Abstract
We explored the genetic basis for intraspecific variation in mycoplasmal sialidase activity that correlates with virulence, and its potentially advantageous linkage to nutrient catabolism. Polymorphism in N-acetylneuraminate scavenging and degradation genes (sialidase, N-acetylneuraminate lyase, N-acetylmannosamine kinase, N-acetylmannosamine-6-phosphate epimerase, N-acetylglucosamine-6-phosphate deacetylase, and glucosamine-6-phosphate deaminase) was evident among eight strains of the avian pathogen Mycoplasma synoviae. Most differences were single nucleotide polymorphisms, ranging from 0.34 ± 0.04 substitutions per 100 bp for N-acetylmannosamine kinase to 0.65 ± 0.03 for the single-copy sialidase gene nanI. Missense mutations were twice as common as silent mutations in nanI; 26% resulted in amino acids dissimilar to consensus; and there was a 12-base deletion near the nanI promoter in strain WVU1853T, supporting a complex genetic basis for differences in sialidase activity. Two strains had identical frameshifts in the N-acetylneuraminate lyase gene nanA, resulting in nonsense mutations, and both had downstream deletions in nanA. Such genetic lesions uncouple extracellular liberation of sialic acid from generation of fructose-6-phosphate and pyruvate via intracellular N-acetylneuraminate degradation. Retention of nanI by such strains, but not others in the M. synoviae phylogenetic cluster, is evidence that sialidase has an important non-nutritional role in the ecology of M. synoviae and certain other mycoplasmas.
Keywords: Mycoplasma synoviae, polymorphism, N-acetylneuraminate catabolism, sialidase, virulence
1. Introduction
Mycoplasma synoviae is a major avian pathogen associated with osteoarthritis, synovitis, and respiratory tract lesions in gallinaceous birds [1, 2, 3]. Infection can produce disease that ranges from subclinical to severe, and clinical outcome can be influenced by co-infection with other agents [4, 5, 6, 7, 8]. The majority of prior studies have focused on cytadherence and/or hemadsorption as pathogenic mechanisms of M. synoviae, with particular attention to antigenically-variable hemagglutinins, although the molecular basis of M. synoviae pathogenicity is still not well-understood [9, 10].
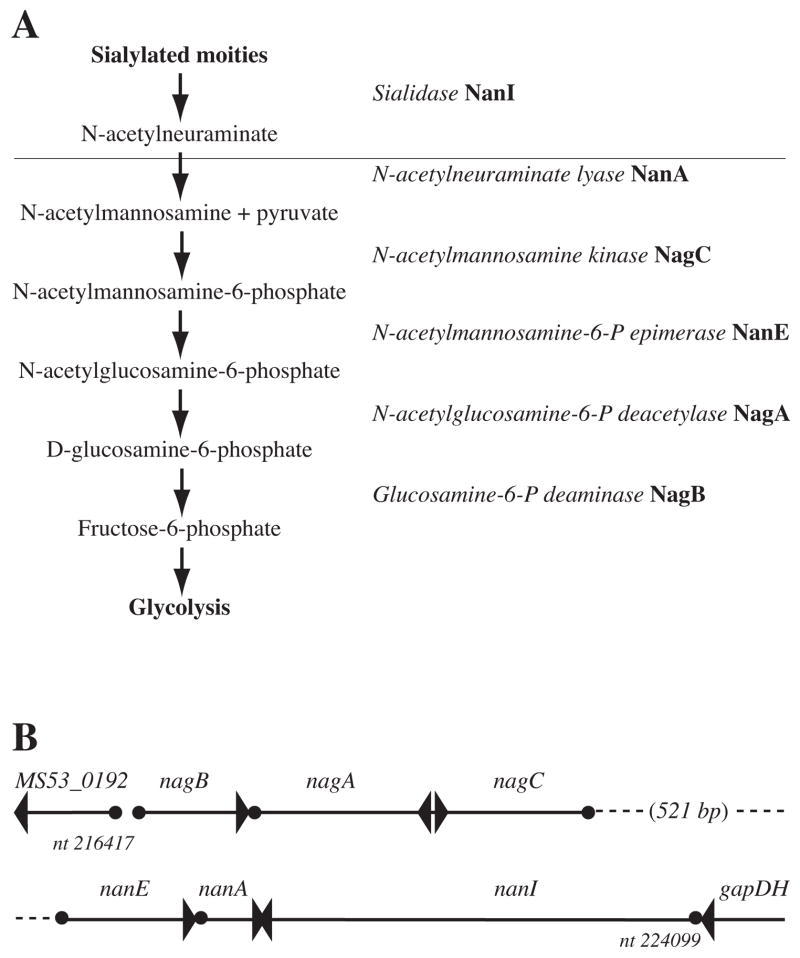
The recently annotated genome of M. synoviae field isolate 53 includes putative sialidase nanI (synonymous with nanH of Gram-negative species and strain 53 GenBank accession no. ABS50356), N-acetylneuraminate lyase nanA, N-acetylmannosamine kinase nagC, N-acetylmannosamine-6-phosphate epimerase nanE, N-acetylglucosamine-6-phosphate deacetylase nagA, and glucosamine-6-phosphate deaminase nagB genes in a locus comprising a canonical sialic acid scavenging and degradation pathway (Figure 1A) [11]. This was unexpected, although sialidase activity is common in other pathogenic bacteria [12], because it is very rare in mycoplasmas, having been described previously only in the lethal pathogen of alligators Mycoplasma alligatoris [13] and an extinct strain of the avian pathogen Mycoplasma gallisepticum [14, 15, 16]. The term sialic acid is the family name covering all derivatives of neuraminic acid [17], the aldol condensation product of D-mannosamine and pyruvic acid, which are potential bacterial nutrients. In vertebrate animals including birds, diverse sialic acid derivatives are involved in recognition processes, cellular connections with extracellular matrix (ECM) components, and intercellular interactions [18]. They protect against hydrolysis of the glycosidic or peptide bonds of oligosaccharides, glycoproteins, glycolipids and gangliosides located on eukaryotic cell surfaces, and against degradation of the ECM. In addition, sialylated lipopolysaccharide and polysialic acid capsules are surface features of certain Gram-negative and Gram-positive bacteria [19].
Figure 1.
(A) The canonical sialic acid degradation pathway [28]. The horizontal line represents the interface between extracellular and intracellular processes. (B) Organization of the 7.9 kb sialic acid degradation locus in the M. synoviae strain 53 genome [11].
Exo-α-sialidases (EC 3.2.1.18) catalyze hydrolysis of α-(2–3)-, α-(2–6)-, and/or α-(2–8)- glycosidic linkages of terminal sialic acid residues on oligosaccharides, glycoproteins, glycolipids, colominic acid (a homopolymer of N-acetylneuraminic acid), and synthetic substrates. Synonyms for sialidase include neuraminidase, α-neuraminidase, and N-acylneuraminate glycohydrolase. Most bacterial sialidases preferentially cleave α-(2–3)-linked sialic acids, and are found in species that live in close contact with vertebrate host cells as commensals or facultative pathogens. Sialidase activity is involved in bacterial colonization and dissemination, ECM degradation, and induced host-cell death [12, 20, 21, 22, 23]. It has also been proposed that bacterial desialylation of host glycoconjugates could expose or form new host antigens to play a role in autoimmune complications of infection [24, 25].
Most recently, in support of the prediction based on the genomic sequence of strain 53 that a functional sialidase gene occurs in M. synoviae, sialidase activity was readily detected in several additional strains [26, 27]. This suggested that an ability to desialylate sialoconjugates present in its environment is important in the ecology of M. synoviae. Strikingly, strains originally isolated from clinically symptomatic birds had significantly more sialidase activity than strains from asymptomatic birds [27], and certain strains lacked detectable sialidase activity [26], suggesting substantial intraspecies genetic heterogeneity and a role for sialidase activity in the virulence of the organism. In bacterial genomes, including that of M. synoviae strain 53, sialidase genes are often part of a locus encoding additional enzymes that enable the import and intracellular catabolism of free sialic acid. This pathway culminates with the production of fructose-6-phosphate for entry to glycolysis [28], constituting a bacterial nutrient stream that might be essential to offset any selective disadvantage of increased virulence attributable to desialylation of host glycoconjugates [20]. In the present study, we explored the genetic basis for the observed intraspecific variation in sialidase activity, and its potentially advantageous linkage to nutrient catabolism, by characterizing the natural DNA sequence diversity within the sialic acid degradation locus among eight strains of M. synoviae.
2. Results
2.1. Amplification of the sialic acid degradation locus
An 8.4 kb PCR amplicon, including the 7.9 kb sialic acid degradation locus as predicted from the genomic sequence of strain 53 [11], was amplified from M. synoviae strains K3344, K4907A, K5395B, MS173, MS178, and WVU1853T [27] using the flanking primers described in section 4.2. Passages 33 and 126 of the FMT strain [3] only generated approximately 8 kb amplicons, suggesting the deletion of a portion(s) of the locus. Homologs of nanA, nanE, nanI, nagA, nagB, and nagC were present in each strain in the same orientations as in strain 53 (Figure 1B). The 521 bp gap between nagC and nanE, annotated for the strain 53 genome as encoding the 151 aa hypothetical protein of unknown function MS53_0196 (GenBank accession no. AAZ43615), was also conserved. When Southern blots of their fragmented genomic DNA were probed to determine nanI copy number, the strains having the highest (WVU1853T) and lowest (K4907A and K5395B) amounts of sialidase activity all exhibited the banding pattern predicted from the whole-genome sequence of strain 53, showing that only a single copy of nanI was present in those strains despite their having almost 100-fold differences in sialidase activity per colony-forming unit (CFU) [27].
2.2. Nucleotide sequence variability
Numerous point mutations with respect to the consensus sequence were observed throughout the locus in each of the eight strains. The mean (± standard error) number of substitutions per 100 bp across the six genes constituting the locus ranged from 0.34 ± 0.09 for strain MS173 to 0.65 ± 0.08 for strain K4907A (Table 1). The mean number of substitutions per 100 bp within each gene ranged from 0.34 ± 0.04 for the 861 bp N-acetylmannosamine kinase nagC to 0.65 ± 0.03 for the 2,817 bp sialidase nanI (MS53_0199, GenBank accession no. YP_278329). In contrast, the number of substitutions per 100 bp within the two 16S rRNA genes of M. synoviae was only 0.04. For comparison to another species of mycoplasma, the number of substitutions per 100 bp in the 1,002 bp signal recognition particle receptor subunit Y gene ftsY ranged from 0.14 in Mycoplasma hyopneumoniae strain J (GenBank accession no. AE017243) to 0.42 in strain 232 (AE017332), and in the 1,392 bp transcriptional dual regulator dnaA gene ranged from 0.2 in M. hyopneumoniae strains J and 232 to 0.3 in strain 7448 (AE017244).
Table 1.
Genetic heterogeneity in the Mycoplasma synoviae sialic acid degradation locusa.
| Strain
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| K4907A | K5395B | WVU1853T | K3344 | FMT | 53 | MS178 | MS173 | Mean ± S.E. | |
| Geneb
|
|||||||||
| nanI | 0.78 | 0.61 | 0.57 | 0.61 | 0.61 | 0.82 | 0.61 | 0.61 | 0.65 ± 0.03 |
| nanA | 0.92 | 0.61 | 0.58 | 0.38 | 0.49 | 0.58 | 0.46 | 0.46 | 0.56 ± 0.06 |
| nagA | 0.64 | 0.55 | 0.55 | 0.73 | 0.55 | 0.55 | 0.46 | 0.46 | 0.56 ± 0.03 |
| nanE | 0.7 | 0.99 | 0.7 | 0.56 | 0.56 | 0.56 | 0 | 0 | 0.51 ± 0.12 |
| nagB | 0.42 | 0.42 | 0.56 | 0.56 | 0.71 | 0.28 | 0.14 | 0.14 | 0.40 ± 0.07 |
| nagC | 0.46 | 0.23 | 0.34 | 0.46 | 0.34 | 0.11 | 0.46 | 0.34 | 0.34 ± 0.04 |
| Mean ± S.E. | 0.65 ± 0.08 | 0.57 ± 0.10 | 0.55 ± 0.05 | 0.55 ± 0.05 | 0.54 ± 0.05 | 0.48 ± 0.10 | 0.36 ± 0.10 | 0.34 ± 0.09 | |
| 16S rDNAc | N/Dd | N/Dd | 0.04 | 0.04 | N/Dd | 0.04 | 0.04 | 0.04 | 0.04 |
Nucleotide substitutions per 100 bases of consensus sequence.
nanI = sialidase MS53_0199 (synonymous with nanH of GenBank accession no. ABS50356); nanA = N-acetylneuraminate lyase; nagA = N-acetylglucosamine-6-phosphate deacetylase; nanE = N-acetylmannosamine-6-phosphate epimerase MS53_0197 (GenBank accession no. AAZ43616); nagB = glucosamine-6-phosphate deaminase; nagC = N-acetylmannosamine kinase.
Both copies.
N/D = not determined.
2.3. Guanine+cytosine content
The guanine+cytosine content (%G+C) was calculated for the sialic acid degradation genes from each strain, individually and for the consensus sequence of each gene (Table 2). Genes of this locus tended to have a slightly higher %G+C than the 27% calculated for the M. synoviae strain 53 genome as a whole [11], ranging from 27.2 ± 0.11% for N-acetylglucosamine-6-phosphate deacetylase nagA to 33.1 ± 0.23% for N-acetylmannosamine-6-phosphate epimerase nanE (MS53_0197, GenBank accession no. AAZ43616).
Table 2.
Guanine+cytosine content of genes in the Mycoplasma synoviae sialic acid degradation locus.
| Strain
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| K4907A | K5395B | WVU1853T | K3344 | FMT | 53 | MS178 | MS173 | Mean ± S.E. | Consensus | |
| Genea
|
||||||||||
| nanI | 29.3 | 29.2 | 29.1 | 29.0 | 29.0 | 29.1 | 29.1 | 29.1 | 29.1 ± 0.04 | 29.3 |
| nanA | 32.4 | 32.4 | 32.6 | 31.9 | 31.9 | 33.2 | 32.8 | 32.8 | 32.5 ± 0.05 | 32.4 |
| nagA | 27.2 | 27.2 | 27.0 | 27.3 | 27.3 | 27.1 | 27.3 | 27.3 | 27.2 ± 0.11 | 27.4 |
| nanE | 33.1 | 32.6 | 32.9 | 33.2 | 33.1 | 33.3 | 33.2 | 33.2 | 33.1 ± 0.23 | 33.2 |
| nagB | 30.4 | 30.2 | 30.1 | 30.2 | 30.2 | 30.2 | 30.6 | 30.6 | 30.3 ± 0.20 | 30.2 |
| nagC | 30.7 | 30.9 | 30.8 | 30.9 | 31.0 | 31.0 | 30.9 | 31.0 | 30.9 ± 0.04 | 31.1 |
nanI = sialidase MS53_0199 (synonymous with nanH of GenBank accession ABS50356); nanA = N-acetylneuraminate lyase; nagA = N-acetylglucosamine-6-phosphate deacetylase; nanE = N-acetylmannosamine-6-phosphate epimerase MS53_0197 (GenBank accession no. AAZ43616); nagB = glucosamine-6-phosphate deaminase; nagC = N-acetylmannosamine kinase. The calculated %G+C for the M. synoviae strain 53 genome as a whole was 27% [11].
2.4. Insertions, deletions, frameshift, and nonsense mutations
There were no insertions, deletions, or frameshift mutations, with respect to the consensus sequence, in the protein-coding sequences of the nanE, nanI, nagA, nagB, and nagC genes of the eight strains examined. However, strain WVU1853T had a 12 bp deletion in the flanking glyceraldehyde-3-phosphate dehydrogenase gapDH-nanI intergenic region, beginning at base minus 41 from the predicted nanI start codon (Figure 2A), that substantially altered the predicted nucleic acid stem-loop secondary structure of the putative nanI promoter region. The 891 bp nanA gene contained a 2 bp insertion, common to strains FMT (passages 33 and 126) and K3344, creating a frameshift with respect to the consensus sequence that resulted in seven premature stop codons and effectively disabled the gene. Also, deletions of 14 and 462 bp were present in strains K3344 and FMT (both passages), respectively, downstream of the nanA nonsense mutations (Figure 2B). The deletion in strain FMT was bracketed by the direct repeats 5′-AAT TTC TTC A-3′, and completely overlapped the deletion in strain K3344 (Figure 2C). The deletions accounted for the notably short PCR amplicon obtained for the locus from both passages of strain FMT. Strain WVU1853T had a nonsense mutation arising from a single nucleotide substitution in nanA, predicted to result in the loss of 17 aa from the carboxyl terminus of the 296 aa NanA of the other strains.
Figure 2.
Deletion mutations in the sialic acid degradation locus. The M. synoviae strains examined are listed (left). Nucleotide numbering is from the M. synoviae strain 53 genome (GenBank accession no. NC_007294) [11]. (A) Nucleotides 224131–224155. A 12 bp deletion occurred in the intergenic sequence immediately upstream of the sialidase gene nanI in strain WVU1853T. (B) Nucleotides 220678–221153. Slashes represent 446 intervening bases. A 462 base deletion occurred in the 891 bp N-acetylneuraminate lyase gene nanA of strain FMT. A direct repeat bracketing the deletion is boxed. (C) Nucleotides 220953–220976. A 14 bp deletion, that was overlapped by the larger deletion in strain FMT, occurred in the nanA gene of strain K3344.
2.5. Missense and silent mutations
Missense mutations with respect to the consensus sequences were approximately twice as common as silent mutations in the nanI and nagA genes (Figure 3). The number of missense and silent mutations was approximately equal in nanA and nagC, and nagB and nanE genes had slightly more silent mutations. For perspective, silent mutations were approximately 2.5 and 15 fold more common than missense mutations in the M. hyopneumoniae genes dnaA and ftsY, respectively.
Figure 3.
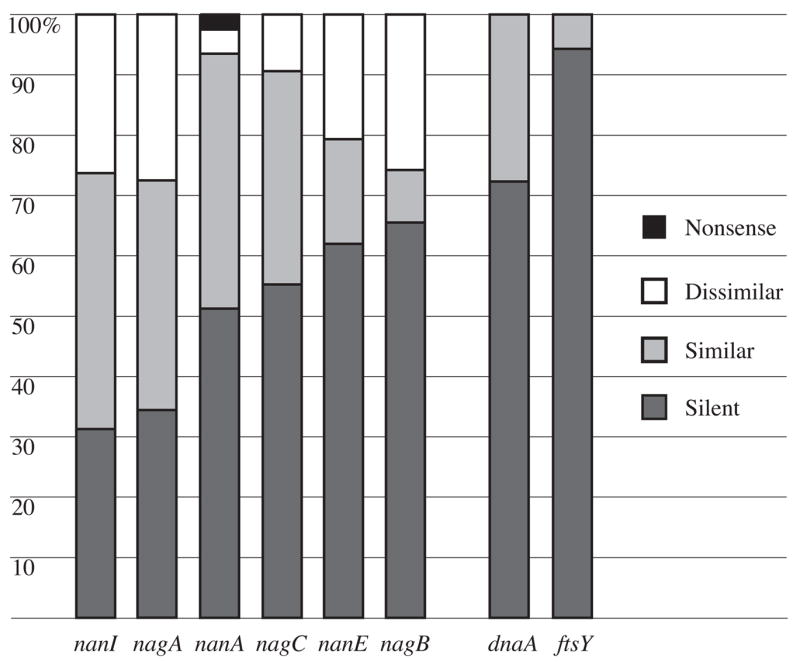
Composition of substitution mutations with respect to consensus sequences. Percentages represent the mean across eight M. synoviae strains for nanI, nagA, nanA, nagC, nanE, and nagB genes of the sialidase locus, and three M. hyopneumoniae strains for housekeeping genes dnaA and ftsY.
Missense mutations resulting in dissimilar amino acids were common throughout the sialic acid degradation locus in all eight M. synoviae strains. They represented 26%, 28%, and 26% of all mutations in NanI, NagA, and NagB, respectively (Figure 3). Approximately half of the dissimilar amino acid substitutions in NanI were in the functional domain defined by the Conserved Domain Database [29]. The signature Arg-Ile-Pro and two Ser-X-Asp-X-Gly-X-Thr-Trp “Asp box” motifs [30], plus Asp box variants Thr-X-Asp-X-Gly-X-Thr-Trp and Ser-X-Asp-X-Gly-X-Asn-Trp, were conserved in NanI. Candidate equivalents of the highly-conserved Arg37, Asp54, Arg56, Asp62, Asp100, Glu230, Arg245, and Tyr347 residues (numbering of the Clostridium perfringens NanI; GenBank accession no. P10481) were readily identified by inspection of local and global sequence alignments, and were conserved across strains, but no equivalent of the Arg312 that is strictly conserved in many other bacterial and eukaryotic sialidases [31] could be recognized in any strain. All of the dissimilar amino acid substitutions of NanA, NanE, NagA, NagB, and NagC were in their broadly-defined functional domains [29]. Dissimilar amino acid changes were least common in NanA, representing just 4% of all mutations in nanA across strains. Missense mutations resulting in dissimilar amino acids were not present in either dnaA or ftsY genes in the three strains of M. hyopneumoniae examined.
3. Discussion and conclusions
The recent discovery by Vasconselos et al. [11] of putative genes for sialidase and the N-acetylneuraminate catabolism pathway in M. synoviae strain 53, when sialidase activity was believed to be extremely rare among mycoplasmas [25, 32], prompted us and others to confirm the annotation by using assays for the enzyme to examine additional strains [26, 27]. The activity was present in most, but not all, M. synoviae strains examined. Unexpectedly, although strain WVU1853T had by far the most activity, there was essentially continuous variation in the amount of sialidase activity per CFU among other strains, which also correlated positively with the degree of strain virulence. In this study, we sought to explain the remarkable intraspecific variation in activity of this candidate virulence factor by measuring nanI copy number in high- and low-activity strains, and by sequencing the sialidase gene nanI and genes constituting the sialic acid degradation locus in multiple strains.
Although the presence and relative spacing of signature motifs and active site residues are conserved in bacterial sialidase catalytic domains, their primary amino acid sequences and lengths are otherwise highly variable [30]. The NanI sialidase of M. synoviae has a theoretical and observed [33] molecular weight of approximately 109 kDa. It consists of an approximately 420 aa N-terminal domain that includes a predicted 29 aa transmembrane region near the N-terminus, and an approximately 520 aa C-terminal six-sheet beta-propellor catalytic domain, Pfam 00064 [34]. NanI has been reported to be extracellular surface-localized in M. synoviae and M. gallisepticum [15, 26], consistent with most other bacterial sialidases that are secreted to effect their actions on the surrounding environment [20]. Negligible activity was found in M. synoviae-conditioned cell-free broth [26, 27]. Since extracellular surface-localization precludes an influence of substrate import on the enzyme’s activity in situ, the quantitative differences in sialidase activity of the magnitude observed for M. synoviae are most simply explained by interstrain variations in the topology of the enzyme. The evidence for a complex genetic basis for the intraspecific variation includes: 1) numerous single nucleotide polymorphisms with respect to their consensus nanI DNA sequence; resulting in 2) a comparatively high frequency of dissimilar missense mutations with respect to the consensus NanI amino acid sequence; 3) presence of only a single copy of nanI regardless of sialidase activity per CFU; and 4) strict conservation of all but one of the several residues believed to constitute the active site [26, 28, 30, 31]. For perspective, individual site-directed mutagenesis of each of the active site residues other than Arg312 reduced C. perfringens sialidase specific activity by 100 to 10, 000 fold [31]. It was noteworthy that the sialidase activity of M. synoviae strain FMT remained quantitatively unchanged after 93 in vitro passages [27]. Since the 12-base deletion in the nanI promoter region in strain WVU1853T was a singular finding among several M. synoviae strains examined [26, 27], and no distinct TATA or ribosome binding sequences were evident in the gapDH-nanI intergenic region adjacent to the putative nanI start codon, a causal relationship between the deletion and that strain’s comparatively high sialidase activity per CFU remains plausible but speculative. It was also remarkable that virulent strain WVU1853T had the least sequence variation in nanI but the highest activity per CFU, whereas avirulent strain K4907A had the most sequence variation in nanI and nearly the least activity per CFU of the strains with quantitated sialidase activity [27].
From a taxonomic standpoint, the capacity to produce sialidase occurs irregularly among bacteria, and sialidases are sometimes produced even by only a single strain within a species [35, 36]. Those findings are most readily explained by horizontal transfer of sialidase genes [30]. The hypothetical protein MGA_0329 of M. gallisepticum strain Rlow (GenBank accession no. NP_853343) shares 94.5% aa identity and 96.5% aa similarity to M. synoviae NanI (hypothetical protein MS53_0199, GenBank accession no. YP_278329). We used the methods described in section 4.1 [27] to confirm that M. gallisepticum strain Rlow does express sialidase activity (our unpublished data). Vasconselos et al. [11] hypothesized a history of nanI homolog transfer between M. synoviae and M. gallisepticum, even though the strain Rlow genome (GenBank accession no. AE015450) lacks all of the genes of the N-acetylneuraminate catabolism pathway [37]. The Arg312 that is conserved in other sialidases including in M. gallisepticum, but missing from M. synoviae, provides evidence of the direction of transfer. The corresponding M. gallisepticum Arg312 codon CGT was substituted in M. synoviae with Gly codons GGT in six of eight strains, or GGC in two strains. Thus a first-position C→G transversion mutation accounts for the loss of the Arg312 residue in M. synoviae and indicates that the direction of transfer was from M. gallisepticum to M. synoviae. In that case, either 1) nanI alone was transferred from M. gallisepticum to M. synoviae or its ancestor, which independently acquired five N-acetylneuraminate catabolism genes in a cluster precisely adjacent to nanI in its chromosome; or, more likely 2) the entire locus was transferred to M. synoviae from a strain of M. gallisepticum or its ancestor whose descendants later lost all of the N-acetylneuraminate catabolism genes. The nanI %G+C, which was similar to the %G+C of the whole genome, did not suggest any other history. Regardless of which history is correct, the implication is that sialidase activity need not remain linked to N-acetylneuraminate catabolism in order to persist in mycoplasmal genomes.
A “neuraminidase-like enzyme” was detected in an unidentified strain of M. gallisepticum [15], and sialidase activity of M. gallisepticum strain TT was characterized in detail [14, 16], but no evidence for sialidase activity was found in M. gallisepticum strains S6 [32] or A5969 [25]. Its genomic context suggests that sialidase activity is important in the ecology of M. gallisepticum strain Rlow independently of its potentially advantageous role in nutrient acquisition [20]. The present study provides further evidence of a separate role for extracellular sialidase also in at least some strains of M. synoviae. Strains FMT and K3344 had identical frameshifts in the 5′-third of nanA, resulting in multiple nonsense mutations, and both strains also had downstream deletions in nanA. Such genetic lesions naturally uncouple extracellular liberation of sialic acid, via sialidase, from generation of N-acetylmannosamine, pyruvate, and eventually fructose-6-phosphate via intracellular N-acetylneuraminate catabolism. We interpret the different deletions as subsequent decay of the gene initially disabled by frameshift in a common ancestor, because it seems less likely that exactly the same 2 bp insertion would occur upstream in the gene independently in two strains if the deletions had occurred first. The deletion in strain FMT completely overlapped the deletion in strain K3344, which implies that K3344 may be an ancestor of FMT, or at least that FMT is less similar to a common ancestor than K3344 is at this locus. In a small sample of other species affiliated with the M. synoviae phylogenetic cluster [38], which we screened for sialidase activity as described in section 4.1 [27], Mycoplasma felis ATCC 23391T, Mycoplasma leonicaptivi ATCC 49890T, and Mycoplasma sturni UC/MFT were negative, but 12 of 13 canine clinical isolates confirmed by PCR-RFLP typing to be the opportunistic mammalian pathogen Mycoplasma canis [39] did express activity (our unpublished data).
An ecological function of sialidase in strains that express activity not linked to N-acetylneuraminate catabolism may be to modulate cytadherence. For example, M. synoviae and M. gallisepticum both utilize sialylated glycoproteins on eukaryotic cell surface membranes as receptors for cytadherence mediated by adhesins such as the vlhA system hemagglutinins [40]. Since receptor desialylation reduces or abolishes cytadherence by M. synoviae, M. gallisepticum, and M. canis [32, 41, 42], it is predictable that a functional balance between the amount of sialidase activity and receptor binding affinity must be essential to promote both colonization and transmission of these mycoplasmas. Strains with comparatively higher sialidase activity would be expected to possess higher-affinity adhesins [43]. The vlhA locus in the M. synoviae strain 53 genome is flanked by homologs of gapDH adjacent to the sialic acid degradation locus. Hypervariability in the VlhA hemagglutinins expressed within and among strains is generated by site-specific recombinations among a large assemblage of vlhA pseudogenes constituting the 69 kb locus [44, 45]. In contrast, there are many potentially independently-transcribed vlhA genes dispersed throughout the M. gallisepticum genome [37, 46], supporting the hypothesis that, like nanI, vlhA also may have been exchanged between these species by horizontal transfer [11, 47]. Results of the present study contribute to a foundation for further work to correlate the probably shifting balances among host immune responses to antigenic adhesins such as VlhA, variations in receptor binding affinity and sialidase activity, and their interplay with cytadherence and pathogenicity in M. synoviae.
4. Materials ands methods
4.1. Mycoplasma synoviae strains and culture conditions
Mycoplasma synoviae strains FMT (passages 33 and 126), K3344, K4907A, K5395B, MS173, MS178, and WVU1853T were cultured in modified Frey’s medium as previously described [27]. The FMT strain, originally isolated from chicken trachea, induced minor respiratory lesions following experimental infection [3]; stocks FMT33 and FMT126 were derived from serial in vitro passages of FMT. Strain K3344 was isolated during an outbreak of apparent reproductive disease in a breeder flock in 1992, but was demonstrated to produce respiratory lesions during experimental infections [48]. Strains MS173 and MS178 were isolated during an outbreak of severe synovitis in Argentina [49]. Lesions from infected birds involved synovial membranes, bursa of Fabricius, liver, kidney, and the lower respiratory tract in breeders [50]. Strain WVU1853T has been most commonly reported to cause airsacculitis and synovitis [2], however, experimental infection studies indicated that this strain is capable of systemic spread and the generation of lesions in multiple tissues [3, 51]. Strains K4907A and K5395B were isolated from clinically normal chickens, and are not suspected to cause significant lesions. A quantitative analysis using using the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid and the sialidase inhibitor 2-deoxy-2,3-didehydro-N-acetylneuraminic acid [27] showed that the units (U) of sialidase activity per CFU varied as much as 65 fold (ANOVA P < 0.0001) among these strains. The highest (Fisher’s Protected Least Significant Difference test P < 0.001) activity observed was for strain WVU1853T (1.3 × 10−7 U/CFU), intermediate amounts were observed for FMT33, FMT126, K3344, and MS178 (1.3–3.9 × 10−8 U/CFU), and low amounts were observed for strains K4907A, K5395B, and MS173 (2.7–6.0 × 10−9 U/CFU). The M. synoviae field isolate 53 was not readily available for phenotypic analysis, and neither clinical data nor sialidase activity have been reported for that strain.
4.2. PCR amplification of the sialic acid degradation locus
Genomic DNA was extracted using Easy DNA reagents (Invitrogen, Carlsbad, California) according to the manufacturer’s instructions. The sialic acid degradation locus (Figure 1B) was amplified from strains FMT33, FMT126, K3344, K4907A, K5395B, MS173, MS178, and WVU1853T using PCR primers designed to anneal in the flanking gapDH (5′-TGT TGA ATC AAA AGA CGG AAG A-3′) and hypothetical gene MS53_0192 (5′-TCA TCG CTT AAT ACT GGG CTT T-3′) open reading frames of strain 53. Amplification reactions were carried out as follows, using the Expand High Fidelity PCR System (Roche Applied Sciences, Indianapolis, Indiana): initial denaturation at 94°C for 2 min, followed by 30 cycles of template denaturation at 94°C for 20 sec, primer annealing at 50°C for 30 sec, and extension at 68°C for 9 min, completed by a final extension at 68°C for 10 min. The expected length of the product was approximately 8.4 kb.
4.3. Nucleotide sequencing and sequence analyses of the sialic acid degradation locus
Nucleotide sequencing for the sialic acid degradation locus of each strain, from the gapDH-nanI intergenic region to the nagB-MS53_0192 junction, was achieved by primer walking using four-dye fluorescent dideoxy labeling methods and the Model 3130 capillary system (Applied Biosystems, Foster City, California). The uncloned amplified DNA described in section 4.2 served as the cycle-sequencing templates. For each strain, 31 reads were required to assemble a contig of reconciled double-stranded sequences using Sequencher version 4.7 software (Gene Codes, Ann Arbor, Michigan). Open reading frames were identified by BLAST alignments with homologs from the M. synoviae strain 53 genome (GenBank accession no. AE017245). The nucleotide sequence of each gene was translated using the ExPASy Translate Tool [52]. The secondary structure of the putative nanI promoter region and structural features of the NanI protein were investigated using tools of the European Molecular Biology Open Software Suite [53, 54]. Nucleotide and amino acid substitutions among strains were mapped using ClustalW alignments [55]. As a benchmark, 1,240 bases of each of the two M. synoviae 16S rRNA genes were sequenced from strains WVU1853T, K3344, MS173, and MS178 using internal primers previously described [56], and compared similarly to the corresponding sequences from strain 53. For another perspective, intraspecies heterogeneity among M. hyopneumoniae strains 232, 7448, and J (GenBank accession nos. AE017332, AE017244, and AE017243) in the sequences of housekeeping genes dnaA and ftsY was also measured.
4.4. Southern blotting to determine nanI copy number
To compare the nanI copy number among strains by Southern blotting, a probe consisting of the 3′ 2.8 kb of nanI was amplified from WVU1853T genomic DNA using PCR primers 5′-TCT CTT CCT TTT TGA GGG CTA-3′ and 5′-GCA AAT CAT CTT AAG AAA AGT CAT T-3′. Amplification conditions, using GoTaq reagents (Promega, Madison, Wisconsin), were as described in section 4.2, with the exception of extension steps of 3 min at 72°C. The amplicons were labeled with digoxygenin (DIG Hi prime, Roche Applied Sciences) according to the manufacturer’s instructions. Genomic DNA from the strains with the highest (WVU1853T) and lowest (K4907A and K5395B) levels of sialidase activity was digested with endonuclease VspI (New England Biolabs, Ipswich, Massachusetts), then separated on a 0.6% agarose gel. The DNA fragments were electrotransferred to nitrocellulose, then cross-linked with shortwave ultraviolet light, using standard methods. Hybridization of the nanI probe and detection of the digoxygenin label were carried out using the DIG EasyHyb system (Roche Applied Sciences) according to the manufacturer’s instructions.
Acknowledgments
The M. synoviae strains FMT (passages 33 and 126), K3344, K4907A, and K5395B were a gift from Dr. Stanley H. Kleven, University of Georgia, Athens, Georgia, USA. Strain K5395B was referred to as K5599A in a preliminary report [27]. Strains MS173 and MS178 were a gift from Dr. Raul Cerda, La Plata University, Buenos Aires, Argentina. The M. gallisepticum strain Rlow was a gift from Dr. Steven J. Geary, University of Connecticut, Storrs, Connecticut, USA. The canine clinical isolates were a gift from Dr. Mary B. Brown, University of Florida, Gainesville, Florida, USA. Cycle sequencing reactions were performed by the Interdisciplinary Center for Biotechnology Research DNA Sequencing Core Laboratory at the University of Florida. This work was supported by Public Health Service grant 1R01GM076584-01A1 from the National Institute of General Medical Sciences (DRB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Meghan May, Email: mmay@ufl.edu.
Daniel R. Brown, Email: BrownD@vetmed.ufl.edu.
References
- 1.Kang MS, Gazdzinski P, Kleven SH. Virulence of recent isolates of Mycoplasma synoviae in turkeys. Avian Dis. 2002;46:102–10. doi: 10.1637/0005-2086(2002)046[0102:VORIOM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 2.Kleven SH, Fletcher OJ, Davis RB. Influence of strain of Mycoplasma synoviae and route of infection on development of synovitis or airsacculitis in broilers. Avian Dis. 1975;19:126–35. [PubMed] [Google Scholar]
- 3.Lockaby SB, Hoerr FJ, Lauerman LH, Kleven SH. Pathogenicity of Mycoplasma synoviae in broiler chickens. Vet Pathol. 1998;35:178–90. doi: 10.1177/030098589803500303. [DOI] [PubMed] [Google Scholar]
- 4.Giambrone JJ, Eidson CS, Kleven SH. Effect of infectious bursal disease on the response of chickens to Mycoplasma synoviae, Newcastle disease virus, and infectious bronchitis virus. Am J Vet Res. 1977;38:251–3. [PubMed] [Google Scholar]
- 5.Kleven SH. Mycoplasmas in the etiology of multifactorial respiratory disease. Poult Sci. 1998;77:1146–9. doi: 10.1093/ps/77.8.1146. [DOI] [PubMed] [Google Scholar]
- 6.Rhoades KR. Turkey sinusitis: synergism between Mycoplasma synoviae and Mycoplasma meleagridis. Avian Dis. 1977;21:670–4. [PubMed] [Google Scholar]
- 7.Springer WT, Luskus C, Pourciau SS. Infectious bronchitis and mixed infections of Mycoplasma synoviae and Escherichia coli in gnotobiotic chickens. I. Synergistic role in the airsacculitis syndrome. Infect Immun. 1974;10:578–89. doi: 10.1128/iai.10.3.578-589.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vardaman TH, Deaton JW, Feece FN. Serological responses of broiler-type chickens, with and without Newcastle disease and infectious Bronchitis vaccine, to experimental infection with Mycoplasma synoviae by foot pad, air sac and aerosol. Poult Sci. 1975;54:737–41. doi: 10.3382/ps.0540737. [DOI] [PubMed] [Google Scholar]
- 9.Noormohammadi AH, Markham PF, Whithear KG, Walker ID, Gurevich VA, Ley DH, et al. Mycoplasma synoviae has two distinct phase-variable major membrane antigens, one of which is a putative hemagglutinin. Infect Immun. 1997;65:2542–2547. doi: 10.1128/iai.65.7.2542-2547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bencina D, Narat M, Dovc P, Drobnic-Valic M, Habe F, Kleven SH. The characterization of Mycoplasma synoviae EF-Tu protein and proteins involved in hemadherence and their N-terminal amino acid sequences. FEMS Microbiol Lett. 1999;173:85–94. doi: 10.1111/j.1574-6968.1999.tb13488.x. [DOI] [PubMed] [Google Scholar]
- 11.Vasconcelos AT, Ferreira HB, Bizarro CV, Bonatto SL, Carvalho MO, Pinto PM, et al. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J Bacteriol. 2005;187:5568–77. doi: 10.1128/JB.187.16.5568-5577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vimr E, Lichtensteiger C. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 2002;10:254–7. doi: 10.1016/s0966-842x(02)02361-2. [DOI] [PubMed] [Google Scholar]
- 13.Brown DR, Zacher LA, Farmerie WG. Spreading factors of Mycoplasma alligatoris, a flesh-eating mycoplasma. J Bacteriol. 2004;186:3922–7. doi: 10.1128/JB.186.12.3922-3927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller HE, Sethi KK. Occurrence of neuraminidase in Mycoplasma gallisepticum. Med Microbiol Immunol. 1972;157:1608. doi: 10.1007/BF02124476. [DOI] [PubMed] [Google Scholar]
- 15.Roberts DH. Neuraminidase-like enzyme present in Mycoplasma gallisepticum. Nature. 1967;213:87–8. [Google Scholar]
- 16.Sethi KK, Muller HE. Neuraminidase activity in Mycoplasma gallisepticum. Infect Immun. 1972;5:260–2. doi: 10.1128/iai.5.2.260-262.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blix FG, Gottshalk A, Klenk E. Proposed nomenclature in the field of neuraminic and sialic acids. Nature. 1957;179:1088. doi: 10.1038/1791088b0. [DOI] [PubMed] [Google Scholar]
- 18.Achyuthan KE, Achyuthan AM. Comparative enzymology, biochemistry and pathophysiology of human exo-α-sialidases (neuraminidases) Comp Biochem Physiol B. 2001;129:29–64. doi: 10.1016/s1096-4959(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 19.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–22. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 20.Corfield T. Bacterial sialidases--roles in pathogenicity and nutrition. Glycobiol. 1992;2:509–21. doi: 10.1093/glycob/2.6.509. [DOI] [PubMed] [Google Scholar]
- 21.Hunt ME, Brown DR. Role of sialidase in Mycoplasma alligatoris-induced pulmonary fibroblast apoptosis. Vet Microbiol. 2007;121:73–82. doi: 10.1016/j.vetmic.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King SJ, Whatmore AM, Dowson CG. NanA, a neuraminidase from Streptococcus pneumoniae, shows high levels of sequence diversity, at least in part through recombination with Streptococcus oralis. J Bacteriol. 2005;187:5376–86. doi: 10.1128/JB.187.15.5376-5386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushita O, Okabe A. Clostridial hydrolytic enzymes degrading extracellular components. Toxicon. 2001;39:1769–80. doi: 10.1016/s0041-0101(01)00163-5. [DOI] [PubMed] [Google Scholar]
- 24.Biberfeld G. Autoimmune reactions associated with Mycoplasma pneumoniae infection. Zentralbl Bakteriol [Orig A] 1979;245:144–9. [PubMed] [Google Scholar]
- 25.Kahane I, Reisch-Saada A, Almagor M, Abeliuck P, Yatziv S. Glycosidase activities of mycoplasmas. Zentralbl Bakteriol [Orig B] 1990;273:300–5. doi: 10.1016/s0934-8840(11)80432-9. [DOI] [PubMed] [Google Scholar]
- 26.Bercic RL, Bencina D, Narat M, Rojs OZ, Slavec B, Dovc P. Neuraminidase activity of pathogenic avian Mycoplasma species. 15th Congr World Vet Poult Assoc; 10–15 September; Beijing, China. 2007. 2007, WVPC2007-03-071 (abstr.) [Google Scholar]
- 27.May M, Kleven SH, Brown DR. Sialidase activity in Mycoplasma synoviae. Avian Dis. 2007;51 doi: 10.1637/7806-120106-Reg. (in press) Published ahead of print on 26 April 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–53. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35(Database issue):D237–40. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roggentin P, Schauer R, Hoyer LL, Vimr ER. The sialidase superfamily and its spread by horizontal gene transfer. Mol Microbiol. 1993;9:915–21. doi: 10.1111/j.1365-2958.1993.tb01221.x. [DOI] [PubMed] [Google Scholar]
- 31.Kleineidam RG, Kruse S, Roggentin P, Schauer R. Elucidation of the role of functional amino acid residues of the small sialidase from Clostridium perfringens by site-directed mutagenesis. Biol Chem. 2001;382:313–9. doi: 10.1515/BC.2001.038. [DOI] [PubMed] [Google Scholar]
- 32.Glasgow LR, Hill RL. Interaction of Mycoplasma gallisepticum with sialyl glycoproteins. Infect Immun. 1980;30:353–61. doi: 10.1128/iai.30.2.353-361.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bercic L, Slavec B, Lavric M, Narat M, Bidovec A, Dovc P, et al. Identification of major immunogenic proteins of Mycoplasma synoviae isolates. Vet Microbiol. 2007 doi: 10.1016/j.vetmic.2007.07.020. (in press) Published ahead of print on 25 July 2007. [DOI] [PubMed] [Google Scholar]
- 34.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popoff MR, Dodin A. Survey of neuraminidase production by Clostridium butyricum, Clostridium beijerinckii, and Clostridium difficile strains from clinical and nonclinical sources. J Clin Microbiol. 1985;22:873–6. doi: 10.1128/jcm.22.5.873-876.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoyer LL, Hamilton AC, Steenbergen SM, Vimr ER. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol Microbiol. 1992;6:873–84. doi: 10.1111/j.1365-2958.1992.tb01538.x. [DOI] [PubMed] [Google Scholar]
- 37.Papazisi L, Frasca S, Jr, Gladd M, Liao X, Yogev D, Geary SJ. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low) Microbiology. 2003;149:2307–16. doi: 10.1099/mic.0.26427-0. [DOI] [PubMed] [Google Scholar]
- 38.Johansson K-E, Pettersson B. Taxonomy of Mollicutes. In: Razin S, Herrmann R, editors. Molecular biology and pathogenicity of mycoplasmas. London: Kluwer Academic Press; 2002. pp. 1–29. [Google Scholar]
- 39.Spergser J, Rosengarten R. Identification and differentiation of canine Mycoplasma isolates by 16S-23S rDNA PCR-RFLP. Vet Microbiol. 2007;125:170–4. doi: 10.1016/j.vetmic.2007.04.045. [DOI] [PubMed] [Google Scholar]
- 40.Allen JL, Noormohammadi AH, Browning GF. The vlhA loci of Mycoplasma synoviae are confined to a restricted region of the genome. Microbiology. 2005;151:935–40. doi: 10.1099/mic.0.27109-0. [DOI] [PubMed] [Google Scholar]
- 41.Gesner B, Thomas L. Sialic acid binding sites: role in hemagglutination by Mycoplasma gallisepticum. Science. 1965;151:590–1. doi: 10.1126/science.151.3710.590. [DOI] [PubMed] [Google Scholar]
- 42.Manchee RJ, Taylor-Robinson D. Utilization of neuraminic acid receptors by mycoplasmas. J Bacteriol. 1969;98:914–9. doi: 10.1128/jb.98.3.914-919.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner R, Matrosovich M, Klenk H-D. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–66. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 44.Noormohammadi AH, Markham PF, Kanci A, Whithear KG, Browning GF. A novel mechanism for control of antigenic variation in the haemagglutinin gene family of Mycoplasma synoviae. Mol Microbiol. 2000;35:911–23. doi: 10.1046/j.1365-2958.2000.01766.x. [DOI] [PubMed] [Google Scholar]
- 45.Hong Y, Garcia M, Leiting V, Bencina D, Dufour-Zavala L, Zavala G, et al. Specific detection and typing of Mycoplasma synoviae strains in poultry with PCR and DNA sequence analysis targeting the hemagglutinin encoding gene vlhA. Avian Dis. 2004;48:606–16. doi: 10.1637/7156-011504R. [DOI] [PubMed] [Google Scholar]
- 46.Glew MD, Browning GF, Markham PF, Walker ID. pMGA phenotypic variation in Mycoplasma gallisepticum occurs in vivo and is mediated by trinucleotide repeat length variation. Infect Immun. 2000;68:6027–33. doi: 10.1128/iai.68.10.6027-6033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markham PF, Duffy MF, Glew MD, Browning GF. A gene family in Mycoplasma imitans closely related to the pMGA family of Mycoplasma gallisepticum. Microbiology. 1999;145:2095–2103. doi: 10.1099/13500872-145-8-2095. [DOI] [PubMed] [Google Scholar]
- 48.Ewing ML, Cookson KC, Phillips RA, Turner KR, Kleven SH. Experimental infection and transmissibility of Mycoplasma synoviae with delayed serologic response in chickens. Avian Dis. 1998;42:230–8. [PubMed] [Google Scholar]
- 49.Cerda RO, Giacoboni GI, Xavier JA, Sansalone PL, Landoni MF. In vitro antibiotic susceptibility of field isolates of Mycoplasma synoviae in Argentina. Avian Dis. 2002;46:215–8. doi: 10.1637/0005-2086(2002)046[0215:IVASOF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 50.Cerda RO, Xavier JA, Petrucelli MA, Everrigaray ME. Aislamento de Mycoplasma Synoviae de pollos parrilleros y gallinas reproducturas primera comunicacion an la Republica Argentina. Analect Vet. 1998;18:41–6. [Google Scholar]
- 51.Hinz KH, Blome C, Ryll M. Virulence of Mycoplasma synoviae strains in experimentally infected broiler chickens. Berl Munch Tierarztl Wochenschr. 2003;116:59–66. [PubMed] [Google Scholar]
- 52.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–8. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 54.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95:1460–5. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown DR, Farley JM, Zacher LA, Carlton JM, Clippinger TL, Tully JG, et al. Mycoplasma alligatoris sp. nov from American alligators. Int J Syst Evol Microbiol. 2001;51:419–24. doi: 10.1099/00207713-51-2-419. [DOI] [PubMed] [Google Scholar]