Abstract
The cell matrix adhesion regulator (CMAR) gene has been suggested to be a signal transduction molecule influencing cell adhesion to collagen and, through this, possibly involved in tumor suppression. The originally reported CMAR cDNA was 464 bp long with a tyrosine phosphorylation site at the extreme 3′ end, which mutagenesis studies had shown to be central to the function of this gene. Since the discovery of a 4-bp insertion polymorphism within the originally reported coding region, further sequence information has been obtained. The cDNA has been extended 5′ by ≈2 kb revealing a 559-bp region showing strong homology to the proposed 5′ untranslated sequence of a murine protein kinase receptor family member, variant in kinase (vik). CMAR genomic sequencing has shown the presence of an intron, the intron/exon boundary lying within this region of homology. An RNA transcript for CMAR of ≈2.5 kb has also been identified. The data suggest complex mechanisms for control of expression of two closely associated genes, CMAR and the vik- associated sequence.
A cDNA encoding the gene named cellular matrix adhesion regulator (CMAR; formerly CAR), isolated from a colorectal cell line library, was reported to enhance binding to the extracellular matrix component, collagen type 1 (1). CMAR was thought to be an intronless gene whose structure, integrin β1 dependence, and probable tyrosine phosphorylation activation, suggested a role in signal transduction (2–4). It has since been mapped to 16q24 (5). Because high levels of allelic loss in this region have been shown in breast (6), liver (7), and prostate cancer (8, 9), it has been proposed that the gene might function as a tumor suppressor.
Evidence for expression of the CMAR gene had been difficult to demonstrate in colorectal cell lines and colon tissues, both tumor and normal.
On partial sequencing of a human CMAR containing cosmid, a 4-bp (CACA) insertion was identified within the putative coding sequence of this gene (5). Thirteen offspring whose parents were both known to be heterozygous for the +4-bp form were studied. The +4-bp variant segregated as expected and its population distribution showed a classical Hardy-Weinberg fit (4, 10). The population frequency of this polymorphism was also examined in 131 Caucasoids, consisting of 54 cancer patients and 77 healthy individuals. Results showed 72% to be homozygote for the original CMAR sequence, 25% to be heterozygote and 3% to be +4-bp homozygote. No significant difference in the frequency of the insertion was found between tumor and normal samples (4), indicating a normal polymorphism in the predicted coding region. It was therefore decided to examine further human and animal CMAR sequences, both cDNA and genomic, to learn more about this potentially important gene.
Seven new human cDNAs have been isolated. Mouse and primate libraries also have been screened for CMAR clones. The sequence of the longest human clone, JY44, ≈2.5 kb long, and partial genomic sequences for chimpanzee, gibbon, African green monkey, baboon, and rhesus monkey have been analyzed by using appropriate database searching and sequence retrieval software.
To obtain data for human CMAR genomic organization, a 5.5-kb EcoRI CMAR positive cosmid fragment has been shotgun-cloned into the vector, M13 mp18. The sequences of derived contigs and the cDNA clone JY44 have been compared. CMAR human genomic and cDNAs have also been examined by using the PCR to look for the presence of introns.
MATERIALS AND METHODS
cDNA libraries from the human B lymphoblastoid cell line JY, from the gibbon lymphosarcoma cell line MLA144 (11), and several mouse and human cDNA libraries were kindly donated by D. Simmons (Cell Adhesion Laboratory, Imperial Cancer Research Fund, Institute of Molecular Medicine, John Radcliffe Hospital, Headington, Oxford, U.K.). Two gridded cDNA libraries, number 508, from human fetal thymus and 510, from adult mouse brain (12) as well as MP1 703 a mouse P1 library and two mouse C3H YAC libraries, 902 and 903, were supplied by the Reference Library Database, Max-Planck-Institut for Molecular Genetics (Berlin).
Large-scale and miniprep plasmid preparations were made by using standard alkaline lysis procedures (13). Messenger RNA was prepared by using the Fast Track 2.0 mRNA kit, (Invitrogen), in some cases after stimulation of B lymphoblastoid cells with phorbol 12-myristate 13-acetate (PMA) (Sigma) or NiSO4, (Fluka). Southern and Northern blot analyses were carried out according to standard methods (13) with Hybond-N membranes (Amersham). Probes used for Southern blot analysis were the 464-bp CMAR insert, the 2,411-bp JY44 insert, and PCR fragment LRF3-LRR1, nucleotides 1,303–1,831 of the JY44 sequence (see Fig. 2). Northern blot analysis probes were CMAR and JY44 insert, with actin serving as a mRNA control.
Figure 2.
Nucleotide sequence of CMAR cDNA, JY44. Nucleotides underlined in bold indicate the originally described CMAR cDNA sequence. The position of the CACA repeat in a CMAR polymorphic variant is indicated by the dots after nucleotide 2,232. The proposed methionine start sites boxed at positions 2,131–2,213 for the variant are indicated with an arrow. Other start codons in translation frame 1 at positions 949–1,111 and in frame 3 at positions 552–1,611 are boxed. The underlined sequence between nucleotides 1,021–1,580 indicates the region showing strong homology with vik. The two blocks of sequence between nucleotides 200–500 and 700–900 indicate the regions containing Alu repeat elements. Regions in which ests have been found, nucleotides 1,200–1,750 and 1,800–2,411 are indicated on the right.
PCR was performed in a Hybaid HBTR2 thermocycler. Reactions were denatured at 94°C for 3 min and cycled 35 times with annealing at 60°C for 1 min, extension at 72°C for 2 min, and denaturation at 94°C for 1 min, with a final extension at 72°C for 10 min. For amplification of PCR products over 2 kb long, an extension time of 4 min was used. Oligonucleotide primers for PCR, defined as follows, were synthesized and where appropriate biotinylated at the Human Genetic Resources Laboratory (Imperial Cancer Research Fund, Clare Hall Laboratories, Hertfordshire, U.K.): JY44R1, 5′-GCCGCGAGAGGAAGTGCAAA; JY44F5P3, 5′-CACCCCCATCGCCTACTCCATG; R2, 5′-AAACCAAGGCATCCCCAACA; F2, 5′-TAGACAGTCGTTCCCAGTGT; Fc, 5′-GCAGAGCATTCAGACTCCAA; and Fx, 5′-TCAGCCACCCTGAGTTGCTT.
Automatic sequencing was carried out by using the Applied Biosystems Prism 377 DNA sequencer with an Applied Biosystems Prism Dye Terminator Cycle Reaction kit (Perkin–Elmer), or the ALF DNA Sequencer with an Autoread Sequencing Kit using Fluor-dATP labeling Mix (Pharmacia). Some hand dideoxynucleotide chain termination reaction sequencing (14) was carried out by using a Sequenase 2.0 kit (United States Biochemical/Amersham). Genomic DNA was sequenced either by direct sequencing of a PCR-amplified fragment with the Applied Biosystems Prism 337 DNA Sequencer, automatic sequencing of a PCR fragment cloned into the plasmid pGem-T (Promega), or hand sequencing of a PCR fragment following strand separation with streptavidin-coated Dynabeads (Dynal, Great Neck, NY) where amplification had taken place with one of the primers biotinylated.
Shotgun cloning was carried out by using the vector M13 mp18 (Stratagene) (15).
Computer sequence analyses were carried out by using GCG software, and databases searched were Genetics Computer Group (Madison, WI) and the Institute for Genomic Research Human cDNA Database (tigr hcd) level 1 (16).
RESULTS
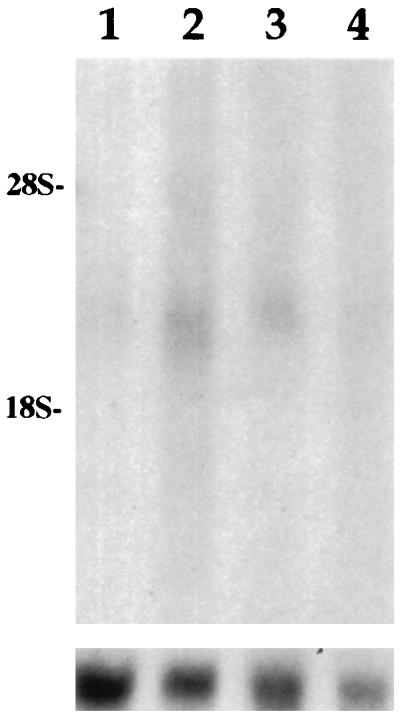
By using Northern blot analysis with the probe JY44, a band of ≈2.5 kb has been identified in human B lymphoblastoid cell lines, Jurkat 6 and JY after stimulation of cells with PMA or NiSO4 (Fig. 1). Very little or no mRNA was seen in the lymphoblastoid cell lines without stimulation. In colorectal cell lines no message was detected by Northern blot analysis before or after PMA or NiSO4 treatment of cells.
Figure 1.
Expression of the CMAR gene shown by Northern blot analysis. Messenger RNA was isolated from Jurkat 6 cells after stimulation for 1 hr with (i) no treatment; (ii) 10 ng/ml PMA and 10−2 M NiSO4; (iii) 10 ng/ml PMA; and (iv) 10−2 M NiSO4. mRNA (5 μg) was separated on a formaldehyde gel, transferred to Hybond-N and probed with insert JY44. The positions of 28S and 18S ribosomal RNAs are shown on the left. The same filter probed with actin as a mRNA control is shown in the lower portion.
The sequences of the two human CMAR cDNA clones, JY44 (2,411 bp) and 15.3 (1,100 bp) have been compared. Where these overlapped they matched exactly except where JY44 had nucleotides TCT at positions 5–7 in the poly(A) tail. The sequence of the largest cDNA isolated, JY44, has been studied in detail (Fig. 2). Examination with GCG Frames showed a number of ORFs in all three translations. The proposed original CMAR AUG and the +4-bp polymorphic variant start sites in frame 1 both fulfill the Kozak consensus sequence GCC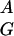 CCAUGG, with strong and weak initiation codons, respectively, the first with G at −3, the variant with G at +4. The most important nucleotide residue, determining a strong initiator codon, is considered to be purine A or G in position −3. With C or T in this position, G at +4 is critical, and surrounding nucleotides come into play, designating a weak initiation codon (17). Other strong start codons were found at position 1111 in frame 1 and positions 552 and 1611 in frame 3. One codon satisfying both −3 and +4 criteria was found at position 949 in an ORF of 126 nucleotides in frame 1. Further GCG sequence analysis tools, codonpreference, testcode, and pepplot were used to analyze sequences of the cDNA JY44 and its three forward peptide translations but none indicated a particular candidate region that might contain a protein.
CCAUGG, with strong and weak initiation codons, respectively, the first with G at −3, the variant with G at +4. The most important nucleotide residue, determining a strong initiator codon, is considered to be purine A or G in position −3. With C or T in this position, G at +4 is critical, and surrounding nucleotides come into play, designating a weak initiation codon (17). Other strong start codons were found at position 1111 in frame 1 and positions 552 and 1611 in frame 3. One codon satisfying both −3 and +4 criteria was found at position 949 in an ORF of 126 nucleotides in frame 1. Further GCG sequence analysis tools, codonpreference, testcode, and pepplot were used to analyze sequences of the cDNA JY44 and its three forward peptide translations but none indicated a particular candidate region that might contain a protein.
JY44 sequence homology searches were made by using tigr hacd level 1 and GCG, fasta, and blast database search software. Two regions covering base pairs ≈200–500 and 700–900 contained strong sequence homology with Alu repeat elements (18) (Fig. 2), the majority falling within the most 5′ region.
By using expressed sequence tags (est) within blast, more than 100 ests having high homology with JY44 were found and these fell broadly within two main regions, nucleotides 1,200–1,750 and 1,800–2,411. They ranged from ≈20–300 bp long. Seventy five percent, which included the longest test sequences, were in the second group at the 3′ end of JY44 that contains the original 464-bp CMAR sequence (Fig. 2). These JY44 high homology est sequences had been derived from several human tissues, namely, brain, breast, colon, epididymis, ovary, skeletal muscle, skin, fetal lung, fetal heart, and fetal liver.
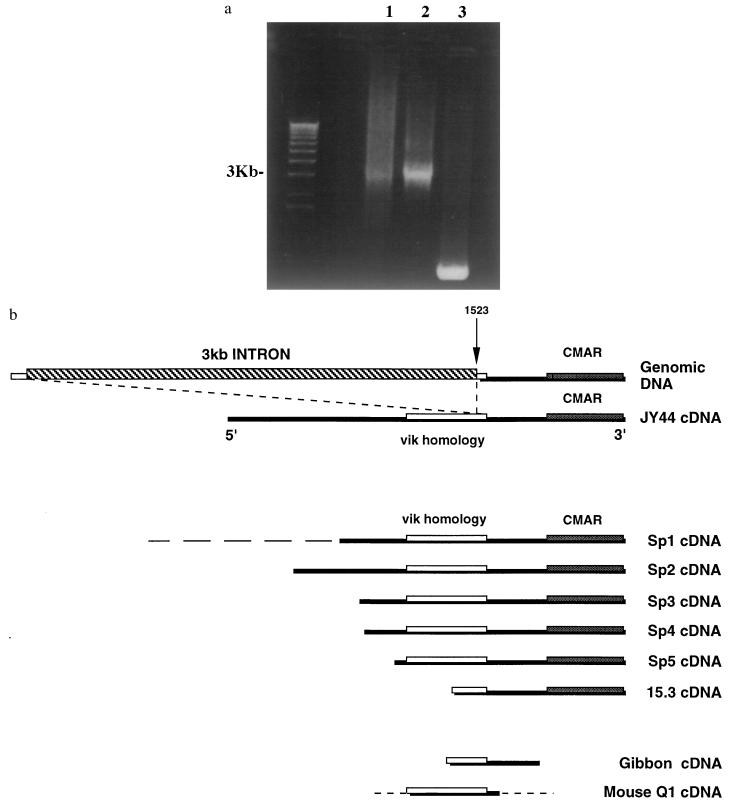
An interesting nucleotide homology was discovered with most of the 5′ noncoding sequence of a murine protein called vik (variant in the kinase). This gene is a putative receptor tyrosine kinase, and shows strong homology at the amino acid level to other receptor protein kinases (19). GCG bestfit analysis over the region of homology with vik in JY44, nucleotides 1,021–1,580 (Fig. 2) showed significant nucleotide identity (84.3%). To learn more about the significance of the vik homology, a human fetal thymus gridded cDNA library was screened with a PCR-derived probe of 523 bp, LRF3-LRR1, which included part of the vik homology region. Five clones, Sp1 ≈3,000 bp, Sp2, (2,036 bp), Sp3, (1,634 bp), Sp4, (1,600 bp), and Sp5, (1,332 bp) were isolated. These all contained the region of vik homology and the original 450-bp CMAR sequence at the 3′ end. The 3,000-bp clone was rearranged 5′ with respect to the vik homology region, but the remaining clones, where they overlapped, all showed complete homology with JY44 (Fig. 4b), apart from different length of polyA tails. These varied between 16 and 50 A residues, and one clone had nucleotides TCT within the poly(A) tail at positions 5–7, as had JY44.
Figure 4.
Genomic organization of CMAR. (a) PCR from (as shown in Fig. 1) genomic DNA, (as shown in Fig. 2) cosmid DNA, and (as shown in Fig. 3) cDNA by using primers JY44F5P3 and JY44R1. Lambda one kilobase markers are shown on the left. (b) Human genomic DNA showing position of intron compared with human cDNAs, JY44, Sp1, Sp2, Sp3, Sp4, Sp5, and 15.3, gibbon cDNA, and mouse cDNA Q1. The broken line at the 5′ end of Sp1 indicates rearranged sequence. The dotted lines in mouse Q1 cDNA indicate no homology with other sequences. The vik homology region and the originally reported CMAR region are shown.
The original CMAR 464-bp probe has, in our hands, been unsuccessful in isolating a mouse CMAR homolog. A gridded P1 mouse library, two YAC mouse libraries, and an adult mouse thymocyte cDNA library were screened by using Southern blot analysis of plated colonies. Eight other mouse cDNA libraries were screened directly by Southern blot analysis. Even under low stringency conditions [50°C, 2× standard saline citrate (0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS] no CMAR positive mouse clones were identified. Because probe LRF3-LRR1, covering the vik homology region, had been successful in isolating several human CMAR cDNAs, this probe was used to screen a gridded adult mouse brain cDNA library. Only one partial cDNA of 1,113 bp named Q1 was isolated. GCG gap analysis showed homology with the JY44 sequence between nucleotides 196–845, spanning the 559 nucleotides of vik homology (see Fig. 4b). When Q1 and the variant in kinase sequences were compared, bestfit analysis showed strong homology of Q1 nucleotides 196–760 with vik nucleotides 8–563, having 81% identity. Surrounding 5′ and 3′ sequences showed no similarity, indicating that the mouse clone Q1 was not part of the variant in kinase gene. In addition, it showed no similarity with mouse or human tyrosine kinase receptors (RYK) (20, 21).
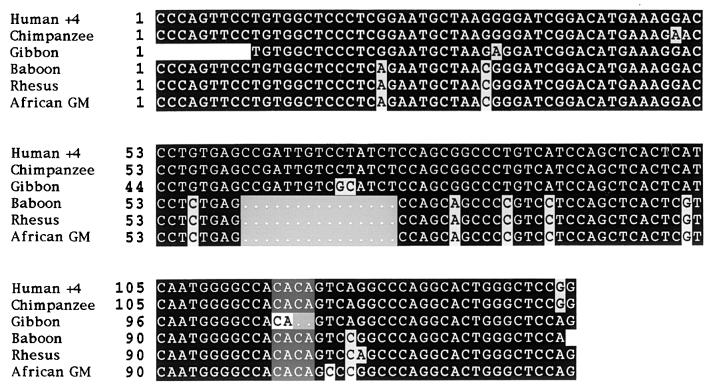
Several partial primate CMAR sequences have been obtained by sequencing PCR products amplified from genomic DNA by using primers R2 and F2. GCG pileup and lineup multiple sequence alignment software comparing human, chimpanzee, gibbon, African green monkey, baboon, and rhesus monkey sequences showed that the CMAR sequence over this limited region is well conserved between various Old World monkey species (Fig. 3). All showed the 4-bp insertion found in the polymorphic variant of CMAR, except gibbon, which had a 2-bp CA insertion at the same position (Fig. 3). Further sequence differences noted between these species included a 15-bp insertion/deletion and scattered single nucleotide transitions and transversions. A partial cDNA gibbon sequence was also obtained. This showed GCG bestfit identity of 95% with nucleotides 1,431–1,982 of JY44 that covered 159 nucleotides of the vik homology region, but did not extend 3′ as far as the polymorphic insertion position (Fig. 4b). The presence of one CA repeat in the gibbon sequence was checked in two additional independent lar gibbon (Hylobates lar) genomic DNA samples derived from the lymphosarcoma cell line MLA 144 and from fresh gibbon liver (obtained from London Zoological Gardens). Two sets of primers, Fx, R2 and F1, R2, designed to span the insertion position and also amplify longer regions of DNA were used. Results confirm that gibbon genomic DNA has only a 2-bp CA insertion, unlike the 4-bp CACA insertion seen in other monkey species and in the human polymorphic variant of CMAR (Fig. 3).
Figure 3.
Comparison of human and primate CMAR sequences. PCR fragments were amplified from the human +4-bp variant, chimpanzee, gibbon, baboon, rhesus, and African green monkey genomic DNA by using primers F2 and R2, and sequences were compared by using GCG pileup/lineup software. Old World monkeys show a 15-bp insertion/deletion at position 61–76 and scattered single nucleotide transitions and tranversions. The gibbon has a 2-bp CA insertion at position 107 in contrast to the 4-bp insertion in the other primate species.
An EcoRI, 5.5-kb CMAR positive cosmid fragment was shotgun cloned into M13 mp18. The sequences of the resulting M13 mp18 clones were analyzed by using GCG sequence assembly that identified three main contigs. Contig 1 of 2945-bp contained the original CMAR cDNA sequence. When contigs were compared with the sequence of cDNA JY44, Gap analysis showed a perfect match between nucleotides 1,046–1,910 in contig 1 and nucleotide 1,524 to the 3′ end in the cDNA, apart from the 4-bp insertion polymorphism in the cosmid. 5′ to this position there was no homology between the cDNAs and genomic sequences. These results indicated the presence of an intron in the cosmid sequence ending at the position equivalent to 1,523 in the cDNA JY44 sequence (Fig. 4b). PCR was carried out on genomic, cosmid, and cDNA templates by using primers, F5P3 and JY44R1, which span the predicted intron/exon boundary. A product of 3 kb was amplified from both genomic templates and one of 500 bp was amplified from the cDNA template, as predicted (Fig. 4a). Direct Applied Biosystems Prism 337 sequencing of the 3-kb cosmid PCR fragment confirmed the presence of an intron and identified GT and AG splice site consensus sequences at exon/intron and intron/exon boundaries, respectively (22).
DISCUSSION
An RNA transcript of ≈2.5k has been identified for the CMAR gene (Fig. 1) but this could be detected only after treatment of B lymphoblastoid cells with PMA (23) or nickel (24). It would appear, however, that the message is widely expressed because, in addition to the original CMAR cDNA derived from colorectal cells, CMAR cDNAs have been isolated from B and T lymphoblastoid (A. Arruffo, Scripps Institute, Seattle, personal communication) and melanoma cells (5), and numerous est sequences with strong homology to JY44 have been found in a wide range of human tissues.
The sequence of cDNA JY44, except 5′ to position 375, has been confirmed in six other independent cDNAs isolated from three different libraries (Fig. 4b). The presence of Alu repeat elements within nucleotides ≈200–500 and 700–900 has been shown. Alu-like sequences appear in about 5% of published cDNA sequences, the great majority, 96% lying within untranslated regions (25).
The region of JY44, nucleotides 1,021–1,580 showing strong identity with the predicted 5′ untranslated region in the murine variant in kinase gene (Fig. 4b), is of particular interest because this level of human/mouse homology suggests there is some biological significance in this sequence conservation. A short gibbon cDNA sequence also includes part of this homologous region (Fig. 4b). Because this is the only region of homology between JY44 and vik it seems possible that these are unrelated genes that share a common controlling region. It was noted that alternative 5′ initiation sites exist in the vik ORF, several with strong Kozak consensus sequences (19). If the vik initiation site favored by Kelman et al. (19) were not the correct one, this would place the vik/JY44 hr within the coding sequence implying the same for the region defined by nucleotides 963–1,580 in JY44. The vik protein as defined in (19), however, is related to the tyrosine-specific protein kinase receptors family showing strong homology with the human and mouse RYK cDNA sequences (21) that would not fit easily with the vik JY44 hr being part of the same protein. Although an alternative initiation site for the mouse RYK has been discussed (20), the predicted start site in the human RYK sequence seems unequivocal. On the basis of this, the predicted start site for mouse vik would seem to be the correct one because a short common region is revealed in comparisons between JY44, vik, mouse RYK and human RYK was 78 bp 5′ to the initiation site in human RYK (Fig. 5). Thus it would appear that the JY44 sequence homologous to vik could indeed lie within the 5′ putative untranslated regions of mouse vik and RYK.
Figure 5.
Comparison of partial sequences showing the relationship of JY44 to protein kinase receptor related molecules, vik, mouse RYK and untranslated 5′ human RYK. A short region of complete homology, marked in bold, 1,571–1,580 in JY44 exists with the vik, mouse RYK, and human RYK sequences. In human RYK the initiation codon is situated a further 78 nucleotides downstream at position 133 in the reported sequence (21). Vik/JY44 homology continues upstream to nucleotide in 1,021 JY44.
The originally proposed CMAR AUG initiation sites at nucleotides 2,131 and 2,213 for the 4-bp insertion variant have not been fully confirmed. Reinitiation has been described where two equally strong 5′ and 3′ codon start sites both may be used if there is a terminator codon in-frame with the first AUG codon and upstream from the second, as occurs in this case, possibly mediating related or different biological functions (17, 26). The possibility of exclusive translation of a downstream ORF by internal ribosome entry segments must also be considered although this would leave an unusually long 5′ untranslated region (27). Furthermore, it is unusual for such controlling regions to show extensive homology across species for such a relatively long sequence. The presence of upstream AUG codons and an intron constitute a strong hint that the cDNA might represent a stable but untranslatable pre-mRNA that requires a final conversion to functional RNA (17). The possibility that two proteins may be made from the same mRNA, CMAR and vik homolgy region, must be considered and emphasizes the need to search for the function of vik homology region. The other potential ORFs in JY44 are all related to Alu sequences and so not relevant to function. Although no mouse CMAR cDNA clone has yet been found, the analysis of the mouse Q1 cDNA at least suggests the existence of a mouse equivalent to the human vik homology region that is adjacent to CMAR on the genome.
Partial CMAR sequences have been found in some Old World monkeys (Fig. 3). As might be expected, many of the sequence differences are split between the Old World monkeys and Great Apes. The presence of the 4-bp insertion in all primate species except gibbon, suggests that this is the original form of CMAR present in a common primate ancestor with gibbon losing one CA repeat and human having a polymorphic variant in which both CA repeats have been lost. The 2-bp insertion in the gibbon sequence interrupts the reading frame at the same position as described for the 4-bp variant in human with yet another frame shift and is so far unexplained. It cannot be ruled out that at such a position, there is slippage of transcription or translation leading to enough normal product for reasonable function. Apart from the alternative use of a CTG start site (28) causing the terminal tyrosine to be retained, we also cannot rule out the possibility that all CMAR variants use the same initiation codon as has been proposed for CMAR (29).
The notion that the original CMAR region is important functionally is supported by our studies with transfectants (4, 5) and a more recent report (30) that supports the original proposal that the gene may function as a tumor suppressor. In this work CMAR transfectants were shown to have reduced metastatic potential in nude mice. In addition, by using reverse transcriptase–PCR, with primers from the 464-bp cDNA, CMAR expression in cancer was found to be less than one-tenth of that in matched noncancerous tissue (30).
Our studies also have provided some information about the CMAR gene structure at the genomic level. Comparisons between genomic and cDNA sequences have shown the original supposition, that CMAR was an intronless gene, to be incorrect. At least one intron of 3 kb has been identified, suggesting some degree of mRNA processing (Fig. 4b). An intron/exon boundary at position 1,523 of the 2.5-kb cDNA, JY44 lies within nucleotides 1,021–1,580, the region showing strong homology with the variant in kinase gene. Sequence analysis suggests that the vik homology region sequence may correspond to a functional protein and that the CMAR message, as represented for example by JY44, is capable of giving rise to two proteins, CMAR and vik homology region. This raises the intriguing question as to what the function of vik homology region, presumably also made by the mouse vik message and its homolog to vik homology region (identified in clone Q1) really is. Clearly, important questions about the mRNA organization of these intriguing sequences still remain to be satisfactorily resolved.
ABBREVIATIONS
- CMAR
cell matrix adhesion regulator
- est
expressed sequence tags
- PMA
phorbol 12-myristate 13-acetate
- vik
variant in kinase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF034795).
References
- 1.Pullman W E, Bodmer W F. Nature (London) 1992;356:529–532. doi: 10.1038/356529a0. ; revision (1993) 361, 564. [DOI] [PubMed] [Google Scholar]
- 2.Juliano R L, Haskill S. J Cell Biol. 1993;120:577–585. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romer L H, Burridge K, Turner C E. Cold Spring Harb Symp Quant Biol. 1992;57:193–202. doi: 10.1101/sqb.1992.057.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Novelli M R, Durbin H, Bodmer W F. In: Topics in Molecular Medicine. Seiss W, Lorenz R, Weber P C, editors. Vol. 1. New York: Raven; 1995. pp. 179–186. [Google Scholar]
- 5.Durbin H, Novelli M, Bodmer W. Genomics. 1994;19:181–182. doi: 10.1006/geno.1994.1038. [DOI] [PubMed] [Google Scholar]
- 6.Sato T, Tanigami A, Yamakawa A, Akiyama F, Kasumi F, Sakamoto G, Nakamura Y. Cancer Res. 1990;50:7184–7189. [PubMed] [Google Scholar]
- 7.Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Harada H, Yagura M, Matsubara K, Nakamura Y. Cancer Res. 1991;51:89–93. [PubMed] [Google Scholar]
- 8.Carter B S, Ewing C M, Ward W S, Treiger B F, Aalders T W, Schalken J A, Epstein J I, Isaacs W B. Proc Natl Acad Sci USA. 1990;87:8751–8755. doi: 10.1073/pnas.87.22.8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunimi K, Bergerheim U S, Larsson I L, Ekman P, Collins V P. Genomics. 1991;11:530–536. doi: 10.1016/0888-7543(91)90059-n. [DOI] [PubMed] [Google Scholar]
- 10.Cavalli-Sforza L L, Bodmer W F. The Genetics of Human Populations. San Francisco: Freeman; 1971. [Google Scholar]
- 11.Kawakami T G, Huff S D, Buckley P M, Dungworth D L, Snyder S P, Gilden R V. Nat New Biol. 1972;235:170–171. doi: 10.1038/newbio235170a0. [DOI] [PubMed] [Google Scholar]
- 12.Lehrach H, Drmanac R, Hoheisel J, Larin Z, Lennon G, Monaco A P, Nizetic D, Zehetner G, Poustka A. In: Genome Analysis. Davies K E, Tilghman S M, editors. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 39–81. [Google Scholar]
- 13.Sambrook J, Frisch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bankier A T, Weston K M, Barrell B G. Methods Immunol. 1987;155:51–93. doi: 10.1016/0076-6879(87)55009-1. [DOI] [PubMed] [Google Scholar]
- 16.Adams, M. D., Kerlavage, A. R., Fleischmann, R. D., Fuldner, R. A., Bult, C. J., et al. (1995) Nature (London) 377, Suppl., 3–174. [PubMed]
- 17.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid C W, Jelinek W R. Science. 1982;216:1065–1070. doi: 10.1126/science.6281889. [DOI] [PubMed] [Google Scholar]
- 19.Kelman Z, Simon C D, Guenet J L, Yarden Y. Oncogene. 1993;8:37–44. [PubMed] [Google Scholar]
- 20.Hovens C M, Stacker S A, Andres A-C, Harpur A G, Ziemiecki A, Wilks A F. Proc Natl Acad Sci USA. 1992;89:11818–11822. doi: 10.1073/pnas.89.24.11818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stacker S A, Hovens C M, Vitali A, Pritchard M A, Baker E, Sutherland G R, Wilks A F. Oncogene. 1993;8:1347–1356. [PubMed] [Google Scholar]
- 22.Lewin B. Genes V. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 23.Lieberman A P, Pitha P M, Shin M L. J Exp Med. 1990;172:989–992. doi: 10.1084/jem.172.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisby S, Muller K M, Jongeneel C V, Saurat J-H, Hauser C. Int Immunol. 1995;7:342–352. doi: 10.1093/intimm/7.3.343. [DOI] [PubMed] [Google Scholar]
- 25.Yulug I G, Yulug A, Fisher E M. Genomics. 1995;27:544–548. doi: 10.1006/geno.1995.1090. [DOI] [PubMed] [Google Scholar]
- 26.Liu C-C, Simonsen C C, Levinson A D. Nature (London) 1994;309:82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- 27.Jackson R J, Kaminski A. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 28.Lock P, Ralph S, Stanley E, Boulet I, Ramsay R, Dunn A R. Mol Cell Biol. 1991;11:4363–4370. doi: 10.1128/mcb.11.9.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molla A, Rouard-Talbot L, Block M R. Biochim Biophys Acta. 1996;1315:6–8. doi: 10.1016/0925-4439(95)00111-5. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto H, Itoh F, Hinoda Y, Imai K. Cancer Res. 1996;56:3605–3609. [PubMed] [Google Scholar]