]methionine (Amersham) and further purified by fractionation on DEAE-cellulose. One to 2 μl of the labeled substrate (25,000–30,000 cpm) were incubated with 5 μl of the lysate in processing buffer (40 mM Tris⋅HCl, pH 7.6/5 mM MgCl2/2 mM DTT) containing ATP/ATP regenerating (0.5 mM ATP/10 mM creatine phosphate/5 μg creatine phosphokinase) or ATP depleting systems (10 mM 2-deoxyglucose/0.5 μg hexokinase). Proteasome inhibitors were added where indicated. Protein breakdown was determined according to standard protocols by trichloroacetic acid (TCA) precipitation after incubation for 2 hr at 37°C or 4°C (14). The precipitates were pelleted for 5 min at 14.000 × g, and the radioactivity in the supernatant was measured in a β-counter (LKB). The percent degradation was calculated as the ratio between the input radioactivity and the radioactivity recovered in the supernatants after subtracting the background of samples incubated at 4°C. The specific degradation corresponds to the difference between degradation in the presence or absence of ATP. The specificity of the reactions was confirmed by SDS-PAGE and densitometry tracing by PhosphorImager (Molecular Dynamics). The formation of ubiquitin/protein conjugates was visualized by performing the reaction in the presence of ATP-γS and ubiquitin aldehyde (11).
Protein Turnover in Vivo.
CV1 cells were infected with recombinant vaccinia viruses expressing EBNA1 or E1-ΔGA (6) at m.o.i. 10 for 8 hr, labeled with [35S]methionine-cysteine mixture for 1 hr, and then chased in medium containing excess cold methionine and cysteine for the indicated time. Cells (5 × 106) were lysed in 1 ml lysis buffer containing 1% Nonidet P-40/150 mM NaCl/50 mM Tris, pH 7.5/1 mM phenylmethylsulfonyl fluoride, and the lysates were precleared overnight with 2 μl normal mouse serum at 4°C. EBNA1 polypeptides were immunoprecipitated with the OT1x mAb, resolved by SDS/PAGE, and visualized by PhosphorImager.
RESULTS
Sensitivity of EBNA4 and EBNA1 to Ubiquitin/Proteasome-Dependent Degradation.
EBNA4, EBNA1, and an EBNA1 deletion mutant lacking the Gly-Ala repeat were compared for their capacity to serve as substrates for ubiquitin/proteasome-dependent degradation in a cell-free system. Processing assays were performed as described using in vitro-labeled substrates and crude reticulocyte lysates or fractionated components of the ubiquitin conjugation system. EBNA4 was degraded in an ATP-dependent fashion as demonstrated by the disappearance of the 35S-labeled polypeptide in SDS/PAGE and by the release of TCA-soluble radioactivity (Fig. 1 A–E and Fig. 2). The ATP-dependent effect required ubiquitin, the ubiquitin conjugating enzymes, and the 26S complex present in fraction II of the rabbit reticulocyte lysate (Fig. 1B). The involvement of ubiquitination was confirmed by the accumulation of high molecular weight EBNA4 species when the assay was performed in the presence of ATP-γS and ubiquitin aldehyde to prevent cleavage of the polyubiquitin chains by C-terminal hydrolases (Fig. 1C). The specificity of the conjugates was demonstrated in reconstitution assays where exogenous ubiquitin and ubiquitin-conjugating enzymes were added in the presence of ATP-γS (Fig. 1D). Finally, the ATP-dependent effect was inhibited by addition of the peptide aldehyde MG132, confirming that the proteasome is required for degradation of the ubiquitinated EBNA4 (Fig. 1E). EBNA4 was also degraded in an ATP-independent manner, probably due to the presence of ATP-independent cytosolic proteases in the crude reticulocyte lysates. The ATP-independent activity exhibited a slower kinetics (data not shown) and was not inhibited by MG132 (Fig. 1E), suggesting that the 20S proteasome is not involved in this effect.
Figure 1.
Processing of EBNA4 in a cell-free system. The 35S-labeled EBNA4 was incubated for 2 hr at 37°C with crude reticulocyte lysate or fractionated components of the ubiquitin conjugation pathway in the presence (+ ATP) or absence (− ATP) of ATP. Control samples were kept on ice. The polypeptides were resolved by 8% SDS/PAGE and visualized by PhosphorImager. The identity of the radiolabeled bands was confirmed by Western blot analysis using specific reagents (data not shown). One representative experiment out of five is shown in the figure. (A) Degradation of EBNA4 by crude rabbit reticulocyte lysate. (B) Reconstitution experiments demonstrating that degradation is dependent on ubiquitin (Ub), ubiquitin-conjugating enzymes, and the 26S proteasome (Fr. II). (C) Accumulation of polyubiquitinated EBNA4 in the presence of ATP-γS. (D) Identity of the polyubiquitinated species demonstrated in reconstitution experiments using ATP-γS and ubiquitin aldehyde (Ubal). (E) Inhibition of ATP-dependent degradation by the proteasome inhibitor MG132. Degradation was assessed by monitoring the release of TCA-soluble radioactivity and percent degradation was calculated as described.
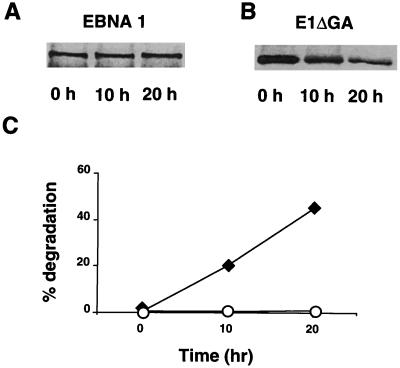
Figure 2.
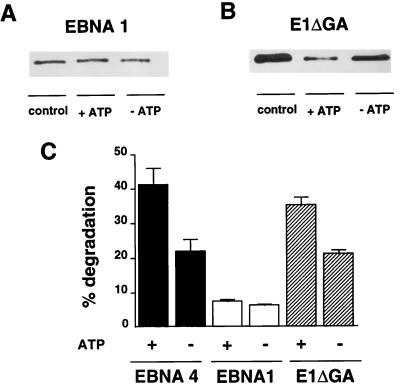
Processing of EBNA1 and E1-ΔGA. In vitro degradation of EBNA1 (A) and E1-ΔGA (B). One representative experiment out of five. (C) Degradation of EBNA4, EBNA1, and E1-ΔGA as monitored by the release of TCA-soluble radioactivity. Results are the mean ± SE of 16 experiments.
We then turned to the EBNA1 protein and found that in vitro translated EBNA1 was resistant to ATP-dependent degradation (Fig. 2A), whereas the E1-ΔGA mutant that lacks the Gly-Ala domain was degraded as efficiently as EBNA4 (Fig. 2B). Ten to 18% specific degradation of the E1-ΔGA polypeptides was demonstrated by monitoring the release of TCA-soluble radioactivity as compared with 1–2% degradation of EBNA1. EBNA1 was also less sensitive than EBNA4 to ATP-independent degradation whereas deletion of the Gly-Ala domain resulted in significant degradation of the E1-ΔGA polypeptide (Fig. 2C), confirming the previous observation that the repeat may influence the sensitivity of EBNA1 to cytosolic proteases (13).
Insertion of Gly-Ala Repeat Prevents the Ubiquitin/Proteasome-Dependent Degradation of EBNA4 Chimeras.
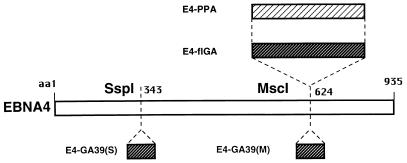
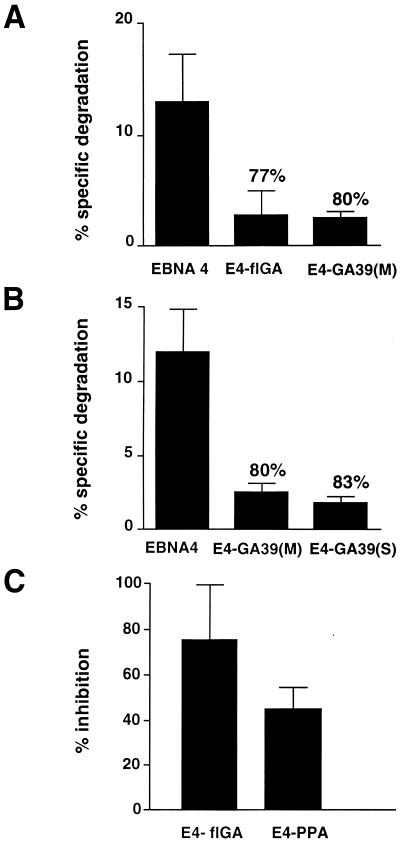
The different behavior of EBNA1 and E1-ΔGA in the cell-free degradation assays suggested that presence of the Gly-Ala repeat may interfere with the ubiquitin/proteasome-dependent processing pathway. To test this possibility and gain some insights on the structural constrains involved in the inhibition, we constructed EBNA4 chimeras containing repetitive sequences of different length or inserted in different sites of the protein (Fig. 3). In-frame insertion of the 239-amino acid-long Gly-Ala repeats of EBNA1 into the MscI site of EBNA4 yielded the E4-flGA chimera that was virtually insensitive to ATP-dependent degradation (Fig. 4A). The flGA sequence had similar inhibitory effect in EBNA1 and EBNA4 as judged by comparing the specific degradation of EBNA1/E1-ΔGA and EBNA4/E4-flGA pairs (Table 1). Similar levels of inhibition were also achieved by inserting a shorter cassette encoding a 17-amino acid-long Gly-Ala polypeptide either in the MscI site or in the N-terminal SspI site of EBNA4 (Fig. 4 A and B).
Figure 3.
Schematic outline of the EBNA4 chimeras. Chimeric EBNA4 proteins were generated by in-frame insertion of sequences coding for the 239-amino acid-long Gly-Ala repeat of EBNA1 (flGA) or a 39-amino acid-long polypeptide containing a Gly-Ala 17 mer flanked by unique EBNA1 residues (GA39) in the MscI site [E4-flGA, E4-GA39(M)] or SspI site [E4-GA39(S)] of EBNA4. Insertion of the flGA cassette in the opposite orientation relative to the EBNA4 sequence yielded the E4-PPA chimera that contains a 238-amino acid-long Pro-Ala repeat.
Figure 4.
Sensitivity of the EBNA4 chimeras to ubiquitin/proteasome dependent degradation. The degradation of EBNA4 was inhibited by insertion of Gly-Ala repeats of different length (A) and in different positions (B). A weaker inhibition was achieved by insertion of a Pro-Ala repeat (C). Specific degradation was calculated from the release of TCA-soluble radioactivity as described in the legend to Fig. 1. Results are the mean ± SD of four experiments. The inhibitory effect of the repeat was calculated relative to the specific degradation of EBNA4.
Table 1.
Effect of the Gly-Ala repeat on the degradation of EBNA1 and EBNA4
| % specific degradation*
|
|||||
|---|---|---|---|---|---|
| Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Mean ± SD | |
| EBNA1 | 1.0 | 2.1 | 2.0 | 1.5 | 1.6 ± 0.4 |
| E1-ΔGA | 10.1 | 13.7 | 18.0 | 13.4 | 13.7 ± 3.3 |
| % Inhibition† | 90 | 85 | 89 | 88 | 88 ± 2 |
| EBNA4 | 29.3 | 33.1 | 18.4 | 13.0 | 23.3 ± 8.7 |
| E4-flGA | 2.4 | 6.3 | 1.2 | 2.2 | 3.1 ± 2.1 |
| % inhibition† | 92 | 82 | 95 | 85 | 88 ± 6 |
Calculated as the difference between the % degradation detected in the presence or absence of ATP.
Calculated as the ratio between the % specific degradation of Gly-Ala-containing and Gly-Ala-deleted polypeptides.
We next asked whether the inhibitory effect was dependent on the amino acid composition of the repeats. Insertion of the flGA cassettes in opposite orientation relative to the EBNA4 sequence resulted in the E4-PPA chimera that contains a 238-amino acid-long Pro-Ala repeat. The E4-PPA polypeptide was degraded less efficiently than EBNA4, although the inhibitory effect of the Pro-Ala sequence appeared to be weaker than that induced by the Gly-Ala repeat (Fig. 4C).
Presence of the Gly-Ala Repeats Does Not Prevent Ubiquitin Conjugation.
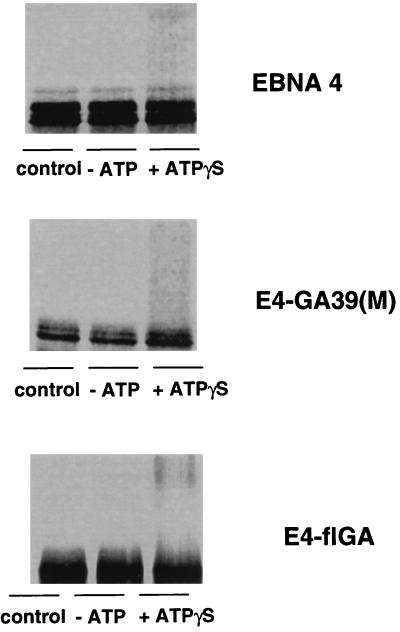
Introduction of foreign sequences may affect the capacity of chimeric proteins to interact with specific ubiquitin-conjugating enzymes and/or ubiquitin protein ligases. To investigate whether this could explain the resistance of the EBNA4 chimeras to ubiquitin/proteasome-dependent degradation, we tested the capacity of the E4-flGA and E4-GA39(M) chimeras to serve as substrate for ubiquitination. Processing assays were performed in the presence of ATP-γS and ubiquitin aldehyde, and the formation of polyubiquitin conjugates was monitored by the appearance of high molecular weight polypeptides. The Gly-Ala-containing chimeras appeared to be ubiquitinated as efficiently as the intact EBNA4 (Fig. 5). Interestingly, we consistently failed to demonstrate ubiquitination of the EBNA1 and E1-ΔGA polypeptides (data not shown). We cannot formally exclude that this may be due to the poor sensitivity of our in vitro ubiquitination assay. However, the different behavior of EBNA4 and E1-ΔGA suggests that the proteasome-dependent degradation of E1-ΔGA may not require ubiquitination.
Figure 5.
Ubiquitination of the Gly-Ala containing EBNA4 chimeras. In vitro-translated EBNA4, E4-flGA, and E4-GA39(M) were incubated for 2 hr at 37°C in crude rabbit reticulocyte lysate containing ATP-γS and ubiquitin aldehyde. Control samples were kept on ice or at 37°C under ATP-depleting conditions. One representative experiment out of three is shown in the figure.
Influence of the Gly-Ala Repeat on the in Vivo Turnover of EBNA1.
Finally, we asked whether presence of the Gly-Ala repeat may influence protein turnover in vivo. A very slow turnover of the endogenous EBNA1 was previously observed in lymphoblastoid cell lines (LCLs), resulting in inefficient metabolic labeling of this protein. Therefore, we turned to a recombinant vaccinia virus system that yields high levels of expression over a short period of time. The half life of the EBNA1 and E1-ΔGA polypeptides was compared in pulse–chase experiments using CV1 cells infected with the corresponding recombinant vaccinia viruses. EBNA1 appeared to be stable over the 20-hr observation time (Fig. 6A). In contrast, E1-ΔGA was degraded in vivo with a half life of approximately 18 hr.
Figure 6.
Influence of Gly-Ala repeat on the in vivo turnover of EBNA1. The in vivo turnover of EBNA1 and E1-ΔGA was monitored in pulse–chase experiments using CV1 cells infected with the corresponding recombinant vaccinia viruses. One representative experiment out of three. (A and B) In vivo-labeled polypeptides were immunoprecipitated using the OT1x mAb and resolved by SDS/PAGE. (C) The percent degradation was calculated from densitometry scans as follows: 1, intensity of the specific band at the indicated time/intensity of the specific band at time 0 × 100. ○, EBNA1; ⧫, E1-ΔGA.
DISCUSSION
The majority of intracellular proteins appear to be degraded via the ubiquitin/proteasome pathway and the antigenic products of this degradation are presented at the cell surface in association with MHC class I molecules (reviewed in refs. 16 and 17). Thus, inhibition of this process is likely to have broad consequences for both the regulation of cellular functions and immunogenicity. In an effort to elucidate the mechanisms by which the Gly-Ala repeat domain of EBNA1 promotes CTL escape, we have studied the effect of the repeat on protein degradation in a cell-free system. We report evidence suggesting that the repetitive domain interferes with ubiquitin/proteasome-dependent degradation and may thereby protect antigenic proteins from immune recognition.
We have shown that EBNA4, a highly immunogenic EBV product that contains several MHC class I-restricted CTL epitopes, is a good substrate for ubiquitin/proteasome-dependent degradation in a cell-free system (Figs. 1 and 2). Up to 50% of an in vitro-labeled EBNA4 polypeptide was degraded to TCA-soluble fragments after incubation for 2 hr with crude rabbit reticulocyte lysate. Most of the hydrolysis was dependent on the presence of ATP (Fig. 1A) and occurred at a linear rate that was proportional to the amount of reticulocyte lysate added to the assay (data not shown). The involvement of the proteasome was confirmed by the almost complete inhibition of ATP-dependent degradation upon addition of the peptide aldehyde MG132 that also inhibits the presentation of EBNA4 epitopes in fibroblasts infected with the relevant recombinant vaccinia virus (Fig. 1E and data not shown). Polyubiquitination appears to be required for degradation of most cellular proteins by the proteasome (reviewed in ref. 18). Accordingly, the ATP-dependent hydrolysis of EBNA4 was inhibited in the absence of ubiquitin and ubiquitin-conjugating enzymes (Fig. 1B) whereas polyubiquitinated species accumulated in the presence of ubiquitin aldehyde that prevents the degradation of ubiquitin conjugates by C-terminal hydrolases (Fig. 1 C and D). In line with the previously reported incapacity of EBNA1 chimeras to present a known CTL target epitope (6), the in vitro-translated polypeptide was resistant to ATP/ubiquitin/proteasome-dependent degradation in the cell-free system (Fig. 2).
In agreement with our previous finding that the Gly-Ala repeat prevents presentation of MHC class I-restricted epitopes, we have now shown that the repeat inhibits processing by the ubiquitin/proteasome pathway. Deletion of the repeat from EBNA1 restored processing both in vitro and in vivo (Figs. 2 and 6) whereas insertion of the repeat into EBNA4 resulted in more than 80% inhibition of ATP-dependent hydrolysis, thus confirming that the in vitro assay may be directly relevant to the mechanisms resulting in the production of antigenic peptides in vivo. Equal levels of inhibition were achieved by insertion of either the 239-amino acid-long repeat of the B95.8 EBNA1 or a shorter repeat containing only 17 Gly-Ala residues. The capacity to act across a wide size range is in line with the observation that the length of the repeat varies in natural EBV isolates, giving rise to EBNA1 polypeptides with characteristic patterns of migration in SDS/PAGE (19). Although the general validity of the inhibitory effect remains to be confirmed for other antigenic proteins, it is of note that a similar Gly-rich domain was shown to regulate the ubiquitin/proteasome-dependent production of NF-κB from its p105 precursor (20). Thus, while the C-terminal portion of p105 is rapidly degraded, sequences upstream of the Gly-rich domain are protected from degradation. We have failed to demonstrate the production of distinct proteolytic products on insertion of the Gly-Ala repeats in two different positions of EBNA4. Although this may be due to the low efficiency of the in vitro processing assay, it is also possible that Gly-rich and Gly-Ala sequences have different effects on the ubiquitin/proteasome pathway, perhaps because of the presence of additional signals that direct the partial degradation of p105.
The mechanism by which the Gly-Ala repeat inhibits ubiquitin/proteasome-dependent degradation still remains obscure. As illustrated by the results obtained with the E4-flGA and E4GA39(M) chimeras, presence of the repeat does not prevent ubiquitination (Fig. 5). This does not exclude the possibility that ubiquitination may be affected. Polyubiquitin chains are built by conjugation of lysines at position 29, 48, and 63 of the ubiquitin molecule (21, 22). Proteasomal degradation of several model proteins was shown to be dependent on polyubiquitination via Lys-48 whereas Lys-63-linked ubiquitin chains serve non proteolytic functions (reviewed in ref. 18). The E2 enzymes involved in this conjugation were implicated in the signal-dependent endocytosis of surface receptors (23). Conceivably, Gly-Ala-containing proteins may be targeted by these E2 enzymes resulting in the formation of polyubiquitinated substrates that are degraded in the endosomal/lysosomal compartment. It is noteworthy that EBNA1 is a dominant target for EBV-specific antibody responses (12) and rare EBNA1-specific MHC class II-restricted CTLs have also been demonstrated (24).
It also possible that presence of the repeat may directly interfere with binding to the 26S complex. The observation that neither EBNA1 nor the E1-ΔGA polypeptide are ubiquitinated in vitro whereas the latter is efficiently degraded by the proteasome provides indirect support to this possibility. Recent evidence demonstrates that Gly-Ala repeat-like domains contribute to the stability of spider silk protein by inducing the formation of β-sheets (25). Similar Ala-rich domains are also found in prion proteins where they may induce protease resistance by promoting the formation of amyloid (26–28). Interestingly, the intact EBNA1 was insensitive to both ATP-dependent and ATP-independent degradation (Fig. 2C), suggesting that presence of the repeat may confer a general resistance to proteases. On the basis of the effect of the repeat on EBNA4, which is clearly a target for both ubiquitination and proteasome degradation, it is tempting to speculate that hydrophobic surfaces generated by the alignment of regularly interspersed Ala residues may interfere with the hydrophobic interaction between the ubiquitin chains and the S5a subunit of the 26S protease (29). Similar hydrophobic surfaces may be generated by the Pro-Ala repeat explaining why the E4-PPA chimera was almost as resistant to degradation as its Gly-Ala containing counterpart (Fig. 4C). Taken together, these finding suggest that certain types of repeat sequences may influence the folding pattern of proteins and affect their capacity to associate with various components of ubiquitin/proteasome pathway, including ubiquitin conjugation enzymes and/or the regulatory subunits of the proteasome. Clearly, a better knowledge of this protein degradation system is required to dissect the mechanism of action of the repeat and determine whether a different branch of the cellular proteolytic machinery may be involved in the degradation of these substrates.
Acknowledgments
We thank J. M. Middeltorp for the kind gift of the OT1x antibody. This investigation was supported by grants from the Swedish Cancer Society, the Petrus and Augusta Hedlunds Stiftelse, the Karolinska Institute, Stockholm, and Therexsys Ltd. to M.G.M. The work in A.C.’s laboratory was supported by grants from the Israeli Academy of Science and Humanities, the Israeli Ministry of Sciences, and the German–Israeli Foundation for Scientific Research and Development. J.L. received an International Cancer Technology Transfer Fellowship (ICRETT), and A.S. was partially supported by a fellowship from the Swedish Institute. Network activities between the laboratories of M.G.M. and A.C. are supported by the European Commission Training and Mobility of Research program on “The central role of the ubiquitin proteasome system in regulatory processes involved in immunological, inflammatory, endocrinological and malignant disorders” contract no. ERBFMRXCT960026.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- EBNA
EBV nuclear antigen
- CTL
cytotoxic T lymphocyte
- MHC
major histocompatibility complex
- TCA
trichloroacetic acid
References
- 1.Masucci M G, Ernberg I. Trends Microbiol. 1994;2:125–130. doi: 10.1016/0966-842x(94)90599-1. [DOI] [PubMed] [Google Scholar]
- 2.Steven N M, Leese A M, Annels N E, Lee S P, Rickinson A B. J Exp Med. 1996;184:1801–1813. doi: 10.1084/jem.184.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray R J, Kurilla M G, Brooks J M, Thomas W A, Rowe M, Kieff E, Rickinson A B. J Exp Med. 1992;176:157–168. doi: 10.1084/jem.176.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanna R, Burrows S R, Kurilla M G, Jacob C A, Misko I S, Sculley T B, Kieff E, Moss D J. J Exp Med. 1992;176:169–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trivedi P, Masucci M, Wimberg G, Klein G. Int J Cancer. 1991;48:794–800. doi: 10.1002/ijc.2910480527. [DOI] [PubMed] [Google Scholar]
- 6.Levitskaya J, Coram M, Levitsky V, Imreh S, Stegerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Nature (London) 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 7.Townsend A, Rothbard J, Gotch F, Bahadur B, Wraith D, McMichael A. Cell. 1986;44:959–968. doi: 10.1016/0092-8674(86)90019-x. [DOI] [PubMed] [Google Scholar]
- 8.Michalek M T, Grant E P, Gramm C, Goldberg A L, Rock K L. Nature (London) 1993;363:552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 9.Rock K L, Gramm C, Rothstein L, Clark R, Stein R, Dick L, Hwang D. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 10.Cerundolo V, Benham A, Braund V, Mukherjee S, Macino B, Neefjes J, Townsend A. Eur J Immunol. 1997;27:336–341. doi: 10.1002/eji.1830270148. [DOI] [PubMed] [Google Scholar]
- 11.Hershko A, Rose I. Proc Natl Acad Sci USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillner J, SternŚs L, Kallin B, Alexander H, Ehlin-Henriksson B, Jörnvall H, Klein G, Lerner R. Proc Natl Acad Sci USA. 1984;81:4652–4656. doi: 10.1073/pnas.81.15.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M-R, Middeltorp J M, Hayward D. J Virol. 1993;67:4875–4885. doi: 10.1128/jvi.67.8.4875-4885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershko A, Ciechanover A, Rose I A. Proc Natl Acad Sci USA. 1979;76:3107–3110. doi: 10.1073/pnas.76.7.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko A, Heller H, Elias S, Ciechanover A. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 16.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 17.York I A, Rock K L. Annu Rev Immunol. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- 18.Hochstrasser M. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 19.Falk K, Gratama J W, Rowe M, Zou J Z, Khanim F, Young L S, Oosterveer M A P, Ernberg I. J Gen Virol. 1995;76:779–790. doi: 10.1099/0022-1317-76-4-779. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, Ghosh S. Mol Cell Biol. 1996;16:2248–2254. doi: 10.1128/mcb.16.5.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 22.Arnason T, Ellison M J. Mol Cell Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicke L, Riezman H. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 24.Khanna R, Burrows S R, Steingerwald-Mullen P M, Thomson S A, Kurilla M G, Moss D J. Virology. 1995;214:633–637. doi: 10.1006/viro.1995.0076. [DOI] [PubMed] [Google Scholar]
- 25.Guerette P A, Ginzinger D G, Weber B H, Gosline J M. Science. 1996;272:112–115. doi: 10.1126/science.272.5258.112. [DOI] [PubMed] [Google Scholar]
- 26.Gasset M, Baldwin M A, Lloyd D H, Gabiel J-M, Holtzman D M, Cohen F, Fletterick R, Pruisner S B. Proc Natl Acad Sci USA. 1992;89:10940–10944. doi: 10.1073/pnas.89.22.10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, Holmes T, Lockshin C, Rich A. Proc Natl Acad Sci USA. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, Rich A. Proc Natl Acad Sci USA. 1997;94:23–28. doi: 10.1073/pnas.94.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Proc Natl Acad Sci USA. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]