Abstract
Insects respond to microbial infection by the rapid and transient expression of several genes encoding potent antimicrobial peptides. Herein we demonstrate that this antimicrobial response of Drosophila is not aspecific but can discriminate between various classes of microorganisms. We first observe that the genes encoding antibacterial and antifungal peptides are differentially expressed after injection of distinct microorganisms. More strikingly, Drosophila that are naturally infected by entomopathogenic fungi exhibit an adapted response by producing only peptides with antifungal activities. This response is mediated through the selective activation of the Toll pathway.
Drosophila, as other insects, relies on both cellular and humoral mechanisms to mount a potent antimicrobial host defense. The hallmark of the humoral defense is the injury-induced secretion of a battery of antimicrobial peptides by the fat body, a functional equivalent of the mammalian liver (1, 2). To date, seven distinct peptides (plus isoforms) have been identified in immune-challenged Drosophila, either by biochemical methods or by molecular cloning techniques (refs. 3–10; for review, see ref. 11). One of them, drosomycin, exhibits antifungal properties, whereas three other peptides—cecropin, drosocin, and defensin—are directed against bacteria. Diptericin and attacins, which have been identified in Drosophila through the corresponding cDNAs, are homologous to antibacterial molecules in other dipteran and/or lepidopteran species. It is anticipated that in Drosophila they exert similar activities. Finally, metchnikowin exhibits both antifungal and antibacterial activities.
We are interested in the control of expression of the genes encoding the antimicrobial peptides in Drosophila and have recently shown by a genetic analysis that at least two regulatory pathways are involved in this process (12, 13). In particular, we have found that the dorsoventral regulatory gene cassette (spaetzle/Toll/cactus referred to as Tl pathway) controls the expression of the antifungal drosomycin gene in Drosophila adults. This regulatory cascade shows striking similarities with the cytokine-induced activation cascade of NF-κB during the inflammatory response, indicating that this activation pathway is an ancient cascade involved in host defense both in insect and mammals (13). We have also found that the antibacterial peptide genes are induced either by a distinct pathway involving the immune-deficiency (imd) gene or by combined activation of both imd and Toll pathways (13).
By taking advantage of the unique situation in Drosophila where several genes encoding antimicrobial peptides with distinct activity spectra have been cloned, we have now examined the possibility that the induction of the humoral defense exhibits a certain degree of specificity. This question had not been addressed in vivo in Drosophila so far and it had been assumed that this response is aspecific, i.e., that a microbial infection indifferently induces the transcription of all genes encoding antibacterial and antifungal peptides. Herein we show that the humoral antimicrobial response of Drosophila is not aspecific but discriminates between various classes of microorganisms and, through activation of distinct regulatory pathways, mounts a response that is, at least for fungal pathogens, adapted to the infection.
MATERIALS AND METHODS
Drosophila Stocks.
OregonR (OrR) flies were used as a standard wild-type strain. The transgenic strain Dipt2.2-lacZ:1 is a ry506 line carrying a diptericin reporter gene on the X chromosome (14). The fusion gene contains 2.2 kb of diptericin upstream sequences fused to the bacterial lacZ coding region and was inserted into the Carnegie 20 vector. The developmental and the inducible expression of the Dipt2.2-lacZ transgene is roughly the same as that of the resident diptericin gene at the adult stage (14).
Tl10b is a dominant gain-of-function ventralizing allele of Toll (Tl) (15). The Tl10b mutation also induces a constitutive activation of the drosomycin gene (13). Tl1-RXA is a null allele of Tl; Tlr632 is a strong loss-of-function allele of Tl when reared at the restrictive temperature (15, 16). The inducibility of the drosomycin gene is strongly reduced in Tlr632/Tl1-RXA adults after bacterial challenge, as compared with wild type (13).
Stocks and crosses were maintained on a standard cornmeal medium.
Microorganisms.
Bacteria were precultured in LB medium. The following bacterial strains were used: Aerococcus viridans, Enterobacter cloacae, Streptococcus faecalis, Pseudomonas aeruginosas, Salmonella typhimurium, Staphylococcus aureus (gifts from H. Monteil, University of Strasbourg); Bacillus subtilis, Bacillus thuringiensis, and Bacillus megaterium (J. Millet and A. Klier, Pasteur Institute of Paris); Serratia marcescens (Db1140 and Db11 strains; H. G. Boman, University of Stockholm); Erwinia carotovora (INRA Angers).
Fungi were grown on malt-agar medium. Spores and hyphae were harvested as in ref.17. The following strains were used: Fusarium oxysporum (MUCL 909), Neurospora crassa (CBS 327–54), and Botrytis cinerea (MUCL 30158) (gifts from W. F. Broekaert, Catholic University of Leuven); Beauveria bassiana (80.2 strain), Paecylomyces fumoroseus (Pfr 319 strain), and Metharizium anasiplae (KVL 131 strain) (from A. Vey, INRA St. Christol les Alès); Aspergillus fumigatus (C. Koenig, University of Strasbourg).
Infecting Experiments.
Two ways of challenging insects were used in this study: pricking with sharpened needles dipped into concentrated cultures of microorganisms or injecting microbial suspensions with a micropipette. For the latter treatment, 10 nl was injected into the thorax of Drosophila adults by using a Nanoject apparatus (Drummond Scientific, Broomall, PA). Both pricking and injecting methods induce an injury that by itself triggers the induction of antimicrobial peptide genes, albeit at a low level (see below). Because of the small size of Drosophila, the use of a micropipette is more traumatic than pricking with a sharpened needle and yielded less reproducible results in our hands. We therefore used the pricking method for immune challenges.
Natural Infection by Entomopathogenic Fungi.
Anesthetized flies were shaken for a few minutes in a Petri dish containing a sporulating fungal culture. Flies covered by spores were then removed to fresh Drosophila medium and incubated at 29°C. Survival experiments were carried out under the same conditions for each genotype tested. Groups of 20 adults, aged 2–4 days, were infected by fungi and transferred to a fresh vial every 3 days.
β-Galactosidase Measurements.
We used the method described in ref. 18.
Northern Blot Analysis.
Total RNA extraction and Northern blot experiments were performed as in ref. 13. The following probes were used: attacin cDNA (8), cecropin A1 cDNA (3), defensin cDNA (6), diptericin cDNA (5), drosocin cDNA (10), drosomycin cDNA (7), metchnikowin cDNA (9), and rp49 cDNA, a PCR fragment of approximately 400 bp generated between two oligonucleotides designed after the rp49 coding sequence (19). The cecropin A1 probe cross-reacts with cecropin A2 transcripts (3).
RESULTS AND DISCUSSION
Comparative Induction of Antimicrobial Peptide Genes After Challenge by Various Microorganisms.
In a first set of experiments, Drosophila adults carrying a diptericin-lacZ reporter gene were pricked with a sterile needle dipped into culture pellets of various living microorganisms (distinct bacterial strains, fungal spores, or hyphae). β-Galactosidase activity was measured 6 h after challenge at 25°C. Under these conditions, the number of bacteria injected (around 103–104 bacteria) remained roughly constant (data not shown). As illustrated in Fig. 1A, the level of induction of the diptericin reporter gene in immune-challenged adults varied strikingly with the microorganism tested. Gram-negative bacteria were potent inducers. In contrast, Gram-positives did not induce expression above the level of a simple injury, with the marked exception of the flagellae-carrying Gram-positive bacilli. It is noteworthy that within a given group of bacteria (Gram-negatives, Gram-positives, and bacilli), the various species that we tested induced a similar level of expression of the reporter gene. Immune challenge with spores or hyphae from a mixture of several fungal strains also resulted in a significant level of expression of the reporter gene (Fig. 1A). In this series, the results were similar when the bacterial or fungal cultures were heat-killed prior to infecting the flies (data not shown), indicating that they do not reflect differential microbial growths within the insects after pricking but rather the variable capabilities of the microorganisms to induce diptericin gene expression.
Figure 1.
Induction of the diptericin gene in Drosophila adults infected by various microorganisms. (A) Drosophila adults (3–5 days old) carrying the Dipt-lacZ reporter gene (14) were pricked with a sterile needle dipped into culture pellets of distinct bacterial strains (OD of the pellet = 100), fungal spores (1010 spores per ml), or hyphae from various fungi. β-Galactosidase activity was measured 6 h after challenge at 25°C. Each bar represents the mean of several independent measurements with confidence interval (P < 5%). (B) Northern blot of total RNA extracted from one and six bacteria- or fungi-challenged wild-type (OregonR) male adults. The blot was successively hybridized with diptericin (Dipt) and ribosomal protein rp49 (Rp49) cDNA. Rp49 was used as an internal control for quantification of RNA. Conditions were as in A. C (control), unchallenged flies; Inj, simple injury; En.c., Enterobacter cloacae (−); S.t., Salmonella typhimurium (−); S.m., Serratia marcescens (Db1140 strain); P.a., Pseudomonas aeruginosas (−); Er.c., Erwinia carotovora; Es. c., E. coli (−); A.v., Aerococcus viridans (+); M.l., M. luteus (+); S.f., Streptococcus faecalis (+); S.a., Staphylococcus aureus (+); B.s., Bacillus subtilis (+); B.t., Bacillus thuringiensis (+) B.m., Bacillus megaterium (+); Fh, hyphal bodies; Fs, fungal spores from a mixture of Fusarium oxysporum, Neurospora crassa, and Botrytis cinerea.
We have corroborated these data by Northern blot experiments. For this, total RNA was extracted from control and 1-h and 6-h immune-challenged adults and probed with diptericin cDNA and rp49 cDNA as an internal control. The results are shown in Fig. 1B and confirm the data obtained with the diptericin reporter gene: all Gram-negative bacteria strongly induced the expression of the diptericin gene; Gram-positive bacteria did not induce the gene above the level of a simple injury, with the exception of Bacillus megaterium that induced a moderate level of expression, comparable to that of fungal spores or hyphae.
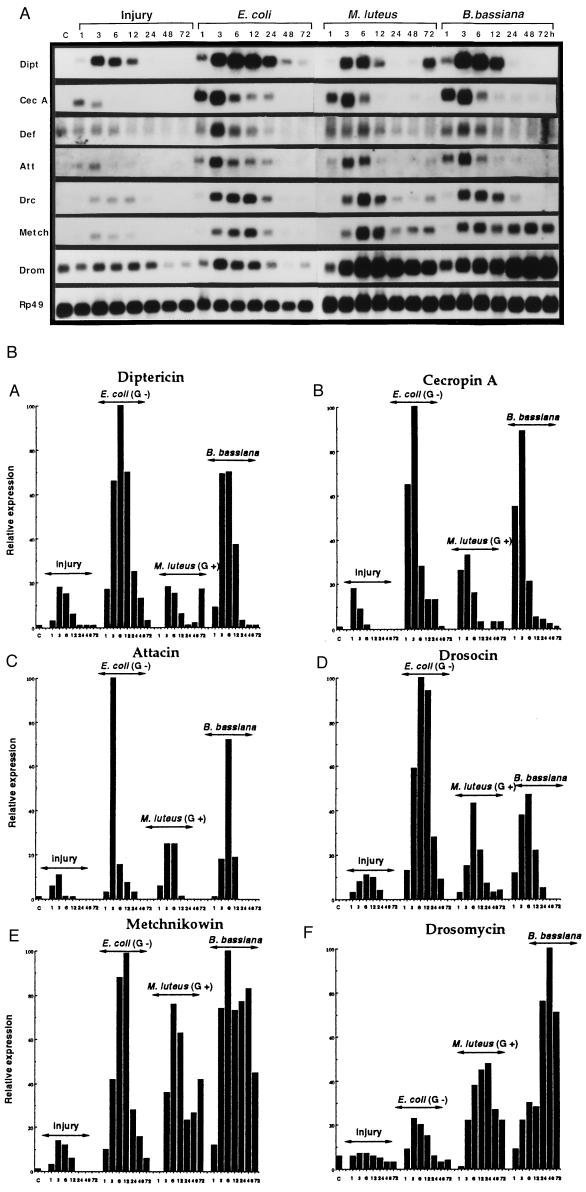
We have extended these experiments to a time-course analysis over a 72-h period. Challenges were restricted to a simple injury and to pricking with either Gram-negative (Escherichia coli) or Gram-positive (Micrococcus luteus) bacteria or with an entomopathogenic fungus (B. bassiana). Flies were kept at 29°C to allow for possible growth of the microorganisms within the infected insects, and RNA was extracted after given time intervals following challenge. Fig. 2 gives the results obtained by probing the Northern blot with a diptericin cDNA. The gene was rapidly induced by all challenges and after peak values at 6–12 h, the signals of the transcripts leveled off. These kinetics, conventionally referred to as acute-phase kinetics (1, 2), were observed for all four types of challenge. Markedly though, the level of induction was strongest with E. coli. It was somewhat lower with fungi and weakest with the Gram-positive M. luteus. These data confirm and extend the results described above.
Figure 2.
Time-course analysis of antimicrobial gene expression after infection by various microorganisms. (A) Northern blot of total RNA extracted from female wild-type (OregonR) adults at different time intervals after challenge (as indicated). Adult flies infected by pricking under the same conditions were kept at 29°C. The blot was successively hybridized with the following cDNA probes: diptericin (Dipt), cecropin A1 (Cec A), attacin (Att), drosocin (Drc), defensin (Def), metchnikowin (Metch), drosomycin (Drom), and rp49 (Rp49). Unchallenged females show a low level of drosomycin gene expression due to the constitutive expression in the sperm storage structures (D. Ferrandon, personal communication). This experiment was repeated several times and yielded similar results. (B) The signals on Northern blots of Fig. 3A were quantified by a Bioimager system. The values were normalized with the corresponding value of rp49. The highest level of expression in a series was normalized as 100, and the results are given in relative activity (percent). Results obtained for defensin were similar to those for diptericin (data not shown).
We have repeatedly dehybridized this Northern blot and successively probed it with cecropin, defensin, attacin, drosocin, metchnikowin, and drosomycin cDNAs. The results, which are presented in Fig. 2, point to two major patterns of induction. A first pattern, which roughly corresponds to that of diptericin, includes the cecropin A, drosocin, defensin, and attacin genes and is marked by a strong inducibility by E. coli and a strong, although somewhat lower, inducibility by fungi. In this pattern, M. luteus is a weak inducer. The second pattern, which is observed for drosomycin, is characterized by a strong inducibility by fungi and (although somewhat weaker) M. luteus; E. coli is a weak inducer. The expression of metchnikowin combines both patterns, as this gene is strongly induced by all microorganisms. We also note for drosomycin and metchnikowin, which encode peptides with antifungal activities, a strong and sustained (2–3 days) expression after infection with the fungus B. bassiana. Although this entomopathogen induces the genes of the first group, their induction levels off mostly between 12 h and 24 h. This discrepancy suggests that the fungus developing in the insect after the initial pricking with spores sustains only the expression of the two genes coding for antifungal peptides (see below).
Altogether, these data indicate that pricking Drosophila adults results in a low but clearly detectable expression of all antimicrobial genes and that these genes are induced above this background level by specific classes of microorganisms. The humoral antimicrobial defense of Drosophila thus appears to respond to a conjunction of the injury and the presence of microorganisms and, most importantly in the present context, the insect appears to be able to discriminate among various classes of microorganisms. These data are evocative of earlier in vitro observations of Samakovlis et al. (20) who had noted that the cecropin gene is strongly inducible in a Drosophila tumorous blood cell line by bacterial lipopolysaccharide and flagellin but only weakly by peptidoglycan. These results pointed to the ability of the blood cells to discriminate between various microbial wall constituents. Interestingly, in most cases, the distinct patterns of induction of the antimicrobial peptide genes appear to correlate with the activity described for the peptides encoded by these genes (for review, see ref. 11). This is illustrated by the observation that Gram-negative bacteria are particularly strong inducers of diptericin, cecropin, attacin, and drosocin, which exhibit preferentially anti-Gram-negative activities, when tested under in vitro conditions. Conversely, drosomycin, which is a potent antifungal peptide, is poorly induced by Gram-negative bacteria but responds strongly to fungal challenge.
Induction of Antimicrobial Genes After Natural Infection by Entomopathogenic Fungi.
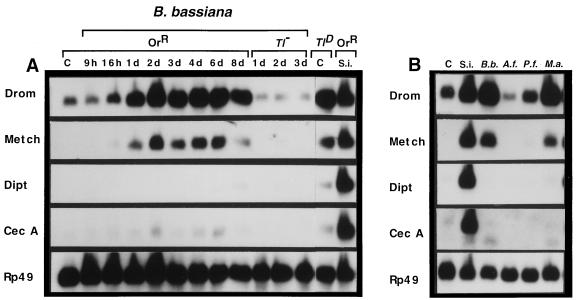
We have further attempted to analyze whether the discrimination among microorganisms is relevant under natural conditions for the insects. To avoid pricking and to mimick a natural infection, we have covered Drosophila adults with spores of B. bassiana, a well-established entomopathogenic fungus. Fungi are important insect pathogens and spores of some entomopathogenic species such as B. bassiana can sporulate on the cuticle and produce chitinases and proteases to penetrate the insect (21, 22). We have monitored the induction of antimicrobial peptides in Drosophila adults covered with B. bassiana spores over a 8-day period by using Northern blot experiments. The two most striking results of these experiments were (i) that this treatment induced a strong and persistent expression of the drosomycin gene and (ii) that the genes encoding antibacterial peptides remained silent (Fig. 3A). The metchnikowin gene followed the expression pattern of the drosomycin gene, albeit with a lower level of intensity of expression. These experiments were repeated independently three times yielding similar results. Our observations indicate that under conditions mimicking a natural infection, the insect host defense has the capacity to identify the fungal aggression and, most importantly, to respond appropriately by producing only peptides with antifungal activities. Note that in the pricking experiments described above, fungal spores induced all antimicrobial peptides, although the effect was strongest for drosomycin and metchnikowin. The discrepancy with the data obtained with a natural infection are most likely linked to the injury induced by pricking.
Figure 3.
Induction of antimicrobial peptide genes in fungi-infected adults. (A) Northern blot analysis of total RNA extracted from wild-type, Tl−, and TlD mutant female adults. Flies were anesthetized and covered with spores of B. bassiana. Flies were placed at 29°C and collected after different time intervals. (B) Northern blot analysis of total RNA extracted from wild-type female adults after natural infection by various fungi. Flies were placed at 29°C and collected 5 days later. A and B were obtained separately. C, control; d, days; Tl−, Tl632/Tl1-RXA female adults; TlD, unchallenged Tl10b female adults; S.i. (septic injury) adults that were challenged by pricking with a needle dipped into a mixture of E. coli and M. luteus and collected 6 h later. B.b., B. bassiana; P.f., Paecylomyces fumoroseus; A.f., A. fumigatus; M.a., Metharizium anasiplae.
We had previously shown that the induction of the drosomycin gene is controlled by the dorsoventral gene cassette spaetzle/Toll (Tl)/tube/pelle/cactus (Tl signaling pathway, ref. 13). In particular, in Tl gain-of-function mutant adults (TlD) in which this pathway is constitutively activated, drosomycin is expressed at a high level. This contrasts with the antibacterial peptide genes whose induction is dependent on an additional regulatory pathway, involving the imd (immune deficiency) gene product (12, 13). The metchnikowin gene is also constitutively expressed in a TlD mutant background, albeit at a lower level than that of drosomycin (Fig. 3A). An attractive hypothesis is that the natural infection by B. bassiana selectively activates the Tl signaling pathway. We have tested this hypothesis by covering Tl-deficient mutants (Tl−) with fungal spores. No induction of the drosomycin and metchnikowin genes was detected in this context (Fig. 3A), indicating that the fungal induction of these antifungal peptide genes is indeed mediated by the selective activation of the Tl signaling pathway. The mechanisms by which the invading fungus activates this pathway is unknown at present. One possibility is that receptors that discriminate between fungal and bacterial outer membrane components are present in the insect (for instance, fungal pattern recognition receptors). Alternatively, the proteases secreted by the fungal pathogen could directly activate the Tl pathway by processing either the putative ligand of Tl (e.g., the spaetzle protein) into its active form or an upstream component of the pathway.
The Tl-mediated antifungal response is essential for resistance in naturally infected insects, as shown by the survival studies. Table 1 illustrates that nearly 90% of infected wild-type adults survived 4 days after being covered with spores of B. bassiana, whereas the Tl-deficient infected insects had succumbed. Eight days after the infection, one out of five wild-type insects still survived the infection of this entomopathogen (Table 1).
Table 1.
Survival after natural infection by entomopathogenic fungi
| Tested genotype | Fungal strain | Flies, n | Survival, %
|
|
|---|---|---|---|---|
| 4 days | 8 days | |||
| + | B.b. | 320 | 87 ± 4 | 18 ± 4 |
| Tl− | B.b. | 253 | 1 ± 1 | 0 |
| + | M.a. | 113 | 76 ± 8 | 5 ± 4 |
| Tl− | M.a. | 54 | 6 ± 6 | 0 |
| + | P.f. | 387 | 92 ± 3 | 82 ± 4 |
| Tl− | P.f. | 206 | 80 ± 6 | 19 ± 6 |
| + | A.f. | 420 | 97 ± 2 | 88 ± 3 |
| Tl− | A.f. | 223 | 79 ± 5 | 40 ± 7 |
Survival rates are given as a percentage. The number of flies tested is indicated. The survival rates were measured 4 and 8 days after the natural infection of wild-type (OregonR) and Tl− (Tl632/Tl1-RXA) adult flies by various fungi. Experiments were performed at 29°C. B.b., B. bassiana; M.a., M. anesiplae; P.f., P. fumoroseus; A.f., A. fumigatus.
We have extended these experiments to three additional fungal strains: Metharizium anisopliae and P. fumoroseus, which are pathogens for some insect species, and A. fumigatus (22). As for B. bassiana, we observed that covering Drosophila adult with spores of M. anisopliae induced the expression of the antifungal peptide genes but the strictly antibacterial peptide genes remained silent (Fig. 3B). In contrast, P. fumoroseus and A. fumigatus did not induce a marked expression of drosomycin. Interestingly, we noted a correlation between the level of expression of the drosomycin gene and the pathogenicity of the various fungi, as determined by the survival rates of infected insects (Table 1). The differential capabilities of the various fungi to elicit antifungal gene expression may reflect their distinct abilities to penetrate through the insect cuticle rather than differences in fungal cell wall components that could interact with insect recognition receptors.
Conclusions.
The present study strongly suggests that the Drosophila host defense can discriminate between various microorganisms. At least for fungal pathogens, we show that this can lead to an adapted response (i.e., production of antifungal peptides) through the selective induction of the Tl pathway. This result gives a biological meaning to our previous observation that distinct regulatory cascades govern the humoral response. The differential levels of induction of the antibacterial peptide genes by Gram-negative and Gram-positive bacteria, although suggestive of some degree of selectivity in the response, are more difficult to interpret at present, probably due to the complex effect of the injury. Unfortunately, we have not yet been able to test natural infections by bacterial strains pathogenic to Drosophila. The Gram-negative species Serratia marcescens, a known pathogen of Drosophila (23), kills the insects within days when introduced into the food, but we have not observed induction of any antimicrobial peptides during such infections; the pathogen probably kills the insects by acting on gut functions. Interestingly however, we note that the genes whose expression levels are most strongly affected by the imd mutation (12, 13) and that code for strictly antibacterial peptides are also those that are most strongly induced by challenge with Gram-negative as compared with Gram-positive bacteria. In contrast, the metchnikowin and drosomycin genes that are strongly induced by Gram-positive bacteria retain most of their inducibility in imd mutants (ref. 13 and B.L., unpublished data).This observation is compatible with the working hypothesis that the imd pathway is preferentially activated by Gram-negative bacteria.
Our data lead us to propose that the humoral antimicrobial response of Drosophila is not aspecific, as hitherto assumed, but discriminates between various classes of microorganisms and, through activation of distinct regulatory pathways, mounts a response that is, at least for fungal pathogens, adapted to the infection. Challenging questions in this field now pertain to the recognition mechanisms that discriminate between various microorganisms and their links to the subsequent activation of the regulatory pathways leading to the expression of the various antimicrobial genes.
Acknowledgments
We are indebted to Dr. Dan Hultmark and Dr. Mitchel Dushay for the gift of attacin and cecropin A1 cDNAs; to Alain Vey for the gift of entomopathogenic fungi; and to Dr. Marie Meister, Dr. Jean Lambert, Dr. Pascale Fehlbaum, Dr. Dominique Ferrandon, Dr. Sarah Ades, and Dr. Alain Vey for stimulating discussions. The technical assistance of Reine Klock, Martine Schneider, and Raymonde Syllas is gratefully acknowledged. This work was supported by Centre National de la Recherche Scientifique, the University Louis Pasteur of Strasbourg, Rhone Poulenc-Agro, and Human Frontier Science Program.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hultmark D. Trends Genet. 1993;9:178–183. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann J A. Curr Opin Immunol. 1995;7:4–10. doi: 10.1016/0952-7915(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 3.Kylsten P, Samakovlis C, Hultmark D. EMBO J. 1990;9:217–224. doi: 10.1002/j.1460-2075.1990.tb08098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tryselius Y, Samakovlis C, Kimbrell D A, Hultmark D. Eur J Biochem. 1992;204:395–399. doi: 10.1111/j.1432-1033.1992.tb16648.x. [DOI] [PubMed] [Google Scholar]
- 5.Wicker C, Reichhart J M, Hoffmann D, Hultmark D, Samakovlis C, Hoffmann J A. J Biol Chem. 1990;265:22493–22498. [PubMed] [Google Scholar]
- 6.Dimarcq J L, Hoffmann D, Meister M, Bulet P, Lanot R, Reichhart J M, Hoffmann J A. Eur J Biochem. 1994;221:201–209. doi: 10.1111/j.1432-1033.1994.tb18730.x. [DOI] [PubMed] [Google Scholar]
- 7.Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert W F, Hetru C, Hoffmann J A. J Biol Chem. 1994;269:33159–33163. [PubMed] [Google Scholar]
- 8.Åsling B, Dushay M S, Hultmark D. Insect Biochem Mol Biol. 1995;25:511–518. doi: 10.1016/0965-1748(94)00091-c. [DOI] [PubMed] [Google Scholar]
- 9.Levashina E A, Ohresser S, Bulet P, Reichhart J M, Hetru C, Hoffmann J A. Eur J Biochem. 1995;233:694–700. doi: 10.1111/j.1432-1033.1995.694_2.x. [DOI] [PubMed] [Google Scholar]
- 10.Bulet P, Dimarcq J L, Hetru C, Lagueux M, Charlet M, Hegy G, Van Dorsselaer A, Hoffmann J A. J Biol Chem. 1993;268:14893–14897. [PubMed] [Google Scholar]
- 11.Hoffmann J A, Reichhart J M. Trends Cell Biol. 1997;7:309–316. doi: 10.1016/S0962-8924(97)01087-8. [DOI] [PubMed] [Google Scholar]
- 12.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemaitre B, Nicolas E, Michaut L, Reichhart J M, Hoffmann J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 14.Reichhart J M, Meister M, Dimarcq J L, Zachary D, Hoffmann D, Ruiz C, Richards G, Hoffmann J A. EMBO J. 1992;11:1469–1477. doi: 10.1002/j.1460-2075.1992.tb05191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider D S, Hudson K L, Lin T Y, Anderson K V. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 16.Gertulla S, Jin Y, Anderson K V. Genetics. 1988;119:123–133. doi: 10.1093/genetics/119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broekaert W F, Terras F R G, Cammue B P A, Vanderleyden J. FEMS Microbiol Lett. 1990;69:55–60. [Google Scholar]
- 18.Lemaitre B, Coen D. Proc Natl Acad Sci USA. 1991;88:4419–4423. doi: 10.1073/pnas.88.10.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connell P, Rosbach M. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samakovlis C, Åsling B, Boman H G, Gateff E, D. Hultmark D. Biochem Biophys Res Commun. 1992;188:1169–1175. doi: 10.1016/0006-291x(92)91354-s. [DOI] [PubMed] [Google Scholar]
- 21.Charnley A K. In: Biotechnology of Fungi for Improving Plant Growth. Whipps J M, Lumsen R D, editors. Cambridge, U.K.: Cambridge Univ. Press; 1989. pp. 86–125. [Google Scholar]
- 22.Vey A, Götz P. In: Hemocytic and Humoral Immunity in Arthropods. Gupta A P, editor. New York: Wiley; 1986. pp. 89–115. [Google Scholar]
- 23.Flyg C, Kenne K, Boman H G. J Gen Microbiol. 1980;120:173–181. doi: 10.1099/00221287-120-1-173. [DOI] [PubMed] [Google Scholar]