Abstract
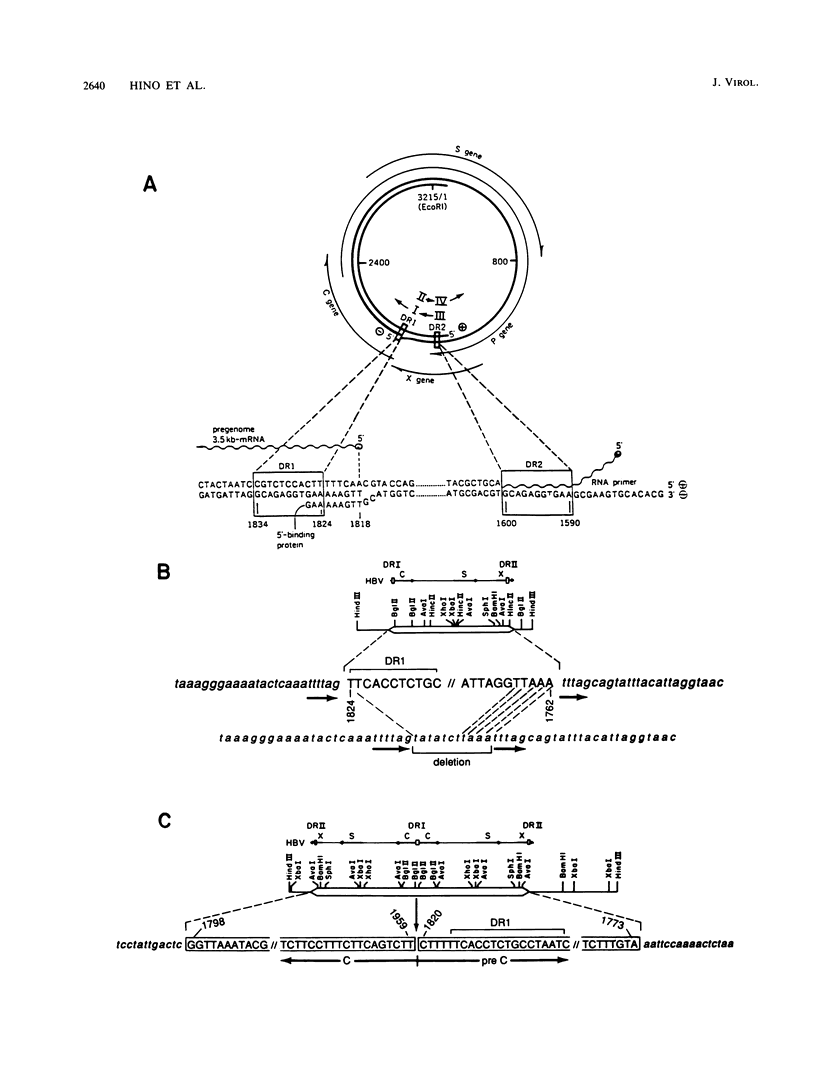
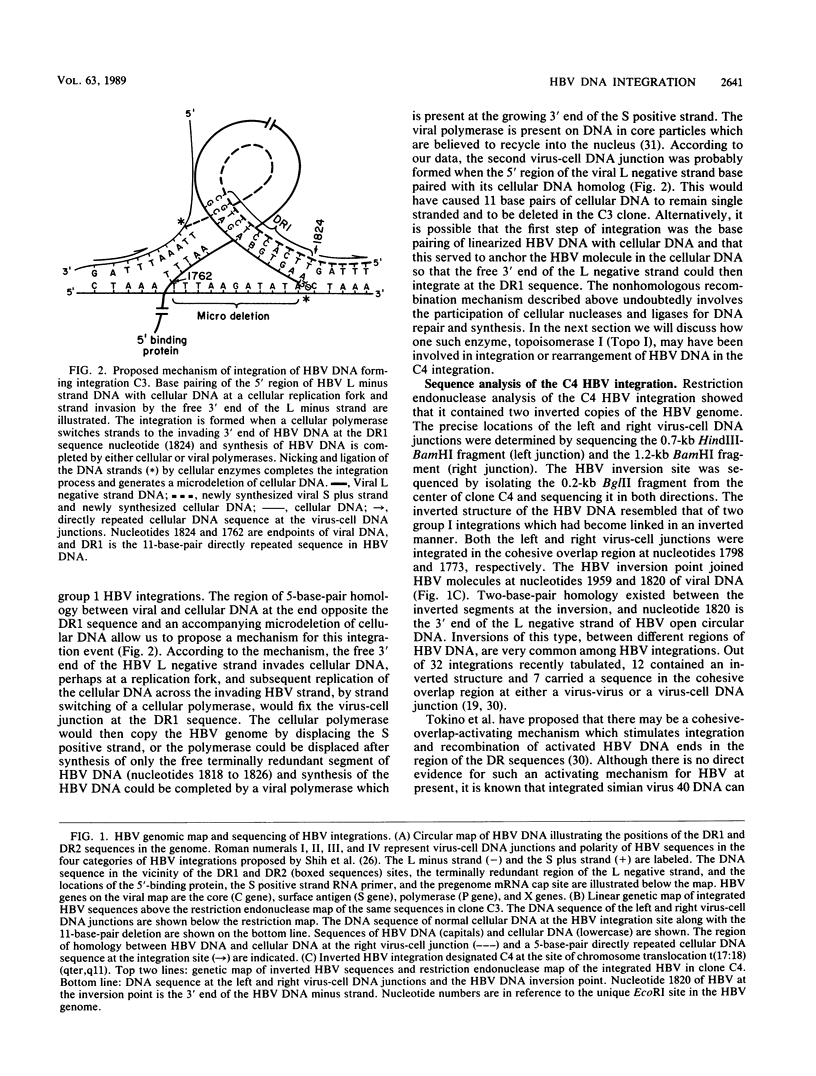
Two integrated hepatitis B virus (HBV) DNA molecules were cloned from two primary hepatocellular carcinomas each containing only a single integration. One integration (C3) contained a single linear segment of HBV DNA, and the other integration (C4) contained a large inverted duplication of viral DNA at the site of a chromosome translocation (O. Hino, T.B. Shows, and C.E. Rogler, Proc. Natl. Acad. Sci. USA 83:8338-8342, 1986). Sequence analysis of the virus-cell junctions of C3 placed the left virus-cell junction at nucleotide 1824, which is at the 5' end of the directly repeated DR1 sequence and is 6 base pairs from the 3' end of the long (L) negative strand. The right virus-cell junction was at nucleotide 1762 in a region of viral DNA (within the cohesive overlap) which shared 5-base-pair homology with cellular DNA. Sequence analysis of the normal cellular DNA across the integration site showed that 11 base pairs of cellular DNA were deleted at the site of integration. On the basis of this analysis, we suggest a mechanism for integration of the viral DNA molecule which involves strand invasion of the 3' end of the L negative strand of an open circular or linear HBV DNA molecule (at the DR1 sequence) and base pairing of the opposite end of the molecule with cellular DNA, accompanied by the deletion of 11 base pairs of cellular DNA during the double recombination event. Sequencing across the inverted duplication of HBV DNA in clone C4 located one side of the inversion at nucleotide 1820, which is 2 base pairs from the 3' end of the L negative strand. Both this sequence and the left virus-cell junction of C3 are within the 9-nucleotide terminally redundant region of the HBV L negative strand DNA. We suggest that the terminal redundancy is a preferred topoisomerase I nicking region because of both its base sequence and forked structure. Such nicking would lead to integration and rearrangement of HBV molecules within the terminal redundancy, as we have observed in both our clones.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beasley R. P., Hwang L. Y., Lin C. C., Chien C. S. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981 Nov 21;2(8256):1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Brand N., Petkovich M., Krust A., Chambon P., de Thé H., Marchio A., Tiollais P., Dejean A. Identification of a second human retinoic acid receptor. Nature. 1988 Apr 28;332(6167):850–853. doi: 10.1038/332850a0. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Dejean A., Brechot C., Tiollais P., Wain-Hobson S. Characterization of integrated hepatitis B viral DNA cloned from a human hepatoma and the hepatoma-derived cell line PLC/PRF/5. Proc Natl Acad Sci U S A. 1983 May;80(9):2505–2509. doi: 10.1073/pnas.80.9.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean A., Sonigo P., Wain-Hobson S., Tiollais P. Specific hepatitis B virus integration in hepatocellular carcinoma DNA through a viral 11-base-pair direct repeat. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5350–5354. doi: 10.1073/pnas.81.17.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D., Varmus H. E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Halligan B. D., Davis J. L., Edwards K. A., Liu L. F. Intra- and intermolecular strand transfer by HeLa DNA topoisomerase I. J Biol Chem. 1982 Apr 10;257(7):3995–4000. [PubMed] [Google Scholar]
- Hino O., Shows T. B., Rogler C. E. Hepatitis B virus integration site in hepatocellular carcinoma at chromosome 17;18 translocation. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8338–8342. doi: 10.1073/pnas.83.21.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan A., Faust E. A. Nonhomologous recombination in the parvovirus chromosome: role for a CTATTTCT motif. Mol Cell Biol. 1986 Aug;6(8):3005–3009. doi: 10.1128/mcb.6.8.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T., Möröy T., Etiemble J., Louise A., Trépo C., Tiollais P., Buendia M. A. Activation of c-myc by woodchuck hepatitis virus insertion in hepatocellular carcinoma. Cell. 1988 Nov 18;55(4):627–635. doi: 10.1016/0092-8674(88)90221-8. [DOI] [PubMed] [Google Scholar]
- Koufos A., Hansen M. F., Copeland N. G., Jenkins N. A., Lampkin B. C., Cavenee W. K. Loss of heterozygosity in three embryonal tumours suggests a common pathogenetic mechanism. Nature. 1985 Jul 25;316(6026):330–334. doi: 10.1038/316330a0. [DOI] [PubMed] [Google Scholar]
- Lien J. M., Aldrich C. E., Mason W. S. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J Virol. 1986 Jan;57(1):229–236. doi: 10.1128/jvi.57.1.229-236.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion P. L., Van Davelaar M. J., Knight S. S., Salazar F. H., Garcia G., Popper H., Robinson W. S. Hepatocellular carcinoma in ground squirrels persistently infected with ground squirrel hepatitis virus. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4543–4546. doi: 10.1073/pnas.83.12.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mizusawa H., Taira M., Yaginuma K., Kobayashi M., Yoshida E., Koike K. Inversely repeating integrated hepatitis B virus DNA and cellular flanking sequences in the human hepatoma-derived cell line huSP. Proc Natl Acad Sci U S A. 1985 Jan;82(1):208–212. doi: 10.1073/pnas.82.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaya T., Nakamura T., Tokino T., Tsurimoto T., Imai M., Mayumi T., Kamino K., Yamamura K., Matsubara K. The mode of hepatitis B virus DNA integration in chromosomes of human hepatocellular carcinoma. Genes Dev. 1987 Oct;1(8):773–782. doi: 10.1101/gad.1.8.773. [DOI] [PubMed] [Google Scholar]
- Ogston C. W., Jonak G. J., Rogler C. E., Astrin S. M., Summers J. Cloning and structural analysis of integrated woodchuck hepatitis virus sequences from hepatocellular carcinomas of woodchucks. Cell. 1982 Jun;29(2):385–394. doi: 10.1016/0092-8674(82)90155-6. [DOI] [PubMed] [Google Scholar]
- Raimondo G., Burk R. D., Lieberman H. M., Muschel J., Hadziyannis S. J., Will H., Kew M. C., Dusheiko G. M., Shafritz D. A. Interrupted replication of hepatitis B virus in liver tissue of HBsAg carriers with hepatocellular carcinoma. Virology. 1988 Sep;166(1):103–112. doi: 10.1016/0042-6822(88)90151-1. [DOI] [PubMed] [Google Scholar]
- Rogler C. E., Sherman M., Su C. Y., Shafritz D. A., Summers J., Shows T. B., Henderson A., Kew M. Deletion in chromosome 11p associated with a hepatitis B integration site in hepatocellular carcinoma. Science. 1985 Oct 18;230(4723):319–322. doi: 10.1126/science.2996131. [DOI] [PubMed] [Google Scholar]
- Rogler C. E., Summers J. Cloning and structural analysis of integrated woodchuck hepatitis virus sequences from a chronically infected liver. J Virol. 1984 Jun;50(3):832–837. doi: 10.1128/jvi.50.3.832-837.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogler C. E., Summers J. Novel forms of woodchuck hepatitis virus DNA isolated from chronically infected woodchuck liver nuclei. J Virol. 1982 Dec;44(3):852–863. doi: 10.1128/jvi.44.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafritz D. A., Shouval D., Sherman H. I., Hadziyannis S. J., Kew M. C. Integration of hepatitis B virus DNA into the genome of liver cells in chronic liver disease and hepatocellular carcinoma. Studies in percutaneous liver biopsies and post-mortem tissue specimens. N Engl J Med. 1981 Oct 29;305(18):1067–1073. doi: 10.1056/NEJM198110293051807. [DOI] [PubMed] [Google Scholar]
- Shih C., Burke K., Chou M. J., Zeldis J. B., Yang C. S., Lee C. S., Isselbacher K. J., Wands J. R., Goodman H. M. Tight clustering of human hepatitis B virus integration sites in hepatomas near a triple-stranded region. J Virol. 1987 Nov;61(11):3491–3498. doi: 10.1128/jvi.61.11.3491-3498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J., Smolec J. M., Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmuness W. Hepatocellular carcinoma and the hepatitis B virus: evidence for a causal association. Prog Med Virol. 1978;24:40–69. [PubMed] [Google Scholar]
- Tokino T., Fukushige S., Nakamura T., Nagaya T., Murotsu T., Shiga K., Aoki N., Matsubara K. Chromosomal translocation and inverted duplication associated with integrated hepatitis B virus in hepatocellular carcinomas. J Virol. 1987 Dec;61(12):3848–3854. doi: 10.1128/jvi.61.12.3848-3854.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttleman J. S., Pourcel C., Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986 Nov 7;47(3):451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- Unoura M., Kobayashi K., Fukuoka K., Matsushita F., Morimoto H., Oshima T., Kaneko S., Hattori N., Murakami S., Yoshikawa H. Establishment of a cell line from a woodchuck hepatocellular carcinoma. Hepatology. 1985 Nov-Dec;5(6):1106–1111. doi: 10.1002/hep.1840050608. [DOI] [PubMed] [Google Scholar]
- Wang H. P., Rogler C. E. Deletions in human chromosome arms 11p and 13q in primary hepatocellular carcinomas. Cytogenet Cell Genet. 1988;48(2):72–78. doi: 10.1159/000132593. [DOI] [PubMed] [Google Scholar]
- Yaginuma K., Kobayashi H., Kobayashi M., Morishima T., Matsuyama K., Koike K. Multiple integration site of hepatitis B virus DNA in hepatocellular carcinoma and chronic active hepatitis tissues from children. J Virol. 1987 Jun;61(6):1808–1813. doi: 10.1128/jvi.61.6.1808-1813.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka O., Omata M., Zhou Y. Z., Imazeki F., Okuda K. Duck hepatitis B virus DNA in liver and serum of Chinese ducks: integration of viral DNA in a hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5180–5184. doi: 10.1073/pnas.82.15.5180. [DOI] [PMC free article] [PubMed] [Google Scholar]


