Abstract
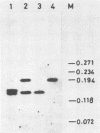
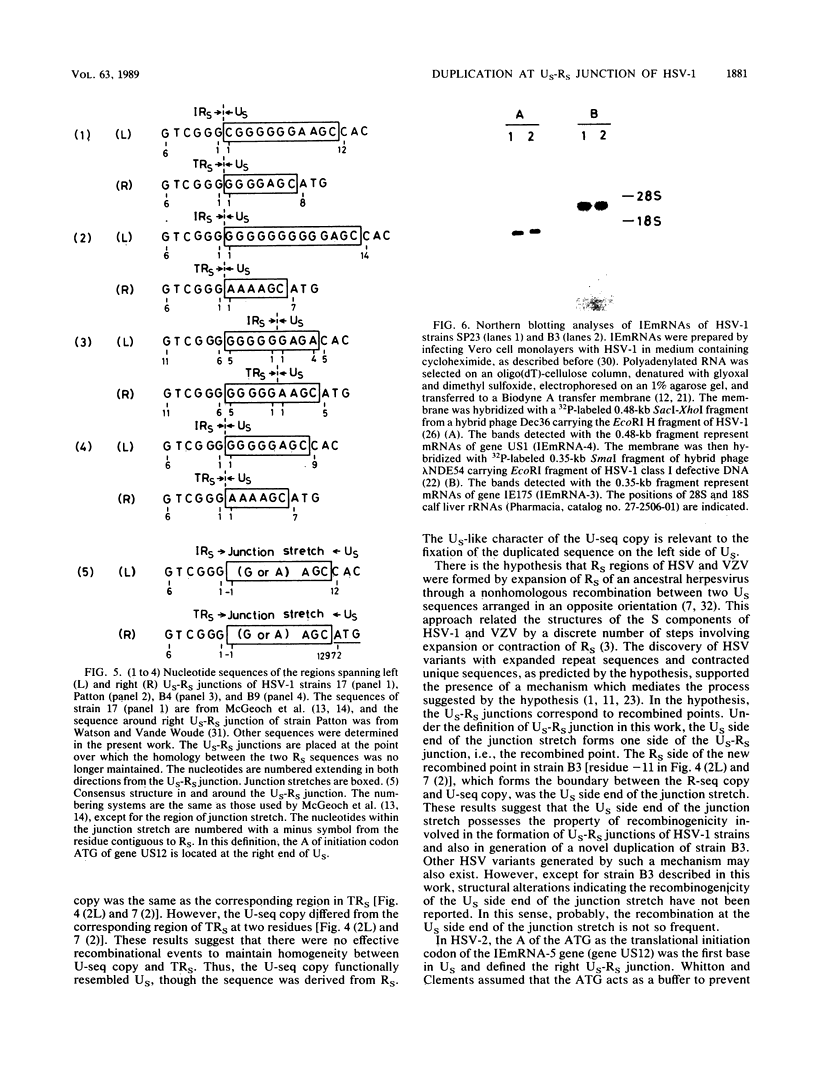
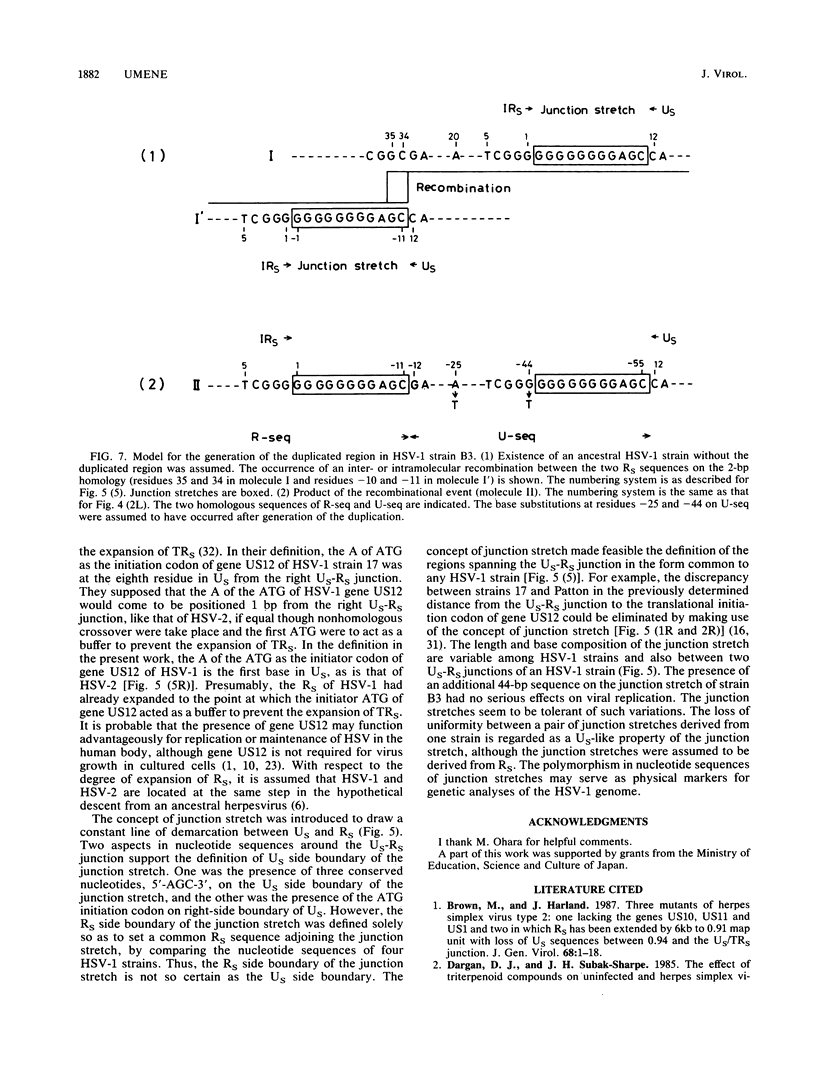
A herpes simplex virus type 1 (HSV-1) strain, B3, was found to have a short duplication on the left junction between the unique sequence (US) and the inverted repeat sequence (RS) in the S component of the genome DNA. A short region of RS contiguous to the left US-RS junction was duplicated in B3. Based on the nucleotide sequences in and around the US-RS junctions of B3 and other HSV-1 strains, a concept of junction stretch was proposed. The organization of junction stretch is RS side 5'-(G or A stretch)AGC-3' US side. Introduction of the concept of junction stretch led to a definition of the structure in and around the US-RS junction, in the form common to HSV-1 strains. The right end of US in the HSV-1 genome was the A of the ATG initiation codon of gene US12, and thus the ATG triplet may act as a buffer to prevent expansion of RS, as is the case with HSV-2. The duplication in B3 was generated by a crossover event between a point on RS and the US side end of the left junction stretch. These observations suggest that the US side end of the junction stretch possesses the property of recombinogenicity, responsible for generation of the duplication in strain B3 and also for the formation of the US-RS junction of HSV.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown S. M., Harland J. Three mutants of herpes simplex virus type 2: one lacking the genes US10, US11 and US12 and two in which Rs has been extended by 6 kb to 0.91 map units with loss of Us sequences between 0.94 and the Us/TRs junction. J Gen Virol. 1987 Jan;68(Pt 1):1–18. doi: 10.1099/0022-1317-68-1-1. [DOI] [PubMed] [Google Scholar]
- Davison A. J., McGeoch D. J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986 Apr;67(Pt 4):597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Inversion of the two segments of the herpes simplex virus genome in intertypic recombinants. J Gen Virol. 1983 Jan;64(Pt 1):1–18. doi: 10.1099/0022-1317-64-1-1. [DOI] [PubMed] [Google Scholar]
- Gentry G. A., Lowe M., Alford G., Nevins R. Sequence analyses of herpesviral enzymes suggest an ancient origin for human sexual behavior. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2658–2661. doi: 10.1073/pnas.85.8.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W. Herpes simplex and 'the herpes complex': diverse observations and a unifying hypothesis. The eighth Fleming lecture. J Gen Virol. 1984 Dec;65(Pt 12):2077–2107. doi: 10.1099/0022-1317-65-12-2077. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Unity and diversity in the herpesviruses. J Gen Virol. 1977 Oct;37(1):15–37. doi: 10.1099/0022-1317-37-1-15. [DOI] [PubMed] [Google Scholar]
- Knipe D. M., Ruyechan W. T., Roizman B., Halliburton I. W. Molecular genetics of herpes simplex virus: demonstration of regions of obligatory and nonobligatory identity within diploid regions of the genome by sequence replacement and insertion. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3896–3900. doi: 10.1073/pnas.75.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J Virol. 1986 May;58(2):583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean A. R., Brown S. M. Deletion and duplication variants around the long repeats of herpes simplex virus type 1 strain 17. J Gen Virol. 1987 Dec;68(Pt 12):3019–3031. doi: 10.1099/0022-1317-68-12-3019. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Donald S., Brauer D. H. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986 Feb 25;14(4):1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Donald S., Rixon F. J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985 Jan 5;181(1):1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Murchie M. J., McGeoch D. J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.978). J Gen Virol. 1982 Sep;62(Pt 1):1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- Rixon F. J., McGeoch D. J. Detailed analysis of the mRNAs mapping in the short unique region of herpes simplex virus type 1. Nucleic Acids Res. 1985 Feb 11;13(3):953–973. doi: 10.1093/nar/13.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Carmichael L. E., Deinhardt F., de-The G., Nahmias A. J., Plowright W., Rapp F., Sheldrick P., Takahashi M., Wolf K. Herpesviridae. Definition, provisional nomenclature, and taxonomy. The Herpesvirus Study Group, the International Committee on Taxonomy of Viruses. Intervirology. 1981;16(4):201–217. doi: 10.1159/000149269. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura T., Umene K., Nishimura J., Fukumaki Y., Sakaki Y., Ibayashi H. Expression of c-myc oncogene during differentiation of human burst-forming unit, erythroid (BFU-E). Biochem Biophys Res Commun. 1986 Mar 13;135(2):521–526. doi: 10.1016/0006-291x(86)90025-2. [DOI] [PubMed] [Google Scholar]
- Umene K. Conversion of a fraction of the unique sequence to part of the inverted repeats in the S component of the herpes simplex virus type 1 genome. J Gen Virol. 1986 Jun;67(Pt 6):1035–1048. doi: 10.1099/0022-1317-67-6-1035. [DOI] [PubMed] [Google Scholar]
- Umene K., Enquist L. W. A deletion analysis of hybrid phage carrying the US region of Herpes simplex virus type 1 (Patton). I. Isolation of deletion derivatives and identification of chi-likes sequences. Gene. 1981 Apr;13(3):251–268. doi: 10.1016/0378-1119(81)90030-5. [DOI] [PubMed] [Google Scholar]
- Umene K., Enquist L. W. Isolation of novel herpes simplex virus type 1 derivatives with tandem duplications of DNA sequences encoding immediate-early mRNA-5 and an origin of replication. J Virol. 1985 Feb;53(2):607–615. doi: 10.1128/jvi.53.2.607-615.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
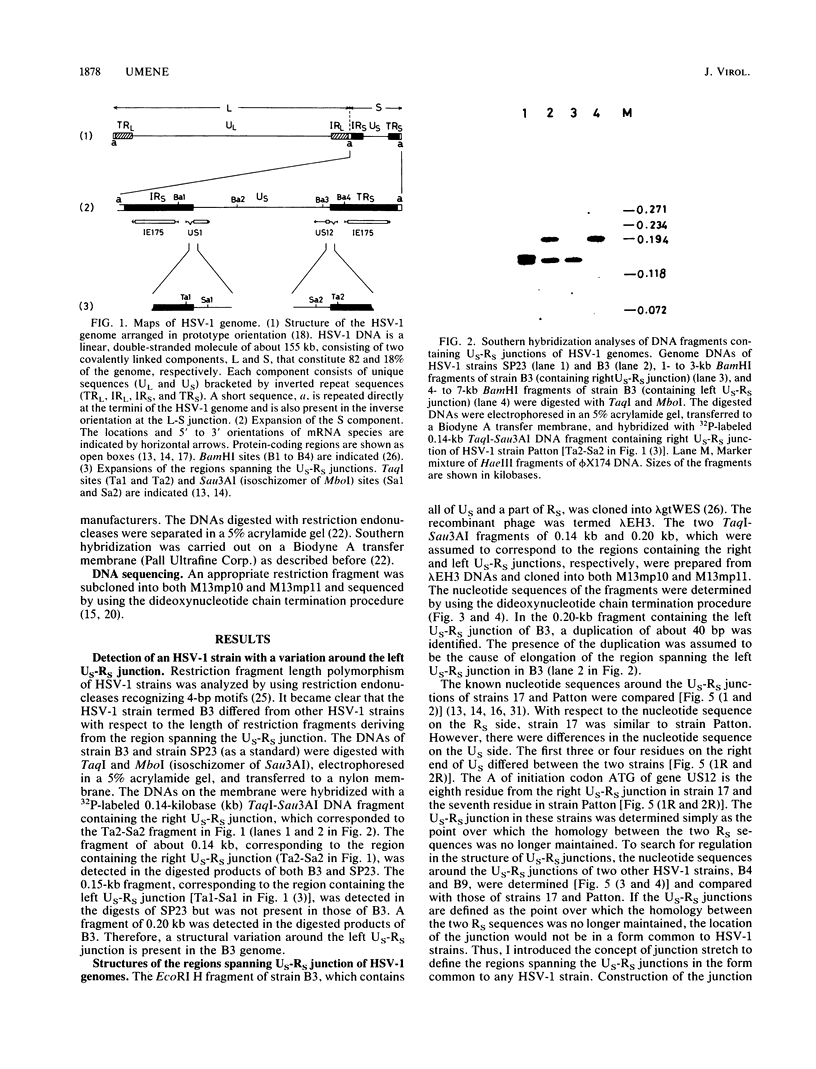
- Umene K. Restriction endonucleases recognizing DNA sequences of four base pairs facilitate differentiation of herpes simplex virus type 1 strains. Arch Virol. 1987;97(3-4):197–214. doi: 10.1007/BF01314421. [DOI] [PubMed] [Google Scholar]
- Umene K. Transition from a heterozygous to a homozygous state of a pair of loci in the inverted repeat sequences of the L component of the herpes simplex virus type 1 genome. J Virol. 1987 Apr;61(4):1187–1192. doi: 10.1128/jvi.61.4.1187-1192.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umene K. Variability of the region of the herpes simplex virus type 1 genome yielding defective DNA: SmaI fragment polymorphism. Intervirology. 1985;23(3):131–139. doi: 10.1159/000149596. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Unstable heterozygosity in a diploid region of herpes simplex virus DNA. J Virol. 1984 Feb;49(2):356–362. doi: 10.1128/jvi.49.2.356-362.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Hyman R. W. Errant processing and structural alterations of genomes present in a varicella-zoster virus vaccine. J Virol. 1985 Oct;56(1):92–101. doi: 10.1128/jvi.56.1.92-101.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. J., Sullivan M., Vande Woude G. F. Structures of two spliced herpes simplex virus type 1 immediate-early mRNA's which map at the junctions of the unique and reiterated regions of the virus DNA S component. J Virol. 1981 Jan;37(1):431–444. doi: 10.1128/jvi.37.1.431-444.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R. J., Vande Woude G. F. DNA sequence of an immediate-early gene (IEmRNA-5) of herpes simplex virus type I. Nucleic Acids Res. 1982 Feb 11;10(3):979–991. doi: 10.1093/nar/10.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L., Clements J. B. The junctions between the repetitive and the short unique sequences of the herpes simplex virus genome are determined by the polypeptide-coding regions of two spliced immediate-early mRNAs. J Gen Virol. 1984 Mar;65(Pt 3):451–466. doi: 10.1099/0022-1317-65-3-451. [DOI] [PubMed] [Google Scholar]