Abstract
The region of human chromosome 22q11 is prone to rearrangements. The resulting chromosomal abnormalities are involved in Velo-cardio-facial and DiGeorge syndromes (VCFS and DGS) (deletions), “cat eye” syndrome (duplications), and certain types of tumors (translocations). As a prelude to the development of mouse models for VCFS/DGS by generating targeted deletions in the mouse genome, we examined the organization of genes from human chromosome 22q11 in the mouse. Using genetic linkage analysis and detailed physical mapping, we show that genes from a relatively small region of human 22q11 are distributed on three mouse chromosomes (MMU6, MMU10, and MMU16). Furthermore, although the region corresponding to about 2.5 megabases of the VCFS/DGS critical region is located on mouse chromosome 16, the relative organization of the region is quite different from that in humans. Our results show that the instability of the 22q11 region is not restricted to humans but may have been present throughout evolution. The results also underscore the importance of detailed comparative mapping of genes in mice and humans as a prerequisite for the development of mouse models of human diseases involving chromosomal rearrangements.
Constitutional and somatic rearrangements of human chromosome 22q11 (HSA22q11) are associated with various clinical disorders. Several tumor-associated chromosomal translocations involving 22q11 have been described including the t(8;22) of Burkitt’s lymphoma (BL, MIM 113970), the t(9;22) of acute lymphocytic leukemia (MIM 159555) and chronic myeloid leukemia (MIM 151410), as well as various 22q11 translocations of malignant rhabdoid tumors (MIM 601607). In the latter tumors, deletions of the 22q11 region also have been described (1). In addition, predisposition to breast cancer has been reported in carriers of the constitutional translocation t(11;22)(q23;q11), the most frequently occurring reciprocal translocation in humans (MIM 600048). Finally, constitutional duplications and deletions of human chromosome 22q11 have been observed in several syndromes involving developmental abnormalities including “cat eye” syndrome (CES, MIM 115470), derivative 22 syndrome (2), Velo-cardio-facial syndrome (VCFS, MIM 192430), and DiGeorge syndrome (DGS, MIM188400).
Analysis of the chromosomal abnormalities in these disorders showed the presence of sites of frequent chromosomal rearrangement and defined the relative location of the regions involved in the rearrangements. About 80% of VCFS and DGS patients exhibit a large, approximately 3 megabases (Mb) interstitial deletion with breakpoints that occur within a defined interval (3–7). In CES patients, extra copies of the 22q11 region are due to the presence of a supernumerary dicentric chromosome characterized as an inv dup(22)(q11) (8). By analyzing the breakpoints of the duplicated regions of CES patients, the CES critical region has been localized immediately proximal to the large deleted region of VCFS/DGS patients (9). Derivative 22 patients have extra copies of the 22q11 region due to a 3:1 missegregation of the constitutional t(11;22) (q23;q11) translocation (2). Because the 22q11 breakpoint of this translocation falls in the distal segment of the VCFS/DGS deleted region, the region duplicated in derivative 22 patients encompasses both the CES critical region and most of the 2.5-Mb VCFS/DGS deleted region (10). Finally, the 22q11 tumor-associated breakpoints seen in BL, acute lymphocytic leukemia, chronic myeloid leukemia, and rhabdoid tumor translocations have all been localized telomeric to the large deleted region of VCFS/DGS patients (1, 11, 12). Thus, it seems that DNA sequences prone to rearrangements are located at several different positions within the 22q11 region. A further indication of the instability of the 22q11 region is found in the fact that the sequences mapping to the sites of frequent chromosomal rearrangements in CES, VCFS/DGS, and various tumors are unstable when cloned in yeast (6, 13). The fact that most cases of VCFS/DGS associated with deletions are sporadic also suggests that the DNA sequences at the breakpoint are inherently unstable. Human chromosome 22q11 has a high gene density, but it also contains several duplicated regions and low-copy repeat families. It has been suggested that the instability of the 22q11 region may be related to the presence of these numerous repeats (14, 15).
Efforts to identify the genes involved in the various disorders associated with 22q11 rearrangements have been successful in a few instances involving balanced translocations, for example in BL and chronic myeloid leukemia. However, identification of the responsible gene(s) in syndromes resulting from aneuploidy of a large chromosomal segment is more complex, and the etiology of the disease in these cases is likely to involve more than a single gene. Therefore, to fully understand the disease etiology in these cases, it will be important to evaluate the consequences of similar rearrangements in other experimentally manipulable mammals such as mice. Techniques have become available recently for generating specific chromosomal translocations and deletions in mice (16–19). As a prelude to such experiments, we compared the organization of the human 22q11 region with the orthologous region in mice. The results show that the genes from 22q11 are distributed on three different mouse chromosomes. Because a large number of genes and other markers have been identified in the region deleted in VCFS/DGS, we further analyzed the corresponding region in mice. Our results show that most of the genes in the VCFS/DGS region are located in one region of mouse chromosome 16 but that the organization of these genes is quite different from that in humans. These differences might reflect an intrinsic property of the DNA sequences in the region to undergo rearrangement.
MATERIALS AND METHODS
Probes.
Mouse probes for Arvcf and Idd genes were prepared from partial cDNA clones isolated by screening at low stringency a mouse newborn brain cDNA library (Stratagene). Sequencing confirmed their similarity to ARVCF and IDD/DGCR2, respectively (20–22). Arvcf (1.8-kb EcoRI fragment) and Idd (1.3-kb BamHI fragment) probes were obtained by digestion of plasmids mp120b and mIdd2.3, respectively (BSJ and AP, unpublished data). Human probes for CLTD, TMVCF, DGS-I, ATP6E, and GP1BB genes were prepared from cDNA clones. TMVCF (1.5-kb HindIII–NotI fragment), CLTD (1.1-kb PstI fragment), GP1BB (600-bp HindIII fragment), and ATP6E (800-bp HindIII fragment) were obtained by digestion of plasmids V2–12, FB13 (23, 24), 295D12, and 214C10, respectively. GP1BB and ATP6E were identified as ESTs from the arrayed NIB library by Blast search using the human gene sequences (25–27). Partial sequencing of the plasmids confirmed their identity to GP1BB and ATP6E sequences, respectively. The human probe for COMT (400-bp EcoRV fragment) was described previously.
Interspecific Mouse Backcross Mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J X Mus spretus) F1 females and C57BL/6J males as described (28). This interspecific backcross mapping panel has been typed for over 2500 loci that are well distributed among all the autosomes as well as the X chromosome (28). C57BL/6J and M. spretus DNAs were digested with several enzymes and analyzed by Southern blot hybridization for informative restriction fragment length polymorphisms (RFLPs) using cDNA probes. A total of 205 N2 mice were used in this study (see figure legends for details). DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, Southern blot transfer, and hybridization were performed essentially as described (29). The presence or absence of M. spretus-specific fragments was followed in backcross mice. Only major restriction fragments detected with each probe are listed below. The fragments that were followed in the backcross analysis are underlined. In all cases, when more than one fragment was followed, the fragments cosegregated. GP1BB detected HincII fragments of 7.4, 7.0, 3.0, 2.3 (C57BL/6J), 4.7, and 3.1 (M. spretus) kb. Idd detected HindIII fragments of 9.8, 5.2, 2.3, 21, 1.9 (C57BL/6J), 10.0, 5.4, 3.2, 2.4, 2.0, and 1.8 (M. spretus) kb. Arvcf detected BglII fragments of 7.6 (C57BL/6J) and 14.5 (M. spretus) kb. COMT detected BglII fragments of 5.1 (C57BL/6J) and 15.0 (M. spretus) kb and TaqI fragments of 6.7 (C57BL/6J) and 6.0 (M. spretus) kb. In this case, the data for BglII and TaqI were combined. TMVCF detected SphI fragments of 7.3 (C57BL/6J) and 13.5 (M. spretus) kb. ATP6E detected SphI fragments of 13.0, 6.8, 2.5, 1.7, 1.3, 0.6 (C57BL/6J), 13.0, 7.7, 4.9, 1.7, 1.3, and 0.6 (M. spretus) kb. CLTD detected EcoRI fragments of 4.7, 2.3 (C57BL/6J), 1.5, and 1.0 (M. spretus) kb. The probes and RFLPs for Ret, M6pr, Glut3, and Cd4 (30, 31), Prm1, Ntan1, Htf9, Cebpd, Igl, Thpo, and Smst (32–34), and Nfl, Scya2, and Nog have been reported previously (35, 36). Each locus was analyzed in pairwise combinations for recombination frequencies using the additional data. Recombination distances were calculated using map manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Physical Mapping on Mouse Chromosome 16.
Mouse YAC clones are from the Whitehead Center for Genome Research (WCGR, http://www-genome.wi.mit.edu/) C57BL/6J library that consists of 40,000 YACs with an average insert size of 820 kb (37). YACs reported as positive for D16Mit143 and D16Mit29 were identified, and the corresponding singly or doubly linked contigs were purchased from Research Genetics. These YACs were analyzed by PCR reactions on template DNA developed from yeast clones as described (6). In the WCGR database, the sequence of primers used to detect D16Mit28 and D16Mit29 are identical, and consequently they are shown as a single locus (D16Mit29/28) on our map. Primers for D22S680E, Ufd1l, Htf9c, Thpo, Igl, Hira, Tbx1, and Gscl were developed from gene sequences deposited in the GenBank database. Primers for D16H22S1742E, Comt, Dgcr6, Gp1bb, Ctp, and Tmvcf were developed from mouse EST sequences that were identified by Blast search and had a similarity greater than 80% to the human or rat gene sequences. Primers for Dgsi and Arvcf were developed from mouse sequences homologous to DGS-I and ARVCF (see probes). Primers for Idd introns and exons were developed from genomic plasmid subclones that were derived from mouse genomic phage clones shown to contain the Idd gene (AP and BSJ, unpublished results). D numbers were assigned for each amplimer, and the corresponding primer sequences were deposited in the Mouse Genome Database (http://www.informatics.jax.org/).
Physical Mapping of and RANBP1 on HSA22q11.
Sequence tagged sites (STSs) were developed as described by Morrow et al. (6) or from sequences retrieved from the GenBank database (genes D22S680E and RANBP1, ESTs D60871 and R25516). Each primer pair was used to perform PCR reactions on template DNA prepared from bacterial clones as described (6). D numbers were assigned for each amplimer, and the corresponding primer sequences were deposited in the Genome data base (http://gdbwww.gdb.org./gdb/gdbtop.html).
RESULTS
Genes from Human Chromosome 22q11 Are Present on Three Different Mouse Chromosomes.
To determine the location of the region in the mouse genome corresponding to the human 22q11 region and to assess its integrity, we performed linkage analysis in mice on seven genes from HSA22q11. These genes had been ordered unambiguously by physical mapping along the human 22q11 chromosomal axis as centromere-ATP6E-IDD-CLTD-TMVCF-GP1BB-COMT-ARVCF-telomere (6, 38). The region containing these genes is estimated to span about 1.5 Mb. The mouse chromosomal locations of the corresponding genes were determined by interspecific backcross analysis using progeny derived from matings of [(C57BL/6J X M. spretus)F1 X C57BL/6J] mice.
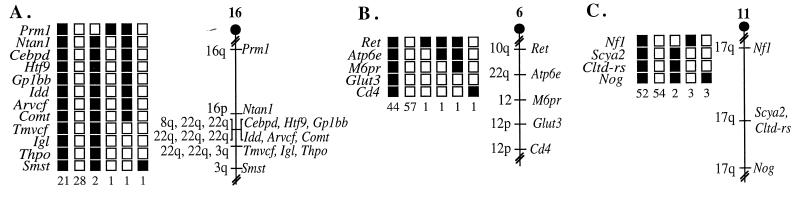
We found that five of the seven genes analyzed, Idd, Gp1bb, Tmvcf, Arvcf, and Comt, are located on mouse chromosome 16 linked to Igl and Htf9, two other genes from HSA22q11 that were already mapped to MMU16 (Fig. 1A) (10, 32). These five genes also were linked to several other genes mapped previously to this region: Prm1, Ntan1, Cebpd, Thpo, and Smst (33, 34) (Fig. 1A). The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci as well as the most likely gene order are centromere-Prm1-4/168-Ntan1-1/166-Cebpd-0/185-Htf9-0/164-Gp1bb-0/141-Idd-0/158-Arvcf-0/180-Comt-1/159-Tmvcf-0/161-Igl-0/176-Thpo-1/121-Smst. These data show that the region of human 22q11 from IDD (centromeric) to IGL (telomeric), corresponding to the VCFS/DGS deleted region, is present in one region of mouse chromosome 16 (Fig. 3). However, we found that two genes from the human 22q11 region, ATP6E and CLTD, were not present on MMU16.
Figure 1.
Genetic mapping of human 22q11 homologous genes on mouse chromosomes. The genes were placed on mouse chromosomes by interspecific backcross analysis. The segregation patterns of human 22q11 genes and flanking genes in backcross animals that were typed for all loci are shown at the left of each panel. For individual pairs of loci, more animals were typed. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J X M. spretus) F1 parent. Shaded boxes represent the presence of a C57BL/6J allele, and white boxes represent the presence of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. Partial chromosome linkage maps showing the location of human 22q11 genes in relation to linked genes are shown at the right of each panel. The positions of loci on human chromosomes, where known, are shown to the left of the chromosome linkage maps. References for the human map positions of loci cited in this study can be obtained from GDB. (A) Gp1bb, Idd, Arvcf, Comt, and Tmvcf map to the proximal region of MMU16. Although 54 mice were analyzed for all markers and are shown in the segregation analysis, up to 185 mice were typed for some pairs of markers. The recombination frequencies expressed as genetic distances in cM ± the standard error are Prm1-2.4 ± 1.2-Ntan1-0.6 ± 0.6-[Cebpd, Htf9, Idd, Gp1bb, Arvcf, Comt]-0.6 ± 0.6-[Tmvcf, Igl, Thpo]-0.8 ± 0.8-Smst. No recombinants were detected between Cebpd and Htf9 in 185 mice typed in common, Htf9 and Gp1bb in 164 mice, Gp1bb and Idd in 141 mice, Idd and Arvcf in 158 mice, Arvcf and Comt in 180 mice, suggesting that the two loci in each pair are within 1.6, 1.8, 2.1, 1.9, and 1.7 cM of each other, respectively (upper 95% confidence limit). In addition, no recombinants were detected between Tmvcf and Igl in 161 mice typed in common or between Igl and Thpo in 176 mice, suggesting the two loci in each of these pairs are within 1.9 and 1.7 cM of each other, respectively (upper 95% confidence limit). (B) Atp6e maps to the distal region of MMU6. Although 105 mice were analyzed for all markers and are shown in the segregation analysis, up to 150 mice were typed for some pairs of markers. The recombination frequencies expressed as genetic distances in cM ± the standard error are Ret-0.7 ± 0.7-Atp6e-0.7 ± 0.7-M6pr-0.7 ± 0.7-Glut3-0.7 ± 0.7-Cd4. (C). Cltd-rs maps to the central region of MMU11. Although 114 mice were analyzed for all markers and are shown in the segregation analysis, up to 151 mice were typed for some pairs of markers. The recombination frequencies expressed as genetic distances in cM ± the standard error are Nfl-4.1 ± 1.6-[Scya2, Cltd-rs]-2.7 ± 1.3-Nog. No recombinants were detected between Scya2 and Cltd-rs in 134 animals typed in common, suggesting that the two loci are within 2.2 cM of each other (upper 95% confidence limit).
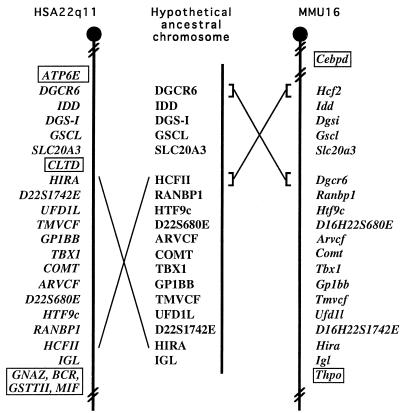
Figure 3.
Comparison of the human VCFS/DGS region with the homologous region in mice. On the left is represented the relative order of genes from the HSA22q11 (this paper and refs. 6, 7, and 10). On the right is represented the relative order of genes in the homologous region in mice as shown by physical and genetic mapping. The distance between each marker is arbitrary. The middle portion of the figure shows the gene order of a hypothetical, ancestral chromosome and possible recombination events leading to the differences in the gene order in mice and humans (see text for details). Gene names within boxes in the map of one species indicate genes that are not present in the homologous region of the other species.
Atp6e, mapped to the distal region of mouse chromosome 6, linked to several genes, Ret, M6pr, Glut3, and Cd4, that had been mapped previously to this region (Fig. 1B) (30, 31). The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci as well as the most likely gene order are centromere-Ret-1/137-Atp6e-1/137-M6pr-1/150-Glut3-1/150-Cd4. Because ATP6E lies at the proximal end of the group of five 22q11 genes that are present in one region of MMU16, the data indicate that the proximal boundary of the HSA22q11/MMU16 homologous region lies between ATP6E and IDD. Likewise, as four genes that map to the telomeric part of human chromosome 22q11, BCR, GNAZ, MIF, and GSTTII, were reported previously to map to mouse chromosome 10 (10, 39–43), the distal boundary of the homologous region must lie between IGL and these four genes. As the distance between ATP6E and BCR has been estimated to be about 6.5 Mb, our data indicate that genes from a relatively small region of human 22q11 are distributed on three different mouse chromosomes.
A Gene from the Central Region of Homology between HSA22q11 and MMU16 Is Not Present on Mouse Chromosome 16.
Although genes from an extensive region of HSA22q11 constituting the VCFS/DGS chromosomal region were found on MMU16, CLTD, a gene localized in the central portion of this region, did not map to mouse chromosome 16. Instead, sequences hybridizing with the CLTD probe seemed to be located in the central region of mouse chromosome 11, linked to genes that had been mapped previously in this region: Nfl, Scya2, and Nog (Fig. 1C) (35, 36). The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci as well as the most likely gene order are centromere-Nfl-6/148-Scya2-0/134-Cltd-rs-4/151-Nog. The central region of mouse chromosome 11 shares a large region of homology with human chromosome 17 (Fig. 1C). Interestingly, a second clathrin heavy chain gene CLTC, which is 84.7% identical to CLTD, maps to 17q11-qter (44). Two cosegregating RFLPs were detected with the CLTD probe. These data suggest that either Cltc and Cltd are tandemly duplicated loci that map to the central region of mouse chromosome 11 or that the mouse genome does not contain a gene corresponding to CLTD. We favor the latter hypothesis, as hybridization of a mouse RNA blot with a human CLTD probe showed an expression pattern more similar to the CLTC gene than to the CLTD gene (45). Because the exact identity of the clathrin-related sequences on mouse chromosome 11 remains to be determined, the locus has been designated Cltd-rs. However, in either case, the absence of a homolog of CLTD on MMU16 suggested that the region of mouse chromosome 16 orthologous to human 22q11 is disrupted.
Multiple Changes in Gene Order in the 22q11/MMU16 Homologous Region.
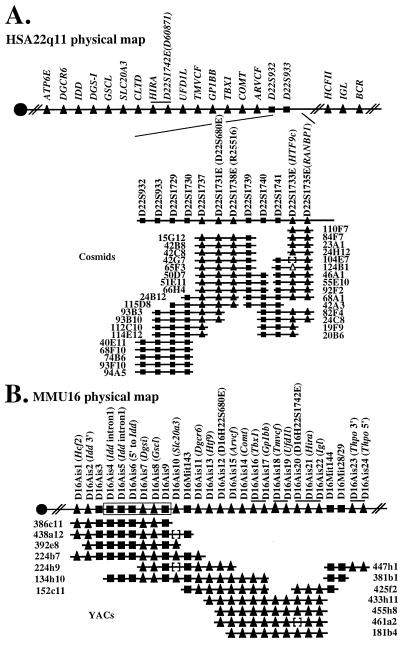
To further assess the integrity of the region in mice orthologous to HSA22q11, we compared the order of genes in this region in the two species. As the Htf9 locus maps to the region in the mouse that is orthologous to HSA22q11 (Fig. 1A) and since its exact location in human 22q11 was unknown, we first integrated its location onto our high resolution physical map of the VCFS/DGS region. The Htf9 locus was isolated as a CpG island that serves as a bidirectional promoter for two genes, Htf9a/Ranbp1 and Htf9c (32, 46). To precisely map RANBP1 and HTF9c, we performed PCR analysis on a cosmid and PAC contig that spans the human VCFS/DGS chromosomal region (6, 7). First, the contig was extended distally, which also allowed us to integrate at a high resolution the location of D22S680E and an EST, R25516 (Fig. 2A). PCR analysis also allowed us to map both RANBP1 and HTF9c genes to this contig and showed that they lie distal to ARVCF, COMT, and D22S680E (Fig. 2A). The two genes map on the same cosmids in the distal part of the VCFS/DGS chromosomal region, showing that, as in the mouse, RANBP1 and HTF9c are extremely close physically. According to our extended physical map, the gene order in human 22q11 is centromere-IDD-TMVCF-GP1BB-COMT-ARVCF-HTF9c-RANBP1-IGL-telomere. Comparing this gene order in humans with the mouse genetic map suggests that the relative order of some of the genes is different in the two species. That is, in mice, [Idd, Gp1bb, Arvcf, Comt, and Htf9] are nonrecombinant, and this cluster maps proximal to [Tmvcf and Igl] (Fig. 1A). On the other hand, in humans, whereas IDD lies proximal to TMVCF, the other members of its recombinant group in mice, namely GP1BB, ARVCF, COMT, and HTF9, map distal to TMVCF, although they remain proximal to IGL (38). A simple explanation of this observation is that during evolution an inversion occurred involving Gp1bb, Arvcf, Comt, and Tmvcf (Fig. 3).
Figure 2.
Physical map of a portion of MMU16 and HSA22q11. The markers used to construct the physical maps are indicated above the line representing either a portion of HSA22q11 (A) or MMU16 (B). For expressed sequences, the name of the gene or EST has been added in parentheses. The YAC and cosmid clones were ordered based upon the presence of genes (triangles) and monomorphic STS markers (squares). Closed symbols represent the markers that were tested and that are present on a particular clone, and open symbols represent markers that were not tested for the particular clone. The distance between each marker is arbitrary and does not reflect the actual physical distance. (A) Partial physical map of HSA22q11. The line at the top presents the relative order of genes from HSA22q11 as determined by high resolution physical mapping (6, 7). The markers used to extend the physical map are indicated above the line representing the region mapped in this study. (B) Physical map of the mouse region homologous with HSA22q11. A line above the symbols identifies genes or markers for which the relative order is not known. Order of markers D16Ais4 to D16Ais9 was inferred from the published 38-kb sequence of a portion of MMU16 (60).
To determine more precisely the boundaries of the inverted region, we established a physical map in this region of the mouse genome. On the Copeland/Jenkins RFLP map (WCGR database), Igl and Htf9 are closely linked to two simple sequence length polymorphisms (SSLPs), D16Mit143 and D16Mit29, respectively. These data allowed us to identify singly and doubly linked YAC contigs reported as containing these SSLP markers (47–49). Primer pairs were developed from exon or intron sequences of 17 homologs of genes from human chromosome 22q11 and from Thpo, a gene linked to Tmvcf and Igl but present on human chromosome 3 (Fig. 1A). PCR analysis was performed on the mouse YAC contigs. As shown on Fig. 2B, we were able to place these 18 genes as well as 3 SSLPs on the different overlapping YACs. A contig was established by minimizing the number of deletions introduced in each YAC. The centromere to telomere orientation of the contig is provided by two independent maps: the Copeland/Jenkins map that places D16Mit143 centromeric to D16Mit29 and a high resolution recombinational map of mouse chromosome 16 that places D16Mit143 centromeric to Comt and Igl, themselves centromeric to D16Mit144 and D16Mit29 (50). The deduced order of genes and markers is centromere-Hcf2-Idd-Dgsi-Gscl-Slc20a3-D16Mit143-Dgcr6-Htf9c-D22S680E-Arvcf-Comt-[Tbx1, Gp1bb]-[Tmvcf, Ufd1l]-[D16H22S1742E, Hira, Igl]-D16Mit144-D16Mit29/28-Thpo-telomere. Our genetic mapping data that places [Idd, Arvcf, Comt, Htf9] centromeric to [Tmvcf, Igl, Thpo] is consistent with the gene order established by physical mapping. The most telomeric YAC, 447h1, was found to contain Thpo (Fig. 2B). This result localizes physically in the mouse an evolutionary breakpoint in the vicinity of markers D16Mit28/29 and D16Mit144. It also shows that the mouse region orthologous to the human VCFS/DGS region is immediately adjacent to a region orthologous to human chromosome 3. From these data, the minimal tiling path containing most of the genes from HSA22q11 consists of only three mouse YACs: 224b7, 152c11, and 181b4. Assuming a mean size of 820 kb for each YAC, the MMU16 region orthologous to HSA22q11 is likely to be about 2.5 Mb. As we have placed 17 genes and 2 SSLPs on this minimal tiling path, it provides a resolution of at least one marker every 130 kb.
A comparison of the mouse and human physical maps confirms the results detected by genetic mapping: the presence of an inverted gene order between mice and humans spanning from HIRA to HTF9 (Fig. 3). In addition to this inversion, our results also show that more complex rearrangements must have occurred in this region during evolution. For example, Hcf2 and Dgcr6 seem to be placed at two completely different positions in the two species (Fig. 3). However, rearrangements of these two genes during evolution does not seem to involve a simple inversion as three genes, Idd, Dgsi, and Gscl, which are located between Hcf2 and Dgcr6, have the same centromere to telomere orientation in mice and humans (Fig. 3). Thus, it seems that two recombination events would be required to account for the observed gene order.
DISCUSSION
The instability of human chromosome 22q11 has been well documented by numerous reports of rearrangements in this region including the most frequently occurring constitutional translocation t(11;22) in humans, various tumor-associated translocations between 22q11 and other chromosomal regions, and rearrangements within the 22q11 region associated with several syndromes involving developmental abnormalities. The sites of these various rearrangements occur throughout 22q11, suggesting that the entire region is susceptible to rearrangement.
Through a combination of genetic and physical mapping techniques, we investigated the chromosomal locations of a large number of genes and ESTs from human 22q11 in the mouse genome. The results show that genes present within a region of 22q11, which we estimate to be about 6.5 Mb in length, are found on three different mouse chromosomes, MMU6, MMU16, and MMU10. By comparison, 18 genes in another part of human chromosome 22 (22q13), which is much larger than 22q11, are all located on mouse chromosome 15 (10, 51–53). These results, together with the fact that the average length of conserved segments between humans and mice was estimated to correspond to 8.8 cM (54), suggest that the region corresponding to 22q11 was prone to rearrangements during evolution.
Further evidence for the evolutionary instability of the 22q11/MMU16 homologous region was found in our detailed comparison of the gene order in the VCFS/DGS region on 22q11 and the corresponding region on mouse chromosome 16. This comparative gene mapping study was facilitated by the very high resolution, human physical map of this region available in our laboratory (7). Although we found that 17 genes and ESTs from the VCFS/DGS region were localized to one region of mouse chromosome 16, we detected three separate segments within the region in which the gene order was different between mice and humans. Furthermore, we found that CLTD, a gene from the central portion of this region, is either located on mouse chromosome 11 or more likely absent from the mouse genome. This seems to be an unusually high rate of evolutionary change in a region that we have estimated in both species to be about 2.5 Mb. Studies of the organization of HSA22q11/MMU16 homologous regions in other mammals are required to determine whether this apparent high rate of change has been present throughout mammalian evolution.
As shown in Fig. 3, the several differences in gene order within the region of HSA22q11/MMU16 homology between mice and humans might be explained by the existence of an ancestral chromosome that diverged in two different ways during evolution. On the one hand, HSA22q11 might have been derived by a simple inversion spanning from HIRA to HCFII. On the other hand, MMU16 may have evolved by an exchange of segments including HCFII and DGCR6. The fact that in humans, VPREB1 is located near the IGL locus and that in mice this gene has been mapped centromeric to D16Mit143, further supports our hypothesis (55, 56).
The reasons for the evolutionary differences and the instability of chromosome 22q11 in humans are not well understood. The 22q11 region contains a large number of highly repetitive as well as intermediate repetitive sequences (10, 13, 14). There also is evidence that several genes may be duplicated in the region (15, 57). The regions in which we have mapped the evolutionary breakpoints of human chromosome 22q11 correspond to regions containing these repetitive sequences. Similarly, Blair et al. (58) described the presence of duplicated sequences in the vicinity of evolutionary breakpoints in the proximal region of the human X chromosome. A correlation might exist between evolutionarily unstable regions and the presence of repetitive sequences. Furthermore, these regions on 22q11 are also the sites of frequent constitutional and somatic rearrangements in man, and the same repetitive sequences might be responsible for this instability. A complete DNA sequence of the region and definition of the breakpoints at the molecular level will be needed to understand the basis for this instability.
The existence of regions of homology to HSA22q11 on MMU6 and MMU10 could suggest that homologs of genes responsible for certain 22q11 associated disorders will also be found on these mouse chromosomes. For example, ATP6E has been reported to delineate the distal boundary of the CES critical region (59). Our finding that Atp6e maps to MMU6 suggests that homologs of genes responsible for CES may be localized on mouse chromosome 6. Likewise, the breakpoints of various tumor-associated translocations reside in the region containing GNAZ, GSTTII, and MIF, lying distal to BCR. As the mouse homologs of these four genes map to mouse chromosome 10, it is possible that genes associated with various malignancies might reside on this mouse chromosome.
Our results also have important implications for efforts to investigate the molecular basis of 22q11 syndromes by genetic manipulations of mice. For example, as most of the VCFS/DGS patients have large deletions encompassing the region from DGCR6 to ARVCF/COMT, the creation of a deletion spanning this region in mice would be an efficient approach for generating a mouse model of VCFS/DGS. However, because of the inverted arrangement of genes in this region, the order of some of the genes as well as their transcriptional orientations differ in the two species. The knowledge gained in the present study will allow us to design more precise genetic manipulations in mice, permitting tests of specific sets of genes in the VCFS/DGS critical region for their role in the etiology of these syndromes. More generally, our results indicate that detailed comparative mapping of genes in mice and humans is essential for the development of mouse models of other human deletion syndromes by generating defined chromosomal deletions in the mouse genome.
Acknowledgments
We thank Hui Xu, Raj K. Pandita, and Deborah B. Householder for excellent technical assistance. A.P. was supported in part by the Fondation Singer-Pollignac. This research was supported in part by National Institutes of Health Grant PO1 HD34980-01 and by the National Cancer Institute, DHHS, under contract with ABL.
ABBREVIATIONS
- MMU16
M. musculus chromosome 16
- HSA22q11
Homo sapiens chromosome 22q11
- VCFS
Velo-cardio-facial syndrome
- DGS
DiGeorge syndrome
- CES
cat eye syndrome
- YAC
yeast artificial chromosome
- EST
expressed sequence tagged site
- BL
Burkitt’s lymphoma
- RFLP
restriction fragment length polymorphism
- SSLP
simple sequence length polymorphism
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. D16Ais1–D16Ais24 and D22S1729–D22S1742).
References
- 1.Biegel J A, Allen C S, Kawasaki K, Shimizu N, Budarf M L, Bell C J. Gene Chromosomes Cancer. 1996;16:94–105. doi: 10.1002/(SICI)1098-2264(199606)16:2<94::AID-GCC3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 2.Iselius L, Lindsten J, Aurias A, Fraccaro M, Bastard C, Bottelli A M, Bui T H, Caufin D, Dalpra L, Delendi N, et al. Hum Genet. 1983;64:343–355. doi: 10.1007/BF00292366. [DOI] [PubMed] [Google Scholar]
- 3.Scambler P J, Kelly D, Lindsay E, Williamson R, Goldberg R, Shprintzen R, Wilson D I, Goodship J A, Cross I E, Burn J. Lancet. 1992;339:1138–1139. doi: 10.1016/0140-6736(92)90734-k. [DOI] [PubMed] [Google Scholar]
- 4.Kelly D, Goldberg R, Wilson D, Lindsay E, Carey A, Goodship J, Burn J, Cross I, Shprintzen R J, Scambler P J. Am J Med Genet. 1993;45:308–312. doi: 10.1002/ajmg.1320450306. [DOI] [PubMed] [Google Scholar]
- 5.Driscoll D A, Salvin J, Sellinger B, Budarf M L, McDonald-McGinn D M, Zackai E H, Emanuel B S. J Med Genet. 1993;30:813–817. doi: 10.1136/jmg.30.10.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrow B, Goldberg R, Carlson C, DasGupta R, Sirotkin H, Collins J, Dunham I, Odonnell H, Scambler P, Shprintzen R, Kucherlapati R. Am J Hum Genet. 1995;56:1391–1403. [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson C, Papolos D, Pandita R K, Faedda G L, Veit S, Goldberg R, Shprintzen R, Kucherlapati R, Morrow B. Am J Hum Genet. 1997;60:851–859. [PMC free article] [PubMed] [Google Scholar]
- 8.Schinzel A, Schmid W, Fraccaro M, Tiepolo L, Zuffardi O, Opitz J M, Lindsten J, Zetterqvist P, Enell H, Baccichetti C, Tenconi R, Pagon R A. Hum Genet. 1981;57:148–158. doi: 10.1007/BF00282012. [DOI] [PubMed] [Google Scholar]
- 9.Mears A J, Duncan A M, Budarf M L, Emanuel B S, Sellinger B, Siegel-Bartelt J, Greenberg C R, McDermid H E. Am J Hum Genet. 1994;55:134–142. [PMC free article] [PubMed] [Google Scholar]
- 10.Budarf M L, Eckman B, Michaud D, McDonald T, Gavigan S, Buetow K H, Tatsumura Y, Liu Z, Hilliard C, Driscoll D, Goldmuntz E, Meese E, Zwarthoff E C, Williams S, McDermid H, Dumanski J P, Biegel J, Bell C J, Emanuel B S. Genomics. 1996;35:275–288. doi: 10.1006/geno.1996.0358. [DOI] [PubMed] [Google Scholar]
- 11.Emanuel B S, Selden J R, Wang E, Nowell P C, Croce C M. Cytogenet Cell Genet. 1984;38:127–131. doi: 10.1159/000132044. [DOI] [PubMed] [Google Scholar]
- 12.Erikson J, Griffin C A, ar-Rushdi A, Valtieri M, Hoxie J, Finan J, Emanuel B S, Rovera G, Nowell P C, Croce C M. Proc Natl Acad Sci USA. 1986;83:1807–1811. doi: 10.1073/pnas.83.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell C J, Budarf M L, Nieuwenhuijsen B W, Barnoski B L, Buetow K H, Campbell K, Colbert A M, Collins J, Daly M, Desjardins P R, et al. Hum Mol Genet. 1995;4:59–69. doi: 10.1093/hmg/4.1.59. [DOI] [PubMed] [Google Scholar]
- 14.Halford S, Lindsay E, Nayudu M, Carey A H, Baldini A, Scambler P J. Hum Mol Genet. 1993;2:191–196. doi: 10.1093/hmg/2.2.191. [DOI] [PubMed] [Google Scholar]
- 15.Morris C, Courtay C, Geurts van Kessel A, ten Hoeve J, Heisterkamp N, Groffen J. Hum Genet. 1993;91:31–36. doi: 10.1007/BF00230218. [DOI] [PubMed] [Google Scholar]
- 16.Ramirez-Solis R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 17.Smith A J H, Desousa M A, Kwabiaddo B, Heppellparton A, Impey H, Rabbitts P. Nat Genet. 1995;9:376–385. doi: 10.1038/ng0495-376. [DOI] [PubMed] [Google Scholar]
- 18.Van Deursen J, Fornerod M, Van Rees B, Grosveld G. Proc Natl Acad Sci USA. 1995;92:7376–7380. doi: 10.1073/pnas.92.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z W, Stark G, Gotz J, Rulicke T, Muller U, Weissmann C. Proc Natl Acad Sci USA. 1996;93:6158–6162. doi: 10.1073/pnas.93.12.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wadey R, Daw S, Taylor C, Atif U, Kamath S, Halford S, O’Donnell H, Wilson D, Goodship J, Burn J, Scambler P. Hum Mol Genet. 1995;4:1027–1033. doi: 10.1093/hmg/4.6.1027. [DOI] [PubMed] [Google Scholar]
- 21.Demczuk S, Aledo R, Zucman J, Delattre O, Desmaze C, Dauphinot L, Jalbert P, Rouleau G A, Thomas G, Aurias A. Hum Mol Genet. 1995;4:551–558. doi: 10.1093/hmg/4.4.551. [DOI] [PubMed] [Google Scholar]
- 22.Sirotkin H, O’Donnell H, DasGupta R, Halford S, St-Jore B, Puech A, Parimoo S, Morrow B, Skoultchi A, Weissman S M, Scambler P, Kucherlapati R. Genomics. 1997;41:75–83. doi: 10.1006/geno.1997.4627. [DOI] [PubMed] [Google Scholar]
- 23.Sirotkin H, Morrow B, DasGupta R, Goldberg R, Patanjali S R, Shi G P, Cannizzaro L, Shprintzen R, Weissman S M, Kucherlapati R. Hum Mol Genet. 1996;5:617–624. doi: 10.1093/hmg/5.5.617. [DOI] [PubMed] [Google Scholar]
- 24.Sirotkin H, Morrow B, Saint-Jore B, Puech A, DasGupta R, Patanjali S R, Skoultchi A, Weissman S M, Kucherlapati R. Genomics. 1997;42:245–251. doi: 10.1006/geno.1997.4734. [DOI] [PubMed] [Google Scholar]
- 25.Kelly M D, Essex D W, Shapiro S S, Meloni F J, Druck T, Huebner K, Konkle B A. J Clin Invest. 1994;93:2417–2424. doi: 10.1172/JCI117249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soares M B, Bonaldo M F, Jelene P, Su L, Lawton L, Efstratiadis A. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baud V, Mears A J, Lamour V, Scamps C, Duncan A M, McDermid H E, Lipinski M. Hum Mol Genet. 1994;3:335–339. doi: 10.1093/hmg/3.2.335. [DOI] [PubMed] [Google Scholar]
- 28.Copeland N G, Jenkins N A. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins N A, Copeland N G, Taylor B A, Lee B K. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan A, Heyner S, Charron M J, Copeland N G, Gilbert D J, Jenkins N A, Thorens B, Schultz G A. Development. 1991;113:363–372. doi: 10.1242/dev.113.1.363. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig T, Ruther U, Metzger R, Copeland N G, Jenkins N A, Lobel P, Hoflack B. J Biol Chem. 1992;267:12211–12219. [PubMed] [Google Scholar]
- 32.Bressan A, Somma M P, Lewis J, Santolamazza C, Copeland N G, Gilbert D J, Jenkins N A, Lavia P. Gene. 1991;103:201–209. doi: 10.1016/0378-1119(91)90274-f. [DOI] [PubMed] [Google Scholar]
- 33.Chang M S, Hsu R Y, McNinch J, Copeland N G, Jenkins N A. Genomics. 1995;26:636–637. doi: 10.1016/0888-7543(95)80193-p. [DOI] [PubMed] [Google Scholar]
- 34.Grigoryev S, Stewart A E, Kwon Y T, Arfin S M, Bradshaw R A, Jenkins N A, Copeland N G, Varshavsky A. J Biol Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- 35.Youn B S, Jang I K, Broxmeyer H E, Cooper S, Jenkins N A, Gilbert D J, Copeland N G, Elick T A, Fraser M, Jr, Kwon B S. J Immunol. 1995;155:2661–2667. [PubMed] [Google Scholar]
- 36.Valenzuela D M, Rojas E, Le Beau M M, Espinosa R R, Brannan C I, McClain J, Masiakowski P, Ip N Y, Copeland N G, Jenkins N A, et al. Genomics. 1995;25:157–163. doi: 10.1016/0888-7543(95)80121-2. [DOI] [PubMed] [Google Scholar]
- 37.Haldi M L, Strickland C, Lim P, VanBerkel V, Chen X, Noya D, Korenberg J R, Husain Z, Miller J, Lander E S. Mamm Genome. 1996;7:767–769. doi: 10.1007/s003359900228. [DOI] [PubMed] [Google Scholar]
- 38.Carlson, C., Sirotkin, H., Pandita, R., Goldberg, R., McKie, J., Wadey, R., Patanjali, S. R., Weissman, S. M., Anyane-Yeboa, K., Warburton, D., Scambler, P., Shprintzen, R., Kucherlapati, R. & Morrow, B. E. (1997) Am. J. Hum. Genet. 61, in press. [DOI] [PMC free article] [PubMed]
- 39.Budarf M, McDonald T, Sellinger B, Kozak C, Graham C, Wistow G. Genomics. 1997;39:235–236. doi: 10.1006/geno.1996.4505. [DOI] [PubMed] [Google Scholar]
- 40.Justice M J, Siracusa L D, Gilbert D J, Heisterkamp N, Groffen J, Chada K, Silan C M, Copeland N G, Jenkins N A. Genetics. 1990;125:855–866. doi: 10.1093/genetics/125.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkie T, Gilbert D, Olsen A, Chen X, Amatruda T, Korenberg J, Trask B, de Jong P, Reed R, Simon M, Jenkins N, Copeland N. Nat Genet. 1992;1:85–91. doi: 10.1038/ng0592-85. [DOI] [PubMed] [Google Scholar]
- 42.Whittington A, Webb G, Baker R, Board P. Genomics. 1996;33:105–111. doi: 10.1006/geno.1996.0165. [DOI] [PubMed] [Google Scholar]
- 43.Bozza M, Kolakowski L, Jr, Jenkins N A, Gilbert D J, Copeland N G, David J R, Gerard C. Genomics. 1995;27:412–419. doi: 10.1006/geno.1995.1071. [DOI] [PubMed] [Google Scholar]
- 44.Dodge G R, Kovalszky I, McBride O W, Yi H F, Chu M L, Saitta B, Stokes D G, Iozzo R V. Genomics. 1991;11:174–178. doi: 10.1016/0888-7543(91)90115-u. [DOI] [PubMed] [Google Scholar]
- 45.Kedra D, Peyrard M, Fransson I, Collins J E, Dunham I, Roe B A, Dumanski J P. Hum Mol Genet. 1996;5:625–631. doi: 10.1093/hmg/5.5.625. [DOI] [PubMed] [Google Scholar]
- 46.Lavia P, Macleod D, Bird A. EMBO J. 1987;6:2773–2779. doi: 10.1002/j.1460-2075.1987.tb02572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dietrich W, Miller J C, Steen R G, Merchant M, Damron D, et al. Nat Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- 48.Dietrich W, Miller J, Steen R, Merchant M, Damron-Boles D, et al. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 49.Copeland N, Gilbert D, Jenkins N, Nadeau J, Eppig J, et al. Science. 1993;262:67–82. doi: 10.1126/science.8211131. [DOI] [PubMed] [Google Scholar]
- 50.Reeves R, Rue E, Citron M, Cabin D. Genomics. 1997;43:202–208. doi: 10.1006/geno.1997.4807. [DOI] [PubMed] [Google Scholar]
- 51.DeBry R W, Seldin M F. Genomics. 1996;33:337–351. doi: 10.1006/geno.1996.0209. [DOI] [PubMed] [Google Scholar]
- 52.Wong L J, O’Brien W E. Genomics. 1995;28:341–343. doi: 10.1006/geno.1995.1152. [DOI] [PubMed] [Google Scholar]
- 53.Takiguchi Y, Kurimasa A, Chen F, Pardington P E, Kyama T, Okinaka R T, Moyzis R, Chen D J. Genomics. 1996;35:129–135. doi: 10.1006/geno.1996.0331. [DOI] [PubMed] [Google Scholar]
- 54.Copeland N G, Jenkins N A, Gilbert D J, Eppig J T, Maltais L J, Miller J C, Dietrich W F, Weaver A, Lincoln S E, Steen R G, et al. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- 55.Mattei M G, Fumoux F, Roeckel N, Fougereau M, Schiff C. Genomics. 1991;9:544–546. doi: 10.1016/0888-7543(91)90422-b. [DOI] [PubMed] [Google Scholar]
- 56.Miller R D, Hogg J, Ozaki J H, Gell D, Jackson S P, Riblet R. Proc Natl Acad Sci USA. 1995;92:10792–10795. doi: 10.1073/pnas.92.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croce C M, Huebner K, Isobe M, Fainstain E, Lifshitz B, Shtivelman E, Canaani E. Proc Natl Acad Sci USA. 1987;84:7174–7178. doi: 10.1073/pnas.84.20.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blair H J, Ho M, Monaco A P, Fisher S, Craig I W, Boyd Y. Genomics. 1995;28:305–310. doi: 10.1006/geno.1995.1146. [DOI] [PubMed] [Google Scholar]
- 59.Mears A J, el-Shanti H, Murray J C, McDermid H E, Patil S R. Am J Hum Genet. 1995;57:667–673. [PMC free article] [PubMed] [Google Scholar]
- 60.Galili N, Baldwin H S, Lund J, Reeves R, Gong W L, Wang Z L, Roe B A, Emanuel B S, Nayak S, Mickanin C, Budarf M L, Buck C A. Genome Res. 1997;7:17–26. doi: 10.1101/gr.7.1.17. [DOI] [PubMed] [Google Scholar]