Abstract
The production of subtle or conditional mutations in mice through the combined use of site-specific and homologous recombination has become an increasingly widespread experimental paradigm in mammalian genetics. Embryonic stem cells containing recombinase transgenes that were expressed in the male germ line, but not in other tissues or in the embryonic stem cells themselves, would substantially simplify the production of such alleles. Here we show that transgenes comprised of the mouse protamine 1 promoter and the Cre recombinase coding sequence mediate the efficient recombination of a Cre target transgene in the male germ line, but not in other tissues. Embryonic stem cell lines generated from one of these transgenic strains were transfected with targeting vectors that included loxP-flanked selectable markers, and homologously recombined alleles containing the marker and functional loxP sites were isolated. These results establish the potential of the system for substantially reducing the time, effort, and resources required to produce homologously recombined alleles in mice that have been secondarily rearranged by a site-specific recombinase.
The analysis of mammalian gene function has increasingly come to require the production of subtle, tissue-specific, and conditional mutations in mice. Although there are a number of methods for engineering subtle mutations in embryonic stem (ES) cells (1, 2), the use of site-specific recombinases to remove the selectable marker that permits isolation of homologously recombined ES cell clones has become increasingly prevalent (3–7). Site-specific recombinases represent the best method for creating tissue-specific and conditional mutations in mice, being employed first to remove the selectable marker to create a functionally wild-type allele, and then to inactivate the allele mosaically in mice by removing some essential component in a tissue-specific or conditional manner (8, 9).
Current protocols for using site-specific recombination to remove selectable markers include transiently transfecting ES cell clones with a recombinase expression vector (6), microinjecting fertilized oocytes containing the recombinant allele with a recombinase expression vector (3, 10), or breeding mice containing the recombinant allele to mice containing a recombinase transgene (5, 11). Each of these approaches requires an investment of some combination of time, resources, and expertise over that required to generate mice with homologously recombined alleles. The most commonly employed method, the secondary transfection of homologously recombined ES cell clones with a recombinase expression vector, additionally requires extended culture time, which may decrease their potential to enter the germ line.
In principle, marker excision would be substantially simplified through the use of ES cells containing recombinase transgenes that were expressed in the male germ line, but not to an appreciable extent in the ES cells themselves or somatic tissues of mice. The lack of ES cell expression would mean that targeting vectors containing selectable markers flanked by recombinase target sites could be used to isolate homologous recombinants without fear that the marker would be excised during culture. Robust recombinase expression in the male germ line would mean that the marker would be excised in at least some of the progeny of ES cell chimeras. Only a single step would be required to isolate subtle mutations and, if two different recombinase systems were employed, conditional and tissue-specific alleles could be produced with similar improvements in efficiency. A germ-line-specific recombinase transgene also could be used to deliver recombined target transgenes to the early embryo (11), as long as the recombined target was not detrimental to the terminal stages of spermatogenesis.
We have sought to determine whether the expression of recombinase transgenes driven by the mouse protamine 1 (Prm1) promoter would be suitably robust and tissue specific for these purposes. The endogenous mouse protamine genes are expressed during the terminal, haploid stages of spermatogenesis (12). Previous reports have shown that expression of transgenes containing the proximal promoter of the Prm1 locus is also restricted to haploid spermatids in mature mice (13–15), although low levels of ectopic expression have been reported with some transgenes (14). Our observations do indeed suggest that ES cells bearing Prm1-Cre recombinase (PrmCre) transgenes will greatly facilitate the production of a variety of mutations in mice.
MATERIALS AND METHODS
DNA Constructs.
A 652-bp fragment of the Prm1 promoter (15) was isolated by PCR by using genomic DNA from CCE ES cells (16) as a template. This fragment was fused to a Cre coding sequence modified to contain a consensus translation start site (17), 11 codons for a human c-myc epitope (18), 7 codons for a minimal simian virus 40 nuclear localization signal (19), and the polyadenylation signal from pIC-Cre (6) in the plasmid pOG304M. The Cre expression plasmid pOG231 was prepared by fusing a Cre coding sequence modified from pIC-Cre, and containing the same translation start and nuclear localization signal, to the synthetic intron and cytomegalovirus promoter of pOG44 (20). A plasmid, pOG277, containing a loxP-flanked neomycin (neo) cassette was prepared by inserting a wild-type loxP site (21) into pBSKS (Stratagene) and then cloning the neo expression cassette from pMC1neo-poly(A) (22) between iterations of this loxP site. The hoxb-1 targeting construct consisted of the phosphoglycerate kinase-thymidine kinase cassette from pPNT (23), and 1.4- and 10.2-kb of sequences 5′ and 3′ to an NruI site 800 bp 5′ to the hoxb-1 transcriptional start site isolated from a 129 strain genomic library (Stratagene) (S.O’G., unpublished results). The loxP-flanked neo cassette from pOG277 was inserted into the NruI site. The targeting construct used to insert the pOG277 neo cassette and a β-galactosidase (β-gal) sequence into the first exon of the large subunit of RNA polymerase II (24) (RP2) to create the P2Bc allele (Fig. 1) will be described in detail elsewhere.
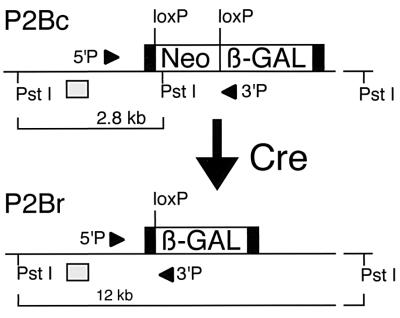
Figure 1.
Schematic of P2Bc and P2Br alleles. Cre-mediated recombination of the P2Bc allele results in the deletion of the neo cassette (Neo) of P2Bc that is flanked by two loxP sites, leaving a single loxP site and fusing the β-gal coding sequence to the initial codons of the RNA polymerase II coding sequence (solid boxes). Recombination increases the size of a PstI fragment recognized by the RP2 probe, which is external to the targeting vector used, indicated by the shaded box below each allele. Positions of the PCR primers used (5′P and 3′P) are indicated on the diagrams of the P2Bc and P2Br alleles.
Production of Transgenic Mice.
Fertilized oocytes obtained from matings of 129/SvJae (25) and BALB/c × C57BL/6 F1 mice were used for pronuclear injections of the protamine-Cre cassette isolated from pOG304M, according to standard protocols (26).
Production of ES Cells and Homologous Recombinants.
Heterozygous PrmCre 129/SvJae mice were intercrossed or backcrossed to 129/SvEms-+Ter?/J mice (25) to produce blastocysts that were cultured to establish ES cell lines (27). The sex (28) and PrmCre status of each line were determined by PCR assays.
Molecular Analyses.
Tail biopsy genomic DNA was used for hybridization assays or PCR assays to identify PrmCre and P2Bc/r mice. PCR reactions for the detection of ectopic Cre activity used 100 ng of genomic DNA as a template to amplify a P2Br-specific product by using a 5′ primer from the RP2 promoter and a 3′ primer from the β-gal coding sequence (Fig. 1). Thirty cycles of amplification were done in a total volume of 100 μl by using 300 ng of each primer, 3 mM MgCl2, 1.5 units of Taq polymerase, and an annealing temperature of 60°C. Southern blots of reaction products were hybridized with a probe specific for the P2Br reaction product.
RESULTS
PrmCre Transgenes Efficiently Recombine Target Alleles.
To determine whether PrmCre transgenes would efficiently recombine a target allele, males were generated that contained a PrmCre transgene and a target for Cre-mediated recombination. These males were then mated to wild-type females, and genomic DNA from the resulting progeny was examined to establish the segregation pattern of PrmCre and target transgenes and determine whether the target transgene was recombined.
A total of nine founder animals with PrmCre transgenes were prepared. Two lines (58 and 70) were derived from injections of 129/SvJae embryos (25), and seven from injections of CB6F2 embryos. The 129/SvJae lines and three randomly selected hybrid lines (71, 75, and 78) were examined in detail. The Cre-recombinase target used for these experiments was a homologously recombined allele of the RNA polymerase II locus (24), designated “P2Bc” for Pol II, β-gal, conditional (Fig. 1). This allele was created by inserting a loxP-flanked neo cassette and a β-gal coding sequence into the first exon of the locus coding for the large subunit of RNA polymerase II (S.O’G., unpublished results). Cre-mediated recombination of the loxP sites was expected to delete the intercalated sequences, creating a “P2Br” allele (Pol II, β-gal, recombined).
When males bearing both a PrmCre transgene and a P2Bc target allele were mated to wild-type females, the large majority of transmitted target alleles were Cre-recombined P2Br alleles and the PrmCre and target alleles segregated independently in the first generation (Table 1 and Fig. 2). Collectively, of 112 target alleles transmitted by males of all 5 lines, 103, or 92%, were recombined. Approximately 50% of mice that inherited a P2Br allele also inherited their male parent’s PrmCre transgene. These data establish that PrmCre transgenes efficiently recombine the P2Bc allele in the male germ line and that the recombined P2Br alleles and PrmCre transgenes segregate in the first generation. The observation that approximately 50% of the progeny inherited the rearranged P2Br allele, rather than 25% as might have been expected, is most likely because of the sharing of gene products by haploid spermatids (29). Approximately one-half of the unrecombined alleles were transmitted by gametes that additionally contain a PrmCre transgene.
Table 1.
Recombination of the P2Bc allele in the progeny of PrmCre mice
| PrmCe line | Born | +P2Bc: +/P2Br | +/PrmCre and +/P2Bc | +/PrmCre and +/P2Br | +/PrmCre |
|---|---|---|---|---|---|
| 58 | 29 | 0:17 | 0 | 10 | 15 |
| 70 | 57 | 3:23 | 1 | 10 | 24 |
| 71 | 33 | 0:18 | 0 | 8 | 13 |
| 75 | 46 | 0:22 | 0 | 8 | 18 |
| 78 | 58 | 6:23 | 4 | 13 | 26 |
| Total | 223 | 9:103 | 5 | 49 | 96 |
Transmission data for recombined (P2Br) and unrecombined (P2Bc) alleles in progeny of males that were heterozygous for a PrmCre allele and a P2Bc target allele. Transmission of the P2Bc/r and PrmCre alleles did not differ significantly from expectation by χ2 test.
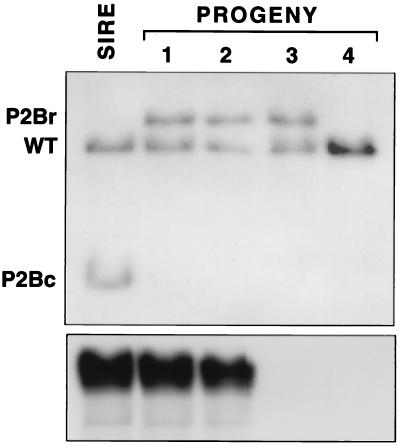
Figure 2.
Southern blot of PstI-digested tail biopsy DNAs from a +/P2Bc, +/PrmCre male (sire) and four of his progeny by a wild-type female probed with an RP2 probe (Upper) and then reprobed with a Cre probe (Lower). All RP2 mutant alleles in the progeny shown (lanes 1–3) were P2Br, and some progeny (no. 3 in this example) inherit a P2Br allele without inheriting a PrmCre transgene. Mouse 4 does not contain a PrmCre transgene and is homozygous wild-type at the RP2 locus.
The progeny of matings between PrmCre males and +/P2Bc females were also examined to determine whether male gametes from PrmCre mice delivered enough Cre to zygotes to effect Cre-mediated recombination of a target sequence. Of 96 progeny examined by Southern blotting, none contained a Cre-recombined P2Br allele.
PrmCre Transgene Expression Is Highly Tissue Specific.
Genomic DNA from 10 different tissues of 5–7-week-old males that contained both a PrmCre transgene and a P2Bc target allele were analyzed in Southern blots (Fig. 3). Only the testis samples showed signal indicating Cre-mediated recombination of the target (Fig. 3 Upper). The intensity of the P2Br signal relative to that of the wild-type allele ranged from 10 to 22% for different PrmCre strains and did not correlate with the PrmCre transgene copy number (Fig. 3 Lower). For example, restriction patterns and densitometric analyses showed that line 58 contained a single copy of the PrmCre transgene, yet showed virtually the same testis recombination signal as the line containing more than 100 copies (78). This variability is similar to results obtained with other Prm1 promoter-driven transgenes (13, 30).
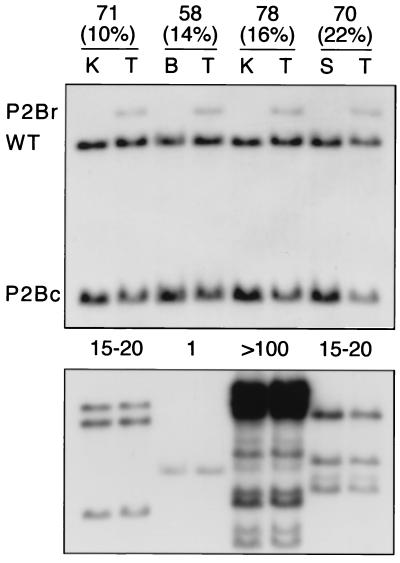
Figure 3.
Recombination in the testes of PrmCre mice. Southern blot of PstI-digested DNA from testes (T) and one other tissue (K, kidney; B, brain; S, spleen) of males heterozygous for one of four PrmCre transgenes (58, 70, 71, 78) and the P2Bc allele. (Upper) Hybridization with an RP2 probe (see Fig. 1). Testis DNA from each male shows a P2Br allele signal, in addition to those generated by the wild-type RP2 (WT) and P2Bc alleles. Other tissues show only the WT and P2Bc signals. (Lower) Rehybridization with a Cre probe. The copy number of PrmCre transgenes (above each pair of lanes) varied among lines showing similar levels of recombination in testis.
As a more sensitive measure of ectopic recombination, PCR amplifications were performed on the same samples. The amplification primers were expected to produce a 325-bp product from the recombined target and a 1.4-kb fragment from the unrecombined allele (Fig. 1). The assay was expected to measure the cumulative level of recombination, because any P2Br alleles formed during transient expression of Cre during development would be preserved and perhaps amplified in descendant cells. Low levels of ectopic recombination product were observed in some tissues of all PrmCre lines except for line 78. The highest level of ectopic activity was observed in cardiac ventricular muscle of mice from strain 71; however, even in these samples, the ectopic signal was more than 100-fold lower than that observed in testis (Fig. 4). Three strains (58, 70, and 75) showed very low levels of recombination in brain tissue, and strain 75 additionally showed ectopic activity in spleen (not shown). Despite the difficulty of quantifying PCR results, these data clearly indicate that ectopic activity occurred at very low levels in most tissues of most PrmCre lines.
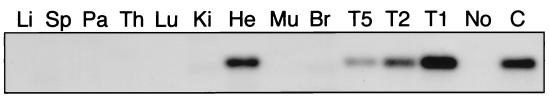
Figure 4.
Southern blot of PCR amplification products of the P2Br allele using tissues from a male heterozygous for the strain 71 PrmCre transgene and the P2Bc allele. DNA from 10 different tissues was amplified by using primers and conditions that produced a 350-bp product from the recombined, P2Br allele (see Fig. 1 and Materials and Methods). Each lane contains 10% of the reactions, except for the testis reactions, which were diluted 500- (T5), 250- (T2), and 100- (T1) fold before loading, and a liver reconstruction control (C), which was diluted 1:100 before loading. (Li, liver; Sp, spleen; Pa, pancreas; Th, thymus; Lu, lung; Ki, kidney; He, heart; Mu, skeletal muscle; Br, brain; No, no DNA control).
Isolation of Homologously Recombined PrmCre ES Cell Clones Using Targeting Vectors with a loxP-Flanked Selectable Marker.
Four male +/PrmCre ES cell lines were established from 129/Sv strain PrmCre transgenic mice (strain 70). In preliminary experiments, passage 5 cells from one of these lines (PC3), which was homozygous for the PrmCre transgene, were used to generate three male chimeras with between 50 and 95% coat color chimerism. In matings with C57BL/6 females, two of these male chimeras have sired a total of 21 pups; 20 pups bore the Agouti coat color signifying germ-line transmission of the ES cell genome, and all 18 of the Agouti pups that were genotyped contained the PrmCre transgene.
To determine whether homologously recombined PrmCre ES cell clones could be isolated by using targeting vectors that contained a loxP-flanked selectable marker, two transfections were done by using variants of a targeting vector in which a loxP-flanked neo cassette was inserted into a NruI site in the hoxb-1 locus promoter (Fig. 5A). In these transfections, 12 of 62 (19%) PC3- and 10 of 56 (18%) PC5-derived clones that were ganciclovir and G418-resistant (31) had recombined homologously (Fig. 5B). In two parallel transfections of CCE cells with the same vectors (16), 32 of 93 (34%) and 15 of 132 (11%) clones had recombined homologously. The total numbers of G418-resistant clones recovered from PrmCre ES cell transfections were reduced relative to the parallel CCE transfections. This may be attributable to both Cre-mediated excision of the neo cassette (see below) and to the fact that the transfections were done under electroporation conditions optimized for CCE cells.
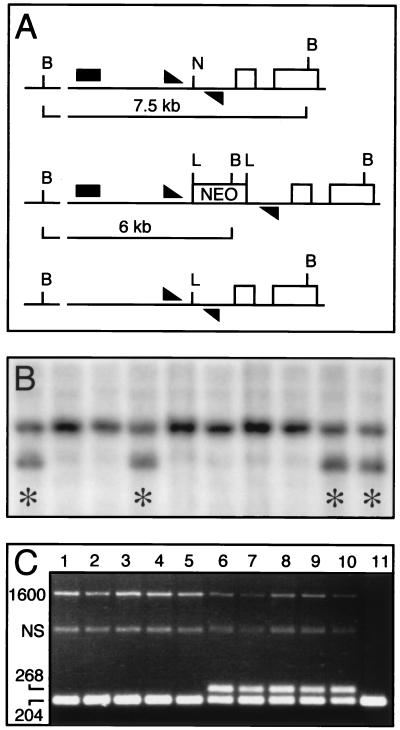
Figure 5.
Targeting of the hoxb-1 locus in PrmCre ES cells by using a targeting vector that contains a loxP-flanked selectable marker. (A Top) Schematic of the wild-type hoxb-1 locus showing the positions of the two exons (open boxes), the position of a 5′ NruI site and flanking BamHI restriction endonuclease sites, and PCR primers (triangles) that amplify a 204-bp product from the wild-type allele that includes the NruI site. (Middle) Predicted organization of homologously recombined hoxb-1 allele in which a neo cassette (NEO), flanked by loxP sites (L), has been inserted into the NruI site shown in Top. The insertion creates a novel BamHI site and the same PCR primers now amplify a 1,600-bp product. (Bottom) Predicted structure of the recombined allele shown in Middle after Cre-mediated excision of the neo cassette to leave a single loxP site in place of the NruI site of the wild-type allele. Amplification with the same primers now yields a 268-bp product. (B) Southern blot of BamHI-digested genomic DNAs harvested from a 96-well plate from 10 doubly selected (31) ES cell clones and hybridized with a probe (A) that is external to the targeting construct. All samples show the 7.5-kb band from the wild-type allele and four clones (∗) additionally show the 6-kb band predicted to result from homologous recombination. (C) PCR products from amplification of five homologously recombined PC3 ES cell clones (lanes 1–10) and the parental PC3 cell line (lane 11) by using the primers shown in A. The cells were either mock transfected (lanes 1–5) or transiently transfected with pOG231 (lanes 6–11). The recombinant clones and parental cell lines show the 204-bp amplification product of the wild-type allele, and the recombinant clones additionally show a 1,600-bp product (1600) resulting from amplification across the neo cassette and a nonspecific 1,100-bp amplification product (NS). The pOG231-transfected recombinant clones show an additional 268-bp product signaling the Cre-mediated excision of the neo cassette from the recombinant alleles of some cells.
Because it was formally possible that the homologously recombined clones contained inactive loxP sites, five recombinant clones were transiently transfected with the pOG231 Cre expression vector. DNA was harvested 48 hr after transfection and used in PCR assays to assess whether the loxP sites in the recombinant clones could be recombined by Cre. In all cases, a clear recombination signal was observed in the pOG231 transfected sample and none were observed in the mock transfected controls, establishing that the loxP sites in these clones were functional (Fig. 5C). Experiments were also done to assess the stability of the loxP-flanked neo cassette in PrmCre ES cells. Five recombinant clones were grown in the presence of G418 for 2 weeks, and then aliquots of each were grown either in the presence or absence of G418 for an additional 10 days. PCR assays were performed to determine whether Cre-recombined alleles were present in any of these samples, and none were found (not shown). These data suggest that there is not enough Cre activity to significantly influence either the ability to isolate recombinant clones or the stability of the selectable markers in those clones.
To determine whether there was any detectable Cre activity in PrmCre ES cells, aliquots of two lines (PC3 and PC5) were transiently transfected with the targeting vector used to create the P2Bc allele. DNA was recovered 48 hr after transfection and used for PCR amplifications of the P2Br plasmid molecules that would be generated by extrachromosomal Cre-mediated recombination. Small amounts of recombination product were seen in both PrmCre ES cell transfections, but none was observed in parallel samples of CCE ES cells (not shown). This shows that the PrmCre ES cell lines express sufficient Cre to recombine some extrachromosomal targets when the latter are present at high copy numbers.
DISCUSSION
The objective of the present work with PrmCre transgenes was to determine the potential of the Prm1 promoter and, by inference, other germ-line-specific promoters, for the implementation of an efficient approach in which site-specific recombinases are used to generate an array of sophisticated mutations in mice. The results show that it is possible to create recombinase transgenes that are expressed at high levels in the male germ line but not to a functionally significant extent in either ES cells or embryonic or adult somatic tissues. Homologous recombinants with a loxP-flanked selectable marker can be isolated in ES cells that contain PrmCre transgenes, and transgenic mice bearing PrmCre transgenes and a target allele transmit the Cre-recombined target to their progeny at high frequencies. Although the data reported here are restricted to a single target allele, we have also found that a loxP-flanked neo cassette in the glutamate receptor R6 subunit locus is efficiently recombined by PrmCre transgenes in mice (B. Vissel, S.O’G., and S. Heinemann, unpublished results), suggesting that the paradigm is likely to be applicable to many different targeted loci.
It remains to be established that PrmCre transgenes will be as active in the gametes of ES cell chimeras as they are in transgenic mice. Barring wholly unanticipated problems at this step, these results establish the principle that mice containing loci that have been homologously recombined and then subsequently site-specifically recombined can be generated simply by using ES cells with a suitable recombinase transgene for the initial targeting. By this mechanism, alleles containing a single recombinase target site and a mutation of interest can be produced in the progeny of ES cell chimeras without any investment of time, expertise, or resources over that required to create an allele that still contains a selectable marker. The paradigm has obvious utility in the production of subtle and conditional mutations that require generation of alleles with minimal structural alterations. Because the presence and transcriptional activity of selectable markers can contribute to phenotypes in an unanticipated and unwanted manner (32, 33), the approach will also be useful for generating null alleles.
Although only 25% of the haploid spermatids of males heterozygous for both PrmCre and target P2Bc transgenes themselves contained both transgenes, most, and in some lines all, transmitted target alleles were recombined. Because expression of the endogenous Prm1 gene (12), and Prm1-driven transgenes (14, 29, 30) is restricted to haploid spermatids, the high frequency of P2Br transmission is consistent with previous observations that both RNA and proteins produced by haploid spermatids are shared via syncytial bridges that connect them (29, 34). It is nonetheless possible that some PrmCre transgene expression occurs at diploid stages of spermatogenesis and effects recombination before segregation. By either mechanism, the result is a highly efficient recombination of target alleles and the segregation of recombinase and target transgenes in the first generation.
Quantitative Southern blot analysis showed that between 10 and 22% of the target alleles in genomic DNA isolated from whole testis were recombined in males of different PrmCre strains. It is difficult to predict the level of P2Br signal that would be observed if Cre-mediated recombination had occurred in all cells that had progressed to or beyond the round spermatid stage at which protamine fusion transgenes typically begin to be expressed (13, 35). Within the fully mature (3-month-old) seminiferous tubule, approximately 70% of cells have reached this point (36). The samples used for these Southern blots were taken from younger animals that would contain more cells at earlier stages of spermatogenesis, and they additionally included extratubular tissues. Neither of these cell populations would be expected to express a Prm1 transgene, and this may contribute to the relatively low intensity of the P2Br signal relative to the P2Bc signal.
Cre-mediated recombination proved to be highly testis specific in PrmCre mice. It is clear that the transgenes are not expressed in the inner cell mass or in other early embryonic tissues. Cells from preimplantation embryos intermingle extensively, and the embryo as a whole is derived from a small number of cells (37, 38). If PrmCre transgenes recombined target sequences during preimplantation stages, at least a few of the cells in many of the tissues would contain the P2Br allele, and Southern blot and PCR analyses showed that this was not the case. The ectopic Cre activity seen in some PrmCre strains probably resulted from low levels of recombinase expression in later embryos or mature tissues, a finding consistent with the expression patterns of other Prm1-driven transgenes. Northern blot analyses have failed to reveal the expression of Prm1-containing transgenes in a variety of mature tissues (13–15, 30), but transgenes containing the Prm1 promoter and the simian virus 40 T antigen led to the consistent development of tumors of the petrosal bone and right cardiac atrium (14). The experiments reported here do not address whether there is any ectopic transcription, but the PCR assays report with high sensitivity whether sufficient levels of Cre protein were produced to effect recombination. Importantly, they measured the cumulative level of recombination, because events that occurred at any stage of development are likely to have been propagated to, and might be amplified by, cell division in descendant populations. The highest level of ectopic recombination was that observed in cardiac ventricular tissue of strain 71, which generated a signal approximately equivalent to that expected if the ratio between recombined and unrecombined alleles were 1:104. The activities observed in other strains were considerably lower than this, and one strain did not show any ectopic activity. None of the strains showed evidence of recombination in the cardiac atria (not shown), and the petrosal bone was not examined. These assays did not rule out the possibility that higher levels of recombination occur in tissues that were not examined or that the low levels of recombination observed in some tissues reflected high levels of recombination in some component cell population.
These low levels of ectopic activity suggest that Prm1-driven recombinase transgenes may be useful for generating embryos containing genetically lethal alleles. Some alleles created by homologous recombination in ES cells will prove to be lethal in heterozygotes, as was the case for an mRNA editing mutation of the GluR2 glutamate receptor subunit (39). Germ-line transmission would be restricted to rare chimeras in which the level of chimerism was low enough in tissues affected by the mutation to allow survival and high enough in the germ line to allow transmission. This problem could be circumvented by creating recombinase-conditional mutations in ES cells bearing Prm1-recombinase transgenes, or by making the same mutations in standard ES cells and then introducing the Prm1-recombinase transgene by breeding. As long as the recombined version of the allele did not adversely affect terminal stages of spermatogenesis, embryos containing the recombined allele could be efficiently produced. Embryos containing recombined transgenes can also be produced through the activity of Cre transgenes that are expressed during early embryogenesis from the human cytomegalovirus minimal promoter (5), the adenovirus EIIa promoter (40), or the zP3 promoter (11). PrmCre and zP3 transgenes have the advantage of delivering a recombined allele to the zygote, guaranteeing that all cells in the derived embryos will contain the recombined allele.
PrmCre ES cells are but one of many different kinds of recombinase-bearing ES cells that could significantly shorten the time and effort required for a wide variety of genetic manipulations in mice. The most obvious of these are complementary PrmFLP ES cells in which the FLP recombinase was derived from Saccharomyces cerevisiae (41) or another species (42). Conceptually distinct from these, but perhaps as generically useful, would be ES cells bearing inducible recombinase transgenes, which would facilitate temporal control of recombinase expression in ES cells, chimeras, and their progeny to generate site-specifically recombined alleles (9, 43–45). Finally, fusion genes that led to recombinase expression in specific tissues could be used to address specific research objectives.
Acknowledgments
S.O’G. would like to thank Drs. Bryce Vissel and Steven Heinemann for permission to cite data on recombination of the GluR6 allele; Drs. M. Alejem, S. P. Linke, and A. Sailer for comments on the manuscript; and Dr. Geoffrey M. Wahl for continued support and insightful discussions. This work was supported by National Institutes of Health Grant HD30255 to S.O’G. and by National Cancer Institute Grant CA14195 to Walter Eckhart.
ABBREVIATIONS
- Prm1
mouse protamine 1
- PrmCre
protamine-Cre
- ES
embryonic stem
- neo
neomycin
- β-gal
β-galactosidase
References
- 1.Hasty P, Ramirez S R, Krumlauf R, Bradley A. Nature (London) 1991;350:243–246. doi: 10.1038/350243a0. [DOI] [PubMed] [Google Scholar]
- 2.Askew G R, Doetschman T, Lingrel J B. Mol Cell Biol. 1993;13:4115–4124. doi: 10.1128/mcb.13.7.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitamoto T, Nakamura K, Nakao K, Shibuya S, Shin R W, Gondo Y, Katsuki M, Tateishi J. Biochem Biophys Res Commun. 1996;222:742–747. doi: 10.1006/bbrc.1996.0814. [DOI] [PubMed] [Google Scholar]
- 4.Fiering S, Kim C G, Epner E M, Groudine M. Proc Natl Acad Sci USA. 1993;90:8469–8473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwenk F, Baron U, Rajewsky K. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu H, Zou Y R, Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 7.Sailer A, O’Gorman S, Schiffer H H, Heinemann S F. In: Taniguchi Symposia on Brain Sciences. Nakanishi S, Silva A J, Aizawa S, Katsuki M, editors. Tokyo: Japan Scientific; 1996. pp. 89–98. [Google Scholar]
- 8.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 10.Araki K, Araki M, Miyazaki J, Vassalli P. Proc Natl Acad Sci USA. 1995;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewandoski M, Wassarman K M, Martin G R. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 12.Hecht N B, Bower P A, Waters S H, Yelick P C, Distel R J. Exp Cell Res. 1986;164:183–190. doi: 10.1016/0014-4827(86)90465-9. [DOI] [PubMed] [Google Scholar]
- 13.Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Proc Natl Acad Sci USA. 1987;84:5316–5319. doi: 10.1073/pnas.84.15.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behringer R R, Peschon J J, Messing A, Gartside C L, Hauschka S D, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1988;85:2648–2652. doi: 10.1073/pnas.85.8.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peschon J J, Behringer R R, Palmiter R D, Brinster R L. Ann NY Acad Sci. 1989;564:186–197. doi: 10.1111/j.1749-6632.1989.tb25897.x. [DOI] [PubMed] [Google Scholar]
- 16.Robertson E J, Bradley A, Kuehn M, Evans M. Nature (London) 1986;323:445–448. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 18.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalderon D, Roberts B L, Richardson W D, Smith A E. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 20.O’Gorman S, Fox D T, Wahl G M. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 21.Hoess R H, Ziese M, Sternberg N. Proc Natl Acad Sci USA. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 23.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 24.Ahearn J M, Bartolomei M S, West M L, Cisek L J, Corden J L. J Biol Chem. 1987;262:10695–10705. [PubMed] [Google Scholar]
- 25.Simpson E M, Linder C C, Sargent E E, Davisson M T, Mobraaten L E, Sharp J J. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 26.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 27.Robertson E J. In: Teratocarcinomas and Embryonic Stem Cells: A Practical Approach. Robertson E J, editor. Oxford: IRL; 1987. pp. 71–112. [Google Scholar]
- 28.King T R, Christianson G J, Mitchell M J, Bishop C E, Scott D, Ehrmann I, Simpson E, Eicher E M, Roopenian D C. Genomics. 1994;24:159–168. doi: 10.1006/geno.1994.1593. [DOI] [PubMed] [Google Scholar]
- 29.Braun R E, Behringer R R, Peschon J J, Brinster R L, Palmiter R D. Nature (London) 1989;337:373–376. doi: 10.1038/337373a0. [DOI] [PubMed] [Google Scholar]
- 30.Zambrowicz B P, Harendza C J, Zimmermann J W, Brinster R L, Palmiter R D. Proc Natl Acad Sci USA. 1993;90:5071–5075. doi: 10.1073/pnas.90.11.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansour S L, Thomas K R, Capecchi M R. Nature (London) 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 32.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 33.Olson E N, Arnold H H, Rigby P W J, Wold B J. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 34.Caldwell K A, Handel M A. Proc Natl Acad Sci USA. 1991;88:2407–2411. doi: 10.1073/pnas.88.6.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braun R E, Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Genes Dev. 1989;3:793–802. doi: 10.1101/gad.3.6.793. [DOI] [PubMed] [Google Scholar]
- 36.Bellve A R, Cavicchia J C, Millette C F, O’Brien D A, Bhatnagar Y M, Dym M. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beddington R S, Morgernstern J, Land H, Hogan A. Development (Cambridge, UK) 1989;106:37–46. doi: 10.1242/dev.106.1.37. [DOI] [PubMed] [Google Scholar]
- 38.Soriano P, Jaenisch R. Cell. 1986;46:19–29. doi: 10.1016/0092-8674(86)90856-1. [DOI] [PubMed] [Google Scholar]
- 39.Brusa R, Zimmermann F, Koh D S, Feldmeyer D, Gass P, Seeburg P H, Sprengel R. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 40.Lakso M, Sauer B, Mosinger B J, Lee E J, Manning R W, Yu S H, Mulder K L, Westphal H. Proc Natl Acad Sci USA. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broach J R, Hicks J B. Cell. 1980;21:501–508. doi: 10.1016/0092-8674(80)90487-0. [DOI] [PubMed] [Google Scholar]
- 42.Araki H, Nakanishi N, Evans B R, Matsuzaki H, Jayaram H, Oshima Y. J Mol Biol. 1992;225:25–37. doi: 10.1016/0022-2836(92)91023-i. [DOI] [PubMed] [Google Scholar]
- 43.No D, Yao T P, Evans R M. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Logie C, Stewart A F. Proc Natl Acad Sci USA. 1995;92:5940–5944. doi: 10.1073/pnas.92.13.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Proc Natl Acad Sci USA. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]