Abstract
Yersiniae, causative agents of plague and gastrointestinal diseases, secrete and translocate Yop effector proteins into the cytosol of macrophages, leading to disruption of host defense mechanisms. It is shown in this report that Yersinia enterocolitica induces apoptosis in macrophages and that this effect depends on YopP. Functional secretion and translocation mechanisms are required for YopP to act, strongly suggesting that this protein exerts its effect intracellularly, after translocation into the macrophages. YopP shows a high level of sequence similarity with AvrRxv, an avirulence protein from Xanthomonas campestris, a plant pathogen that induces programmed cell death in plant cells. This indicates possible similarities between the strategies used by pathogenic bacteria to elicit programmed cell death in both plant and animal hosts.
Keywords: bacterial pathogenesis
Yersinia spp. pathogenic to humans (Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis) all harbor a highly conserved 70-kb plasmid (pYV) that is essential for virulence (1). This plasmid contains ≈50 virulence genes encoding an elaborate type III secretion system (ysc) and several proteins called Yops that are secreted upon contact with eukaryotic host cells. Some of the Yops (including YopE, YopH, YopO, and YopM) are delivered into the host cell cytosol where they damage the cytoskeleton and disrupt the signaling network (2–6). These Yop effectors are translocated across the eukaryotic cell membrane by a specialized apparatus made up of several other Yops including YopB and YopD (2, 6, 7). Secretion of Yop effectors and translocators requires specific bacterial chaperones, called Syc proteins (8). Related type III secretion systems have been encountered in various other animal pathogenic bacteria such as Pseudomonas aeruginosa (Exs/Psc), enteropathogenic Escherichia coli (Sep), Shigella spp. (Mxi/Spa), and Salmonella spp. (Inv/Spa), as well as plant pathogenic bacteria such as Xanthomonas campestris (Hrp), Pseudomonas syringae (Hrp), and Erwinia spp. (Hrp) (9, 10). In all of these bacteria, type III secretion of virulence proteins is required for successful host–pathogen interactions.
In this paper, we show that Y. enterocolitica induces apoptosis in the mouse monocyte–macrophage cell line J774A.1 and that this effect depends on YopP, which is presumably translocated by the pathogen into the target cell.
MATERIALS AND METHODS
Bacterial Strains and Methods.
Y. enterocolitica O:9 E40(pYV40) (4) and nonpolar knockout mutants thereof (construction described below) were used throughout this study. Y. enterocolitica W22703(pYV227) and transposon mutants yopE (pGC1256), yopH (pGC1152), yopM (pBM15), yopOP (pGC559), yopP (pGB107), and yopBD (pGC153) (11) as well as nonpolar yscN (pSW2276) (12) and sycE (pPW2254) (13) mutants were used to support data obtained with strain E40. Bacterial growth, conjugations, temperature induction of the yop virulon in a Ca2+-deficient medium, and Yop protein analysis were as described (3).
yop Mutant Constructions, Plasmids, and Nucleotide Sequencing.
Nonpolar mutations in the following genes in strain E40 virulence plasmid (pYV40) have been described previously: yscN (pMSL41) (4), yopB (pPW401) (6), and sycD (pAB41) (4). Additional mutations in yop genes of the virulence plasmid pYV40 were constructed as follows: The yopD mutant E40(pMSL44) was made by an in-frame deletion of codons 121–165. The yopE mutant E40(pAB4052) was constructed using the yopE mutator pPW52 (13). The yopH mutant E40(pSI4008) was constructed by deleting the first 352 codons. The yopM mutant E40(pAB408) has a stop codon after 23 codons of yopM. The yopO mutant E40(pAB406) was made by an in-frame deletion of codons 65–558. The yopP mutant E40(pMSK41) contains a deletion of a central 514-bp region of yopP between two BamHI sites. All mutants were obtained by allelic exchange (14).
To overexpress YopP in Y. enterocolitica, yopP was PCR amplified using oligonucleotide primers MIPA 495 (5′-CCTGAATAAGGATAAACATATGATTGGGCCA) and MIPA 494 (5′-CCATACTGGAGCAAGCTTTCCAAAGTACATTA) and cloned downstream of the strong yopE promoter in pCNR26 vector to make pMSK13. Plasmid pCNR26 is a mobilizable derivative of pTZ19R, containing the promoter of gene yopE and an optimized Shine Dalgarno sequence (C. Neyt and G.R.C., unpublished work).
Nucleotide sequencing of yopP was performed by the dideoxy-nucleotide chain-termination method (15) using a Taq cycle sequencing kit (Amersham) and an automated sequencer (Li-Cor, Lincoln, NE). Sequence homology searches were performed using blastp (16).
Cell Culture and Macrophage Infection.
Mouse monocyte–macrophage J774A.1 cells (ATCC TIB 67) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (GIBCO) at 37°C under 8% CO2. Unless otherwise indicated, macrophages were seeded at 5 × 105 cells/12-mm glass coverslip in 24-well tissue culture plates 15 h in advance of infection. Macrophages were infected with 10 bacteria per cell with Y. enterocolitica strains grown under conditions for moderate Yop induction at 37°C (17). After a 30-minute infection period, cells were incubated with gentamicin at 30 μg/ml to kill extracellular bacteria. Where indicated, cytochalasin D (Sigma), at a final concentration of 2.5 μg/ml, was added to the J774A.1 cells 30 minutes before infection and was maintained throughout the experiment.
Assessment of Apoptosis by Terminal Deoxyribonucleotidyl Transferase-Mediated dUTP-Digoxigenin Nick End-Labeling (TUNEL) Reaction and Epifluoresence Microscopy.
Immunofluorescence detection of fragmented genomic DNA (characteristic of apoptosis) was accomplished using the TUNEL reaction followed by the addition of fluorescein isothiocyanate-conjugated anti-digoxigenin antibodies. Y. enterocolitica-infected or uninfected J774A.1 cells were fixed for 20 minutes in 2.5% paraformaldehyde, extracted with 2:1 ethanol:acetic acid for 5 minutes at −20°C, and processed for epifluorescence microscopy using the Apoptag In Situ Apoptosis Detection Kit (Oncor Apoptag S7110-KIT) according to the manufacturer’s instructions. Processed cells were evaluated by phase contrast and epifluorescence (480 nm) microscopy of three random fields of view (100–175 cells per field) to determine the percentage of TUNEL-positive nuclei at ×400 magnification. We confirmed that 95–100% of the cells had bacteria associated with them as determined by indirect immunofluorescent staining of Y. enterocolitica using rabbit anti-O:9 Y. enterocolitica antiserum (gift of G. Wauters) followed by the addition of Texas Red-conjugated goat anti-rabbit antibodies (Molecular Probes).
Analysis of DNA Fragmentation.
Oligonucleosomal length DNA fragmentation in Y. enterocolitica-infected or uninfected J774A.1 cells was detected by agarose gel electrophoresis as described (18). In brief, 107 J774A.1 cells were seeded into 100-mm tissue culture dishes and infected using a multiplicity of infection of either 10 or 100 bacteria per cell as described above. J774A.1 DNA was isolated from cells 2 h after infection in 5 ml of lysis buffer (8 mM EDTA/10 mM Tris, pH 7.2/0.2% Triton X-100/15 μg/ml proteinase K) as described (18).
Electron Microscopy.
J774A.1 cells (20 × 106) were seeded into 100-mm tissue culture dishes and infected as described above. Cells were fixed with 1% glutaraldehyde and processed for electron microscopy in pellets as described (19). Semi-thin sections were stained with toluidine blue and examined by light microscopy.
RESULTS
Y. enterocolitica Induces Apoptosis in Infected Macrophages.
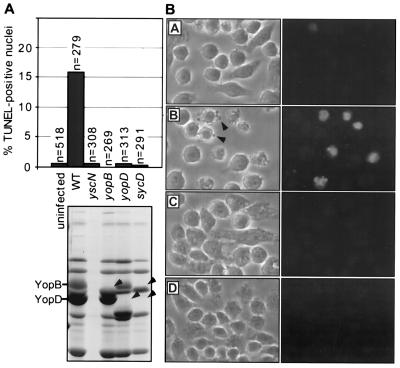
In preliminary experiments, we have found by phase contrast microscopy that the wild-type Y. enterocolitica strain E40 causes cytotoxicity in J774A.1 cells whereas the type III secretion mutant, yscN, has no such effect. These results indicated that a secreted Yop was likely to be involved in macrophage cytotoxicity. However, a mutant unable to secrete cytotoxin YopE was even more cytotoxic than the wild-type, indicating that YopE was not involved. Infected macrophages displayed general features of apoptosis such as membrane blebbing (apoptotic body formation) and nuclear and cellular shrinkage (data not shown). Results presented in Fig. 1A show that DNA fragmentation occurred in a significant number of macrophage nuclei, as revealed by the TUNEL reaction (20). Morphological observations confirming these findings are shown in Fig. 1B.
Figure 1.
A Y. enterocolitica Yop effector induces apoptosis in macrophages. (A) Percentage of TUNEL-positive nuclei in J774A.1 cells 4 h postinfection. n values refer to the number of total cells evaluated per treatment. All strains were tested a minimum of three times. WT, wild-type. Nonpolar mutations in genes required for secretion (yscN) and translocation (yopB, yopD and sycD) were confirmed by 12% SDS/PAGE (lanes correspond with mutants indicated above the gel). Arrowheads indicate the position of the deleted protein. The correct positions of Yops are indicated at left. (B) Phase contrast (left) and fluorescence (right) images depicting the results of the infection (cell morphology) and TUNEL reaction (nuclear morphology), respectively. (A) Uninfected cells, (B) wild-type E40, (C) yscN mutant, and (D) yopB mutant. Arrowheads indicate J774A.1 membrane blebbing induced by wild-type E40 infection.
Y. enterocolitica-Induced Apoptosis Requires Secretion and Translocation of One or More Effector Proteins.
The yscN secretion mutant and the yopB, yopD, and sycD translocation mutants (4, 6) failed to elicit macrophage apoptosis (Fig. 1 A and B). It thus appeared that apoptosis induction requires functional secretion and translocation systems. These findings suggest strongly that apoptosis induction is mediated by one or more bacterial proteins translocated into the cytosol of the target cell. Similar results were obtained with a different macrophage cell line, PU5.1–8 (ATCC TIB 61)(data not shown).
The YopP Protein Is Required for Apoptosis Induction.
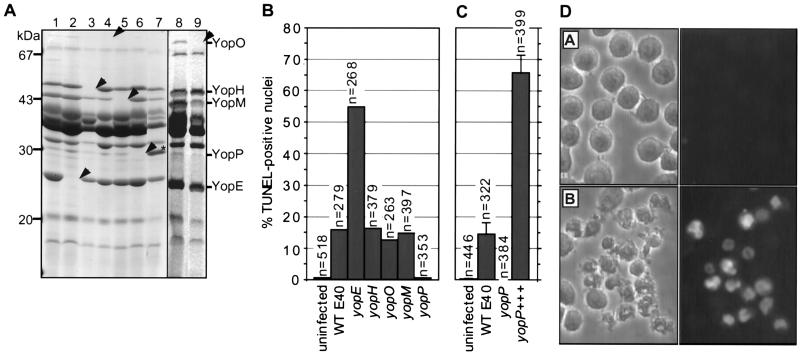
To identify the Yop effector protein(s) required for apoptosis, nonpolar mutations were engineered in the known effector genes yopE, yopH, yopO, and yopM, as well as in the gene yopP, which codes for a 30-kDa secreted Yop protein of unknown function (11) (Fig. 2A). These mutants were used to infect J774A.1 macrophages. A series of eight mutants constructed previously in Y. enterocolitica W22703 (11–13) also was included in this study.
Figure 2.
YopP, a new Y. enterocolitica Yop effector, is required to induce apoptosis in macrophages. (A) 12% SDS/PAGE of secreted proteins from isogenic nonpolar yop mutants and overexpression of YopP in the yopP mutant. Lanes: 1, wild-type E40; 2, yopE; 3, yopH; 4, yopO; 5, yopM; 6, yopP; and 7, yopP+++ (yopP mutant overexpressing YopP). Arrowheads indicate the position of the deleted protein. Asterisk (∗) denotes overexpressed YopP. To clearly show the secretion profile of the yopO mutant, samples were overloaded with supernatants of (lane 8) wild-type E40 and (lane 9) yopO mutant cultures. Loading was based on equivalent numbers of bacteria. Molecular masses are shown at left, and positions of Yops are shown at right. (B) Percentage of TUNEL-positive nuclei in J774A.1 cells infected with wild-type strain E40 or isogenic nonpolar Yop effector mutants (4 h postinfection). n values refer to the number of total cells evaluated per treatment. All strains were tested a minimum of three times. (C) Percentage of TUNEL-positive nuclei in J774A.1 cells infected with wild-type (WT) E40, yopP mutant, or yopP+++. Error bars represent the SEM for 1 field of view (≈100–150 cells) from three separate coverslips. (D) Phase contrast (left) and fluorescence (right) images depicting the results of the infection (cell morphology) and TUNEL reaction (nuclear morphology), respectively in J774A.1 cells infected with (A) yopP mutant and (B) yopP+++.
It is shown in Fig. 2B that yopP is the only E40 mutant that failed to induce apoptosis. A similar result was obtained with the W22703 mutants (data not shown). Gene yopP from Y. enterocolitica then was cloned downstream from the strong yopE promoter (plasmid pMSK13) and introduced in trans into the yopP mutant. After infection of macrophages with the complemented strain (yopP+++), the percentage of apoptotic nuclei was 4-fold higher than that observed after infection with the wild-type strain (Fig. 2 C and D), showing that yopP is the gene required to cause apoptosis in macrophages that is missing in the yopP mutant.
The role of YopP is unlikely to be in internalization of Yop effectors because the yopP mutant is as efficient as the wild-type in translocating proteins YopE, YopM, and YopO (data not shown). On the other hand, because functional translocation is necessary to obtain the effect (Fig. 1 for strain E40 and not shown for W22703), it is likely that YopP must be translocated to be active and, therefore, is a new effector protein.
In both strains, the yopE mutant was more efficient in inducing apoptosis than the wild-type pathogen. The explanation for this may be that the YopE protein competes physically with YopP for internalization into host cells.
The Y. enterocolitica wild-type E40 strain and the yopP+++ strain did not induce nuclear DNA fragmentation in HeLa cells, even at a multiplicity of infection up to 500 bacteria per cell (data not shown). Thus, Yersinia-induced apoptosis presents some cell type specificity. However, the wild-type strain and the yopP mutant were cytotoxic to HeLa cells, as previously reported for Y. pseudotuberculosis (21, 22), which indicates that apoptosis is different from YopE-mediated cytotoxicity.
Extracellular Y. enterocolitica Induces Apoptosis.
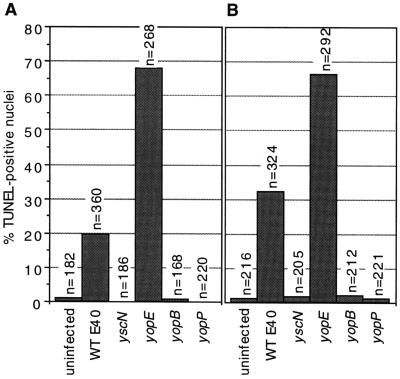
To know whether the induction of apoptosis in macrophages requires internalization of the bacteria, we infected J774A.1 cells with E40 and some mutants in the presence or absence of 2.5 μg/ml cytochalasin D (inhibitor of phagocytic bacterial uptake). According to the gentamicin protection assay (23), cytochalasin D prevented uptake of all of the mutants tested, including the yscN mutant, which is unable to secrete the antiphagocytic YopH protein. Wild-type E40 and its yopE mutant induced nuclear DNA fragmentation equally well in the absence (Fig. 3A) and in the presence of cytochalasin D whereas secretion and translocation mutants did not (Fig. 3B). Thus, the bacteria need not be internalized to induce apoptosis.
Figure 3.
Extracellular Y. enterocolitica induce apoptosis. J774A.1 cells were infected in the presence or absence of 2.5 μg/ml cytochalasin D and processed for the TUNEL reaction at 4 h postinfection. n values indicate the total number of cells evaluated per treatment. Wild-type strain E40 and the yopE mutant were able to induce apoptosis as efficiently in the absence (A) or presence (B) of cytochalasin D (inhibitor of phagocytic bacterial uptake). Uninfected cells, and yscN, yopB, and yopP mutant-infected cells, were negative for the TUNEL reaction. All strains were tested a minimum of three times.
Size of DNA Fragments Isolated from Y. enterocolitica-Infected J774A.1 Cells.
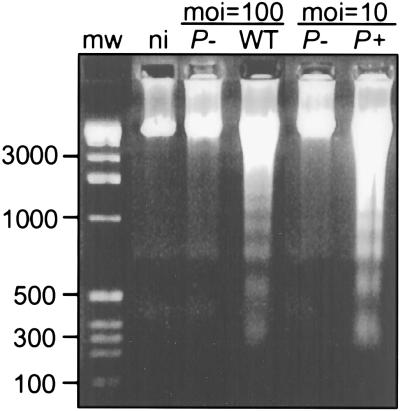
DNA was prepared from macrophages infected with wild-type E40, yopP mutant, or yopP+++ bacteria and analyzed by agarose gel electrophoresis. DNA fragmentation was seen in samples from macrophages infected with wild-type and yopP+++ bacteria but not in samples from uninfected macrophages or macrophages infected with yopP mutant bacteria. Fragmented DNA formed ladders of ≈200 bp (Fig. 4), suggesting that cleavage occurred at the internucleosomal regions, as happens when cells undergo apoptosis.
Figure 4.
DNA fragmentation of Y. enterocolitica-infected J774A.1 cells. Oligonucleosomal length DNA fragmentation was observed in cells infected with wild-type (WT) and P+ (yopP+++) but not in noninfected (ni) cells or cells infected with P- (yopP mutant). J774A.1 cells were infected as described in Materials and Methods using a multiplicity of infection (moi) of either 10 or 100 bacteria per cell as indicated. J774A.1 DNA was isolated 2 h after infection and subjected to agarose gel electrophoresis (18). Molecular weights (in base pairs) are indicated at left.
Structural Evidence for Apoptosis.
As a final check, we examined the structure of Y. enterocolitica-infected macrophages (24). Thin sections from J774A.1 cells infected with wild-type or yopP+++ bacteria revealed classic morphological signs of apoptosis at 4 h postinfection (Fig. 5 A and D). No such signs were observed in cells infected with the yscN secretion mutant or the yopP mutant (Fig. 5 B and C). Wild-type, yopP mutant, and yopP+++ bacteria were extracellular (Fig. 5 A, C, and D) whereas many yscN mutant bacteria were observed within vacuoles (Fig. 5B). Transmission electron micrographs of cells infected with yopP+++ displayed convolution of the nuclear outline (Fig. 5E) and segregation of condensed chromatin in crescents at the periphery of the nuclear envelope (Fig. 5E), in contrast with good preservation of cytoplasmic organelles. These features are characteristic of apoptosis (24).
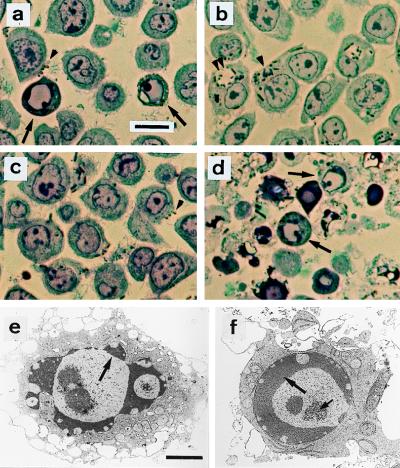
Figure 5.
Structural evidence for Y. enterocolitica-induced apoptosis. Semi-thin sections were stained with toluidine blue and examined by light microscopy (a–d). (a) Wild-type strain E40; apoptotic nuclei (arrows) and cell surface-associated bacteria (brown particles, arrowhead). (b) yscN secretion mutant; no apoptotic cells are detected. Abundance of internalized bacteria either in tight (single arrowhead) or spacious phagosomes (double arrowhead). (c) yopP effector mutant; no apoptotic cells are detected. Bacteria at the cell surface (arrowhead). (d) yopP+++, apoptotic cells (arrows). Ultrastructural analysis of cells infected with yopP+++ from d is shown in e and f. Typical features of apoptosis include (i) peripheral chromatin condensation in crescents, except in the vicinity of nuclear pores (large arrows); (ii) bulging of nuclear crescents into the cytoplasm, best seen in e; and (iii) appearance of central clusters of small particles of unknown nature, typical of apoptosis (small arrow at f). Nuclear and plasma membrane alterations contrast with a good ultrastructural preservation of cytoplasm, particularly of endoplasmic reticulum and mitochondria. (a–d, Bar = 10 μm; e–f, Bar = 2 μm).
Kinetics of Apoptosis.
To further characterize the apoptosis induced by Y. enterocolitica, we recorded the events occurring in the course of the infection of J774A.1 cells by Y. enterocolitica yopP+++. By 1-h postinfection, we already observed many apoptotic bodies (membrane blebbing), but only a small fraction of the nuclei was weakly TUNEL-positive. By 2 h, apoptotic bodies were associated with the vast majority of cells, ≈50% of the nuclei were TUNEL-positive, and nuclear chromatin was beginning to condense at the periphery of the nucleus. By 4–6 h, cellular and nuclear shrinkage was evident in most cells, the majority of the nuclei were TUNEL-positive with chromatin condensed in crescents, and nuclear collapse was clearly visible (data not shown). These observations match classic descriptions of apoptosis (24).
YopP Sequence Analysis.
YopP (288 residues) appeared to be the counterpart of YopJ, a 264-residue Yop protein of unknown function from Y. pseudotuberculosis (25). YopP and YopJ are 92% identical (95% similar) in their first 241 residues. After residue 241, they diverge significantly because of a change in the reading frame (Fig. 6). YopP and YopJ share 25% identity and 41% similarity with AvrRxv from X. campestris (26, 27). Although YopP and YopJ diverge after residue 241, the similarity between YopP and AvrRxv extends to the C terminus of YopP (Fig. 6).
Figure 6.
Alignment between YopP (Y. enterocolitica), YopJ (Y. pseudotuberculosis), and avirulence protein AvrRxv from X. campestris. Identical amino acids are in red, and conserved amino acids are in blue.
DISCUSSION
Yersinia secretes and injects into the cytosol of eukaryotic target cells an arsenal of Yop effectors that undermine the host cellular immune response (1). For example, delivery of tyrosine phosphatase YopH into macrophages leads to the inhibition of phagocytic bacterial uptake (28) and oxidative burst (29). In this report, we demonstrate that type III secretion also induces apoptosis in infected macrophages and that a newly identified Yop effector, YopP, is involved in this phenomenon. There was some cell type specificity; Y. enterocolitica did not induce apoptosis in an epithelial cell line. These observations, which suggest that Yersiniae can eliminate macrophages without inducing an inflammatory response, complete our understanding of the mechanisms by which Yersiniae can proliferate in lymphoid tissues.
There have been reports of Shigella- and Salmonella-induced cytotoxicity in macrophages in which type III secretion also is required (18, 30, 31). In the case of Salmonella, no specific effector protein has been identified (30). However for Shigella, it has been shown that IpaB, a secreted protein that is required for invasion of nonphagocytic cells, induces macrophage apoptosis by binding directly to interleukin 1B-converting enzyme (32). To induce apoptosis, Shigella must be internalized by macrophages and have access to the host cell cytoplasm (18). The mechanism by which Yersinia induces apoptosis appears to be different. First, Yersinia induces apoptosis from outside the host cell. Second, the proteins involved are very different. Indeed, IpaB from Shigella shares some similarity to the Yersinia YopB but not to the effector protein YopP (32). In the case of Yersinia, YopB also was required for apoptosis but presumably only indirectly because of its translocator role. Thus, the mechanism by which Yersinia induces macrophage apoptosis appears to be quite different from that used by Shigella. In contrast, it evokes that used by cytotoxic T lymphocytes, which deliver granzyme B into the cytosol of their target cells thereby inducing apoptosis (33). One of the virulence functions of Yersiniae appears thus to mimic a physiological process of their host.
Not surprisingly, sequence analysis revealed that YopP is nearly identical to YopJ, its homologue from Y. pseudotuberculosis (25). More interesting, YopP (and YopJ) shares a high level of similarity with AvrRxv from X. campestris. AvrRxv is one of many avirulence proteins identified in plant pathogenic bacteria that mediates the hypersensitive response, a process that is likely to result from the activation of a programmed cell death pathway (26, 34). It recently has been proposed that AvrBS3, also from X. campestris, is translocated into plant cells via the Xanthomonas Hrp type III secretion system (35). Additional recent studies have shown that transient expression of avrPto (from plant pathogen Pseudomonas syringae) in plant leaf tissue also results in hypersensitive response, suggesting that AvrPto may also be a translocated effector (36, 37). Given that Yersinia and Xanthomonas share related type III secretion systems and that type III secretion is required for delivery of their respective cell death-inducing proteins, it is likely that the homology between YopP and AvrRxv has functional relevance. Thus animal and plant pathogens not only use related secretion systems to deliver effectors into the cytosol of target cells but the similarity between YopP and AvrRxv indicates that they also deliver related effectors to induce similar answers. AvrBS3 has been shown to require functional nuclear localization signals for eliciting the hypersensitive reaction, suggesting that nuclear factors are involved in AvrBS3 recognition by the plant (35). No such potential nuclear localization motifs were detected in YopP.
In conclusion, we have shown that Y. enterocolitica specifically induces apoptosis in macrophages and that this phenomenon requires type III secretion, Yop translocators, and YopP, which appears to be a novel effector. Most important, YopP shares significant homology with AvrRxv from X. campestris. Animal and plant pathogenic bacteria thus share a type III secretion-dependent effector to elicit programmed cell death (apoptosis) in their respective hosts.
Acknowledgments
We thank J. Kronstad, L. Matsuuchi, R. Fernandez, R. DeVinney, and P. Courtoy for critical reading of the manuscript. We gratefully acknowledge C. Neyt and I. Stainier for contributing unpublished plasmids pCNR26 and pSI4008 to this study. B.B.F. is supported by a Howard Hughes International Research Scholar Award, and the Medical Research Council of Canada. G.R.C. and P.V.D.S. are supported by the Belgian Fonds de la Recherche Scientifique Médicale, the Communauté Française de Belgique, and the Belgian State. A.B. is supported as a research assistant from the Belgian Fonds National de la Recherche Scientifique.
ABBREVIATIONS
- TUNEL
terminal deoxyribonucleotidyl transferase-mediated dUTP-digoxigenin nick end labeling
- yopP+++
yopP(pMSK13)
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF023202 (yopP)].
References
- 1.Cornelis G R, Wolf-Watz H. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 2.Rosqvist R, Magnusson K-E, Wolf-Watz H. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sory M-P, Cornelis G R. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 4.Sory M-P, Boland A, Lambermont I, Cornelis G R. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Håkansson S, Galyov E E, Rosqvist R, Wolf-Watz H. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 6.Boland A, Sory M-P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 7.Håkansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 8.Wattiau P, Woestyn S, Cornelis G R. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 9.Finlay B B, Cossart P. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 10.Lee C A. Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis G, Vanootegem J-C, Sluiters C. Microb Pathog. 1987;2:367–379. doi: 10.1016/0882-4010(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 12.Woestyn S, Allaoui A, Wattiau P, Cornelis G R. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wattiau P, Cornelis G R. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 14.Sarker M, Cornelis G R. Mol Microbiol. 1997;23:409–410. doi: 10.1046/j.1365-2958.1997.t01-1-00190.x. [DOI] [PubMed] [Google Scholar]
- 15.Sanger F, Nicklens S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Rosqvist R, Bölin I, Wolf-Watz H. Infect Immun. 1988;56:2139–2143. doi: 10.1128/iai.56.8.2139-2143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zychlinsky A, Prevost M C, Sansonetti P J. Nature (London) 1992;358:167–169. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]
- 19.Nassogne M-C, Louahed J, Evrard, Courtoy P. J Neurochem. 1997;68:2442–2450. doi: 10.1046/j.1471-4159.1997.68062442.x. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Sasson S A, Sherman Y, Gavrieli Y. Methods Cell Biol. 1995;46:30–39. [PubMed] [Google Scholar]
- 21.Rosqvist R, Forsberg A, Rimpiläinen M, Bergman T, Wolf-Watz H. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 22.Rosqvist R, Forsberg A, Wolf-Watz H. Infect Immun. 1991;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isberg R R, Falkow S. Nature (London) 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 24.Kerr J F R, Gobé G C, Winterford C M, Harmon B V. Methods Cell Biol. 1995;46:1–27. doi: 10.1016/s0091-679x(08)61921-4. [DOI] [PubMed] [Google Scholar]
- 25.Galyov E E, Håkansson S, Wolf-Watz H. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whalen M C, Wang J F, Carland F M, Heiskell M E, Dahlbeck D, Minsavage G V, Jones J B, Scott J W, Stall R E, Staskawicz B J. Mol Plant–Microbe Interact. 1993;6:616–627. doi: 10.1094/mpmi-6-616. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson K, Carballeira N, Magnusson K E, Persson C, Stendahl O, Wolf-Watz H, Fällman M. Mol Microbiol. 1996;20:1057–1069. doi: 10.1111/j.1365-2958.1996.tb02546.x. [DOI] [PubMed] [Google Scholar]
- 29.Bliska J B, Black D S. Infect Immun. 1995;63:681–685. doi: 10.1128/iai.63.2.681-685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L M, Kaniga K, Galan J E. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 31.Monack D M, Raupach B, Hromockyj A E, Falkow S. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Smith M R, Thirumalai K, Zychlinsky A. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L, Mai S, Israels S, Browne K, Trapani J A, Greenberg A H. J Exp Med. 1997;185:855–866. doi: 10.1084/jem.185.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittler R, Lam E. Trends Microbiol. 1996;4:10–15. doi: 10.1016/0966-842x(96)81499-5. [DOI] [PubMed] [Google Scholar]
- 35.van den Ackerveken G, Marois E, Bonas U. Cell. 1996;87:1307–1316. doi: 10.1016/s0092-8674(00)81825-5. [DOI] [PubMed] [Google Scholar]
- 36.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 37.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]