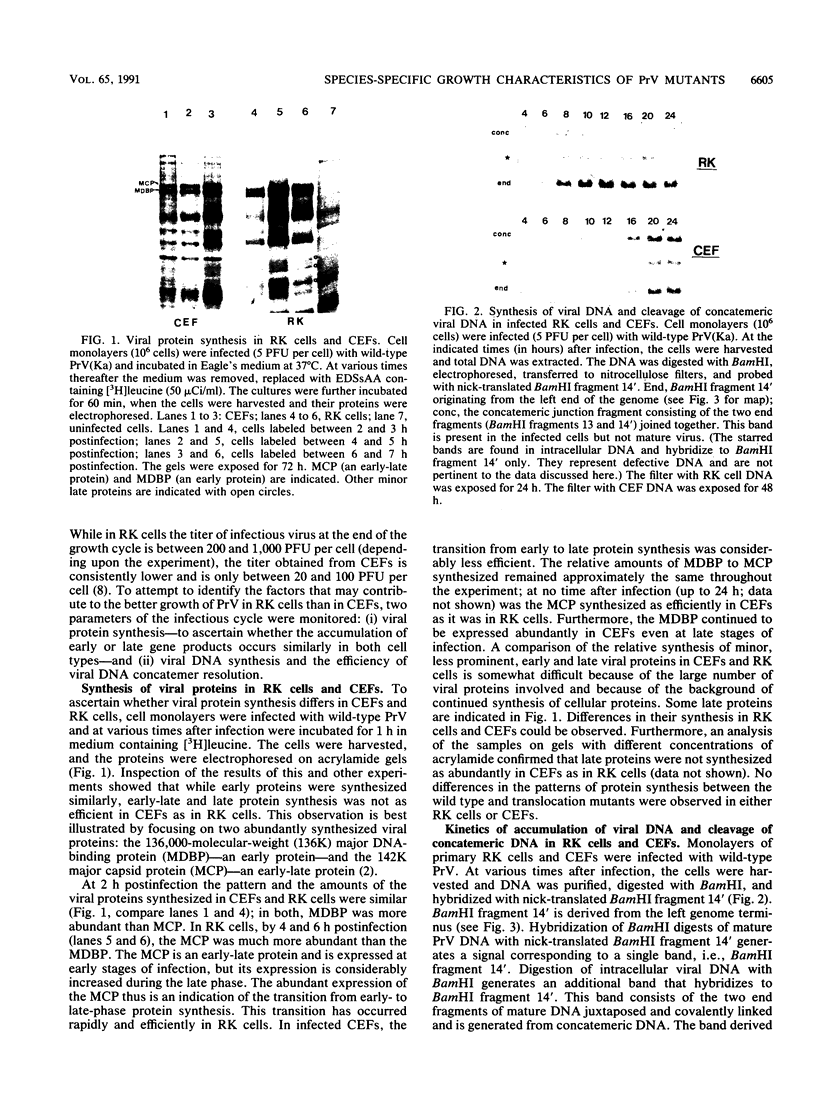
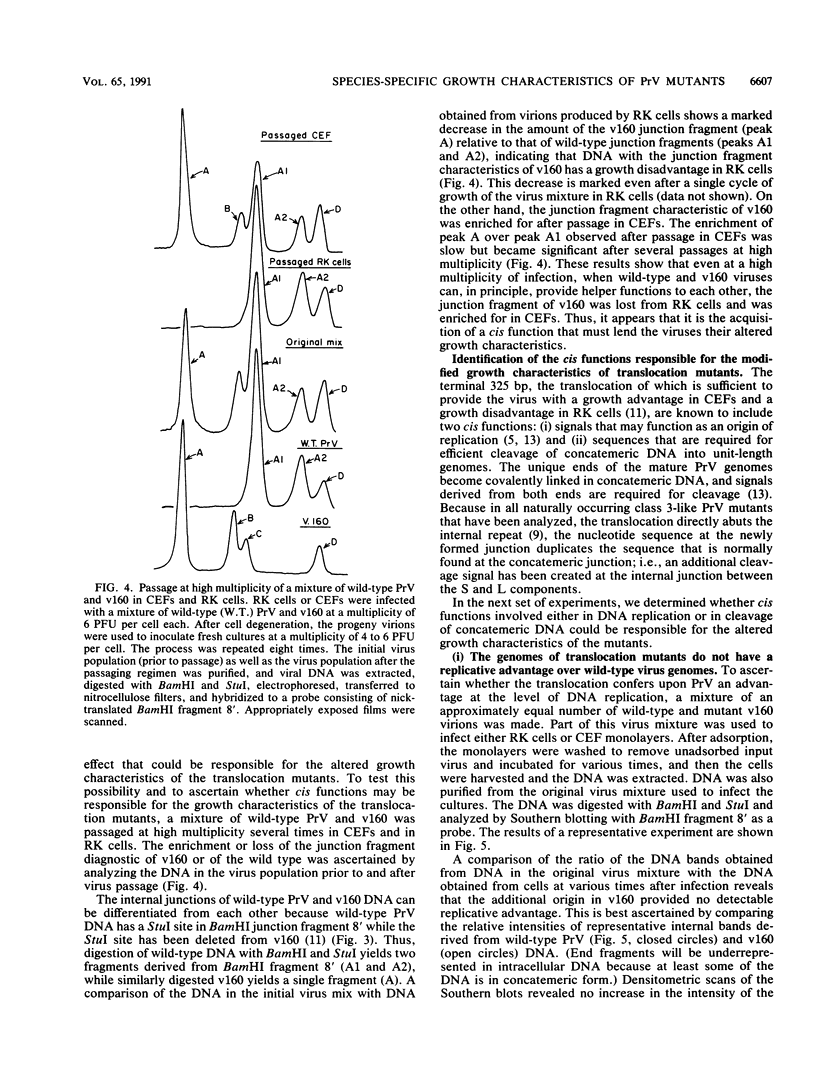
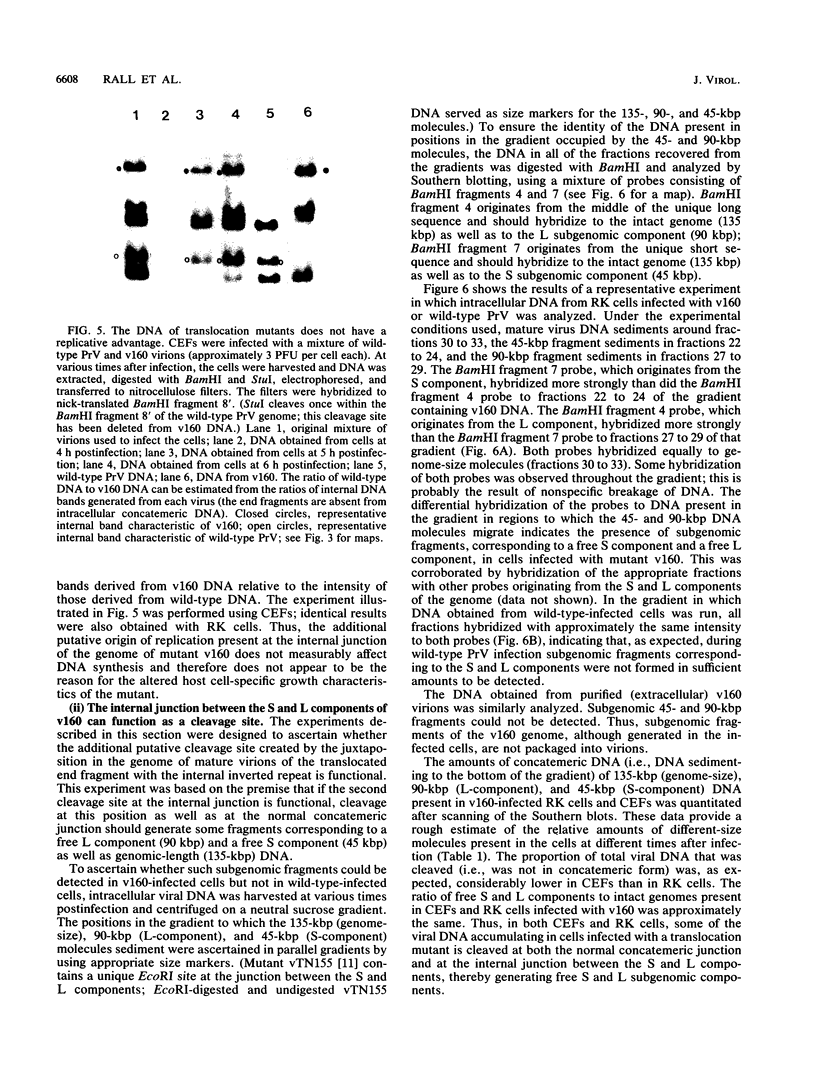
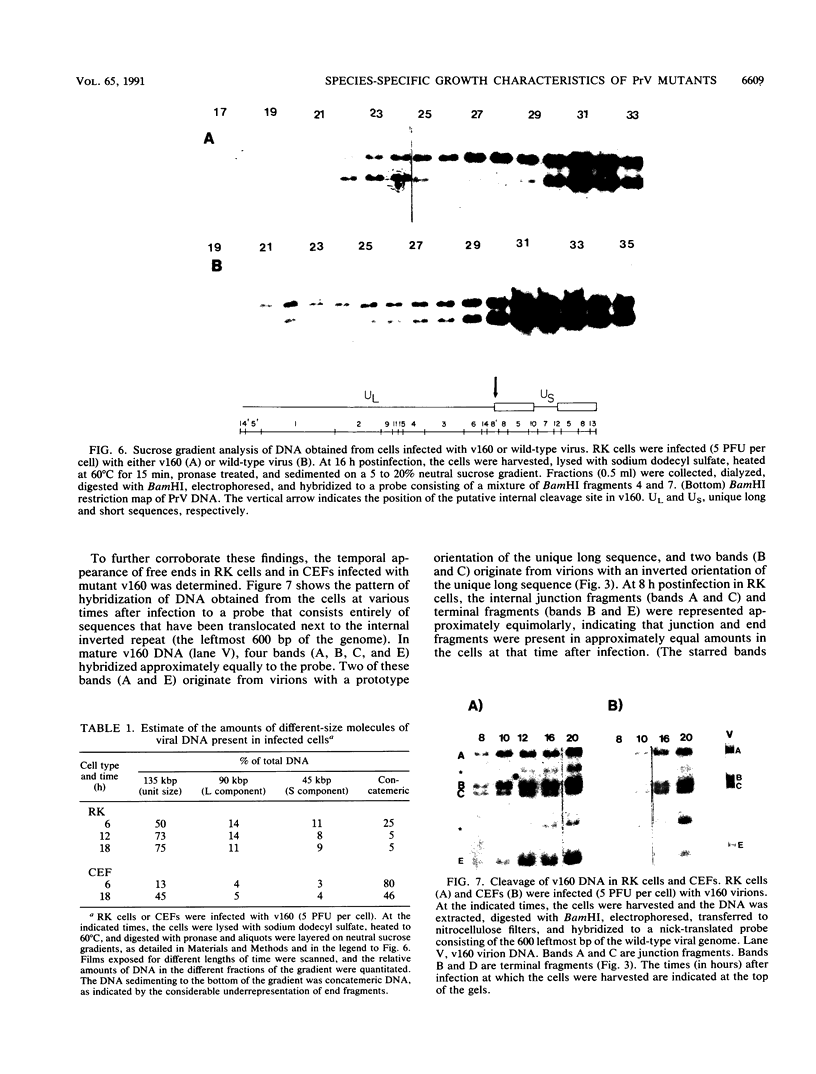
Abstract
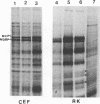
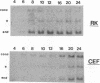
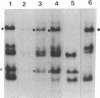
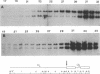
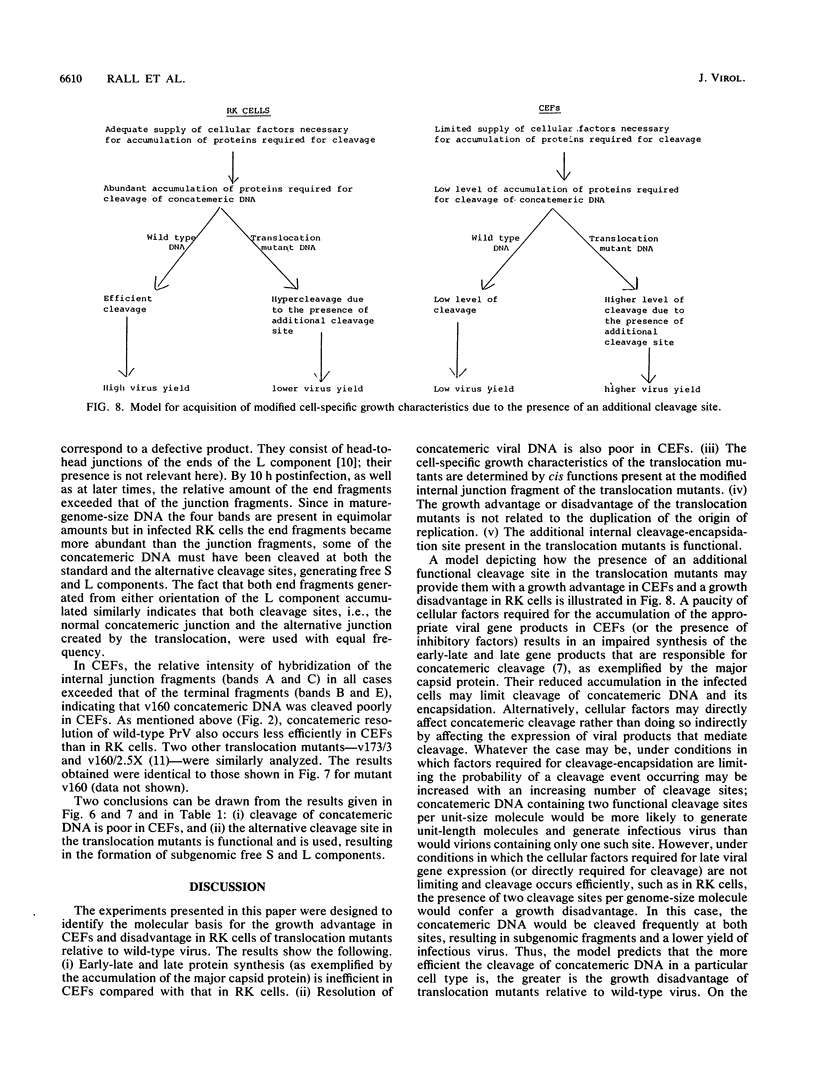
The translocation of the 325 leftmost bp of the genome of pseudorabies virus (PrV) to the internal junction between the L and S components confers upon the virus a growth advantage relative to wild-type PrV in chicken embryo fibroblasts (CEFs) and chickens and a growth disadvantage in rabbit kidney (RK) cells and mice. To clarify the molecular basis for the species-specific growth characteristics of the translocation mutants, we have compared several parameters of the virus growth cycle in CEFs and RK cells infected with wild-type PrV and with translocation mutants. The salient findings are as follows. (i) The synthesis of early-late and late proteins is not as effective in CEFs as it is in RK cells, and these proteins, in particular, the major capsid proteins, accumulate less abundantly in CEFs than in RK cells. (ii) Cleavage of concatemeric DNA to genome-size molecules is also not as effective in CEFs as it is in RK cells. (iii) The internal junction present in translocation mutants is a functional cleavage site. (iv) In RK cells, translocation mutants are hypercleaved and a significant proportion of the total viral DNA is cleaved into subgenomic fragments. (v) In CEFs infected with translocation mutants, subgenomic fragments also accumulate but most of the viral DNA remains in concatemeric form. A model which postulates that the cell-specific growth advantage or disadvantage of the translocation mutants is related to the presence of a second cleavage site within their genomes and is affected by the efficiency of cleavage of concatemeric DNA in particular infected cell types is presented. The significance of these findings as they relate to the evolution of herpesviruses with class 2- and class 3-like genomes is discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., Kaplan A. S., Stehn B., Rubenstein A. S. Concatemeric forms of intracellular herpesvirus DNA. Virology. 1976 Feb;69(2):547–560. doi: 10.1016/0042-6822(76)90484-0. [DOI] [PubMed] [Google Scholar]
- DeMarchi J. M., Lu Z. Q., Rall G., Kupershmidt S., Ben-Porat T. Structural organization of the termini of the L and S components of the genome of pseudorabies virus. J Virol. 1990 Oct;64(10):4968–4977. doi: 10.1128/jvi.64.10.4968-4977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatly A. M., Ben-Porat T. Relation between the levels of mRNA abundance and kinetics of protein synthesis in pseudorabies virus-infected cells. Virology. 1985 Jun;143(2):558–568. doi: 10.1016/0042-6822(85)90394-0. [DOI] [PubMed] [Google Scholar]
- Hampl H., Ben-Porat T., Ehrlicher L., Habermehl K. O., Kaplan A. S. Characterization of the envelope proteins of pseudorabies virus. J Virol. 1984 Nov;52(2):583–590. doi: 10.1128/jvi.52.2.583-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladin B. F., Blankenship M. L., Ben-Porat T. Replication of herpesvirus DNA. V. Maturation of concatemeric DNA of pseudorabies virus to genome length is related to capsid formation. J Virol. 1980 Mar;33(3):1151–1164. doi: 10.1128/jvi.33.3.1151-1164.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomniczi B., Kaplan A. S., Ben-Porat T. Multiple defects in the genome of pseudorabies virus can affect virulence without detectably affecting replication in cell culture. Virology. 1987 Nov;161(1):181–189. doi: 10.1016/0042-6822(87)90184-x. [DOI] [PubMed] [Google Scholar]
- Lu Z. Q., DeMarchi J. M., Harper L., Rall G. F., Ben-Porat T. Nucleotide sequences at recombinational junctions present in pseudorabies virus variants with an invertible L component. J Virol. 1989 Jun;63(6):2690–2698. doi: 10.1128/jvi.63.6.2690-2698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyaizu N., Chirmule N., Ohnishi Y., Kalyanaraman V. S., Pahwa S. Human immunodeficiency virus type 1 envelope glycoproteins gp120 and gp160 induce interleukin-6 production in CD4+ T-cell clones. J Virol. 1991 Nov;65(11):6277–6282. doi: 10.1128/jvi.65.11.6277-6282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly L. M., Rall G., Lomniczi B., Mettenleiter T. C., Kuperschmidt S., Ben-Porat T. The ability of pseudorabies virus to grow in different hosts is affected by the duplication and translocation of sequences from the left end of the genome to the UL-US junction. J Virol. 1991 Nov;65(11):5839–5847. doi: 10.1128/jvi.65.11.5839-5847.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wu C. A., Harper L., Ben-Porat T. cis Functions involved in replication and cleavage-encapsidation of pseudorabies virus. J Virol. 1986 Aug;59(2):318–327. doi: 10.1128/jvi.59.2.318-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]