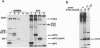Abstract
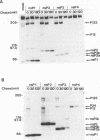
A heterologous system expressing functional Sindbis virus nonstructural proteins (nsPs) has several possible uses for studying Sindbis virus-specific RNA replication and transcription in vivo and in vitro. Of the many possible approaches, vaccinia virus offers an attractive transient expression system given that Sindbis virus replication can occur in cells which have been previously infected by vaccinia virus. In this report, a vaccinia virus recombinant (called vSINNS), which contains the cDNA encoding the Sindbis virus nsPs under the control of either the vaccinia virus 7.5K promoter or the bacteriophage T7 promoter, has been constructed and characterized. Upon infection of several cell types with vSINNS, Sindbis virus nsP precursors and processed forms, including nsP1, nsP2, and both phosphorylated and nonphosphorylated forms of nsP3, were synthesized. Proteins containing the putative RNA-dependent RNA polymerase domain (nsP4 and nsP34), which are normally produced in small amounts by readthrough of an opal termination codon, were not detected in vSINNS-infected cells. However, all nsP functions necessary for Sindbis virus-specific RNA synthesis must have been expressed, since both replication and subgenomic mRNA transcription of an engineered Sindbis virus defective interfering RNA in cells infected with vSINNS was observed. Furthermore, vSINNS could be used as a helper virus to amplify, to relatively high titers, a replication-defective Sindbis virus mutant containing an in-frame deletion in the conserved N-terminal domain of nsP3. These data, as well as the observation that normal yields of parental Sindbis virus are produced in cells which have been previously infected with vSINNS, indicate that expression of Sindbis virus nsPs, in the absence of Sindbis virus-specific RNA replication, is not sufficient to block the formation of active RNA replication complexes by superinfecting Sindbis virus.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. H., Brown D. T. BHK cells expressing Sindbis virus-induced homologous interference allow the translation of nonstructural genes of superinfecting virus. J Virol. 1985 May;54(2):351–357. doi: 10.1128/jvi.54.2.351-357.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D. J., Sawicki S. G., Sawicki D. L. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J Virol. 1988 Oct;62(10):3597–3602. doi: 10.1128/jvi.62.10.3597-3602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge B. W., Pfefferkorn E. R. Complementation between temperature-sensitive mutants of Sindbis virus. Virology. 1966 Oct;30(2):214–223. doi: 10.1016/0042-6822(66)90097-3. [DOI] [PubMed] [Google Scholar]
- COOPER P. D. RESCUE OF ONE PHENOTYPE IN MIXED INFECTIONS WITH HEAT-DEFECTIVE MUTANTS OF TYPE 1 POLIOVIRUS. Virology. 1965 Mar;25:431–438. doi: 10.1016/0042-6822(65)90064-4. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Brechling K., Moss B. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985 Dec;5(12):3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran M. A., Puckett C., Moss B. In vitro mutagenesis of the promoter region for a vaccinia virus gene: evidence for tandem early and late regulatory signals. J Virol. 1985 Apr;54(1):30–37. doi: 10.1128/jvi.54.1.30-37.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M. X., Schlesinger M. J. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology. 1989 Jul;171(1):280–284. doi: 10.1016/0042-6822(89)90539-4. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Felgner P. L., Gadek T. R., Holm M., Roman R., Chan H. W., Wenz M., Northrop J. P., Ringold G. M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Blinov V. M., Donchenko A. P., Koonin E. V. An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J Mol Evol. 1989 Mar;28(3):256–268. doi: 10.1007/BF02102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., Donchenko A. P., Blinov V. M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989 Jun 26;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Levis R., Raju R., Huang H. V., Rice C. M. A cis-acting mutation in the Sindbis virus junction region which affects subgenomic RNA synthesis. J Virol. 1989 Dec;63(12):5216–5227. doi: 10.1128/jvi.63.12.5216-5227.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Grakoui A., Rice C. M., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol. 1989 Mar;63(3):1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J Virol. 1989 Jul;63(7):3142–3150. doi: 10.1128/jvi.63.7.3142-3150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Hahn Y. S., de Groot R. J., Strauss E. G., Strauss J. H. Synthesis and processing of the nonstructural polyproteins of several temperature-sensitive mutants of Sindbis virus. Virology. 1990 Jul;177(1):199–208. doi: 10.1016/0042-6822(90)90473-5. [DOI] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J Virol. 1988 Mar;62(3):998–1007. doi: 10.1128/jvi.62.3.998-1007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989 Nov;63(11):4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruby D. E., Guarino L. A., Kates J. R. Vaccinia virus replication. I. Requirement for the host-cell nucleus. J Virol. 1979 Feb;29(2):705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. S., Palese P., Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990 Nov;64(11):5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. E., Smith J. F. Selection for accelerated penetration in cell culture coselects for attenuated mutants of Venezuelan equine encephalitis virus. Virology. 1988 Feb;162(2):437–443. doi: 10.1016/0042-6822(88)90484-9. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keränen S., Käriäinen L. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J Virol. 1979 Oct;32(1):19–29. doi: 10.1128/jvi.32.1.19-29.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. J., Hong Z., Strauss J. H. Mutagenesis of the 3' nontranslated region of Sindbis virus RNA. J Virol. 1990 Apr;64(4):1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Lemm J. A., Durbin R. K., Stollar V., Rice C. M. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol. 1990 Jun;64(6):3001–3011. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Schlesinger S., Huang H. V. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J Virol. 1990 Apr;64(4):1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Weiss B. G., Tsiang M., Huang H., Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986 Jan 17;44(1):137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- Li G. P., La Starza M. W., Hardy W. R., Strauss J. H., Rice C. M. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology. 1990 Nov;179(1):416–427. doi: 10.1016/0042-6822(90)90310-n. [DOI] [PubMed] [Google Scholar]
- Li G. P., Rice C. M. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J Virol. 1989 Mar;63(3):1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald M. R., Takeda J., Rice C. M., Krause J. E. Multiple tachykinins are produced and secreted upon post-translational processing of the three substance P precursor proteins, alpha-, beta-, and gamma-preprotachykinin. Expression of the preprotachykinins in AtT-20 cells infected with vaccinia virus recombinants. J Biol Chem. 1989 Sep 15;264(26):15578–15592. [PubMed] [Google Scholar]
- Mackett M., Smith G. L. Vaccinia virus expression vectors. J Gen Virol. 1986 Oct;67(Pt 10):2067–2082. doi: 10.1099/0022-1317-67-10-2067. [DOI] [PubMed] [Google Scholar]
- Mi S., Durbin R., Huang H. V., Rice C. M., Stollar V. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology. 1989 Jun;170(2):385–391. doi: 10.1016/0042-6822(89)90429-7. [DOI] [PubMed] [Google Scholar]
- Niesters H. G., Strauss J. H. Defined mutations in the 5' nontranslated sequence of Sindbis virus RNA. J Virol. 1990 Sep;64(9):4162–4168. doi: 10.1128/jvi.64.9.4162-4168.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattnaik A. K., Wertz G. W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990 Jun;64(6):2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim J. S., Cho H. Y., Huebner R. J. Non-producer human cells induced by murine sarcoma virus. Int J Cancer. 1975 Jan 15;15(1):23–29. doi: 10.1002/ijc.2910150104. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Franke C. A., Strauss J. H., Hruby D. E. Expression of Sindbis virus structural proteins via recombinant vaccinia virus: synthesis, processing, and incorporation into mature Sindbis virions. J Virol. 1985 Oct;56(1):227–239. doi: 10.1128/jvi.56.1.227-239.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D. L., Sawicki S. G. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology. 1985 Jul 15;144(1):20–34. doi: 10.1016/0042-6822(85)90301-0. [DOI] [PubMed] [Google Scholar]
- Sawicki D. L., Sawicki S. G., Keränen S., Käriäinen L. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J Virol. 1981 Aug;39(2):348–358. doi: 10.1128/jvi.39.2.348-358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D., Barkhimer D. B., Sawicki S. G., Rice C. M., Schlesinger S. Temperature sensitive shut-off of alphavirus minus strand RNA synthesis maps to a nonstructural protein, nsP4. Virology. 1990 Jan;174(1):43–52. doi: 10.1016/0042-6822(90)90052-s. [DOI] [PubMed] [Google Scholar]
- Scheidel L. M., Stollar V. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology. 1991 Apr;181(2):490–499. doi: 10.1016/0042-6822(91)90881-b. [DOI] [PubMed] [Google Scholar]
- Schmaljohn A. L., Kokubun K. M., Cole G. A. Protective monoclonal antibodies define maturational and pH-dependent antigenic changes in Sindbis virus E1 glycoprotein. Virology. 1983 Oct 15;130(1):144–154. doi: 10.1016/0042-6822(83)90124-1. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Levinson R., Rice C. M., Dalrymple J., Strauss J. H. Nonstructural proteins nsP3 and nsP4 of Ross River and O'Nyong-nyong viruses: sequence and comparison with those of other alphaviruses. Virology. 1988 May;164(1):265–274. doi: 10.1016/0042-6822(88)90644-7. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VALENTINE R. C., ENGELHARDT D. L., ZINDER N. D. HOST-DEPENDENT MUTANTS OF THE BACTERIOPHAGE F2. II. RESCUE AND COMPLEMENTATION OF MUTANTS. Virology. 1964 Jun;23:159–163. doi: 10.1016/0042-6822(64)90278-8. [DOI] [PubMed] [Google Scholar]
- Xiong C., Levis R., Shen P., Schlesinger S., Rice C. M., Huang H. V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989 Mar 3;243(4895):1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Hardy W. R., Shirako Y., Strauss J. H. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990 Aug;9(8):2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]