Summary
Despite the wealth of different actin structures formed, only two actin nucleation factors are well established in vertebrates, the Arp2/3 complex and formins. Here we describe a novel nucleator, Cordon-bleu (Cobl). Cobl is a brain-enriched protein using three Wiskott-Aldrich syndrome protein homology 2 (WH2) domains for actin binding. Cobl promotes non-bundled, unbranched filaments. Filament formation relies on barbed end growth and requires all three Cobl WH2 domains and the extended linker L2. We suggest that the nucleation power of Cobl is based on the assembly of three actin monomers in cross-filament orientation. Cobl localizes to sites of high actin dynamics and modulates cell morphology. In neurons, induction of both neurites and neurite branching is dramatically increased by Cobl expression - effects that critically depend on Cobl's actin nucleation ability. Correspondingly, Cobl depletion results in decreased dendritic arborization. Thus, Cobl is a novel actin nucleator controlling neuronal morphology and development.
Keywords: actin nucleation factor, actin polymerization, actin cytoskeleton, F-actin assembly, neuronal morphology control, neuritogenesis, dendritic branching, Cordon-bleu, development
Introduction
Assembly, organization and turnover of filamentous actin structures are crucial for metazoan life. Proper cell morphology control in developing neurons is a prerequisite for formation of neuronal networks and for cell/cell communication. These cells undergo dramatic changes in morphology and cytoskeletal organization. The nascent, round cells extend several processes, called neurites, the longest of which will finally become the axon. The remaining neurites evolve into dendrites and form the strongly branched postsynaptic compartment. Constantly, neurites explore the environment by sending out filopodia and novel branches from the existing arbor and thereby contribute to the formation of functional neuronal networks (da Silva and Dotti, 2002). For a deeper understanding of these complex rearrangements of neuronal morphology a more complete knowledge of molecular effectors of the cytoskeleton is urgently required.
The rate-limiting step in the actin filament formation is the assembly of actin nuclei onto which further monomers are then added rapidly. Two types of actin nucleators are well established in vertebrates, the Arp2/3 complex and formins. Recently, the fly protein Spire has also been shown to nucleate actin (Quinlan et al., 2005). The nucleators show striking differences in molecular mechanism, regulation and actin superstructures produced. Formins are implicated in the assembly of the unbranched filaments in yeast actin cables, cytokinetic contractile rings, filopodia, and adherens junctions. Formins surround the fast growing barbed end of filaments and remain processively associated with them (reviewed in Higgs, 2005; Kovar, 2006).
The Arp2/3 complex is a stable assembly of two actin-related proteins (Arp2 and 3) and five further proteins (reviewed in Welch and Mullins, 2002). In contrast to formins, it serves as a template for monomer addition by mimicking the barbed end of a filament. Since the Arp2/3 complex also interacts with the sides of existing filaments, it promotes nucleation of actin filaments as 70° branches on the sides of mother filaments. The Arp2/3 complex thereby remains associated with the pointed end leaving the barbed end free to elongate.
The basic activity of the Arp2/3 complex is relatively low until it is activated by a member of the Wiskott-Aldrich Syndrome protein (WASP) family. These different activators are thought to specifically involve the Arp2/3 complex in different cellular processes requiring the formation of branched actin superstructures, such as those found at the leading edge of moving cells (Scar/WAVE proteins) or at motile membrane compartments (N-WASP) (reviewed in Welch and Mullins, 2002; Qualmann and Kessels, 2002; Vartiainen and Machesky, 2004; Stradal and Scita, 2006). On the molecular level, Arp2/3 complex activation is achieved by bringing Arp2 and Arp3 close together, such that they adopt a filament-like orientation, i.e. mimic a dimer of actin in a cross-filament configuration (Robinson et al., 2001). This activation critically relies on the C-termini of WASP proteins containing WASP homology 2 (WH2) domains and an acidic region.
WH2 domains are G-actin binding modules present in a wide range of cytoskeletal proteins (reviewed in Paunola et al., 2002; Paavilainen et al., 2004). Despite the similarities of the WH2 domains in different proteins, they differ significantly in regulating the actin cytoskeleton. β-thymosins for example very effectively sequester G-actin but proteins, such as N-WASP, WASP, WIP and ciboulot rather seem to bind to G-actin in order to feed it into filament formation processes (Paunola et al., 2002; Hertzog et al, 2004).
Here we report on the discovery of a novel, powerful actin nucleator termed cordon-bleu (Cobl). Cobl is a thus far uncharacterized protein that uses three WH2 domains to nucleate actin filaments. Cobl was originally identified in lacZ gene trap experiments for genes expressed in embryonic stem cells (Gasca et al., 1995). In the forming embryo, Cobl mRNA is enriched in the gastrula organizer and in the axial midline (Carroll et al., 2003). Our analyses suggest that Cobl plays a key role in controlling cellular morphogenesis by regulating cytoskeletal dynamics. Cobl-induced actin nucleation strongly influences cellular morphology and motility processes and in neurons, is crucial for neuritogenesis and dendritic branching. In addition to these cell biological studies, our biochemical and biophysical examinations provide detailed insights into the molecular mechanisms by which Cobl interacts with actin, assembles actin nuclei and forms actin filaments.
Results
Identification of Cobl as a potential mediator of actin dynamics
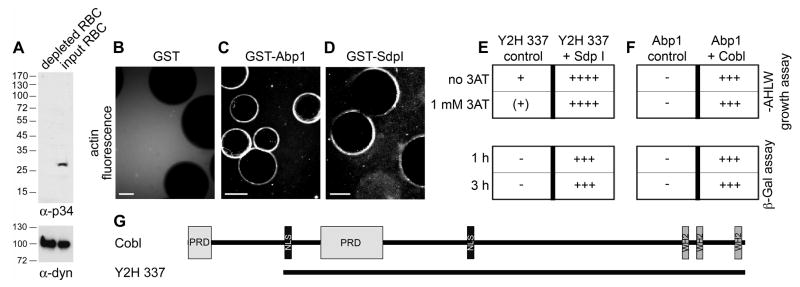
Syndapins and Abp1 function in dynamic actin cytoskeletal processes that involve the Arp2/3 complex actin nucleation machinery and N-WASP (Qualmann and Kelly, 2000; Kessels et al., 2000; Kessels and Qualmann, 2002; Kessels and Qualmann, 2006; Pinyol et al., 2007) but new data indicate that their signaling to the actin cytoskeleton reaches beyond modulation of Arp2/3 complex activity. In rat brain extracts depleted for the Arp2/3 complex (Figure 1A), syndapin as well as Abp1 immobilized on beads were still able to induce F-actin halos around the beads (Figure 1B-D and S1A-F).
Figure 1. Identification of Cobl.
(A) Rat brain cytosol was depleted for the Arp2/3 complex, as demonstrated by anti-p34 immunoblotting and using the anti-dynamin signal as loading control. (B-D) Both immobilized GST-Abp1 (C) and syndapin I (D) but not GST alone (B) were still able to elicit fluorescent F-actin halos in Arp2/3 complex-depleted cytosol endowed with Alexa Fluor™ 568 labeled G-actin. Scale bars, 25 μm.
(E and F) Directed yeast-2-hybrid analyses of the isolated clone 337 (Y2H 337) as a prey with syndapin I as a bait (E) as well as Y2H 337 as a bait and Abp1 as a prey (F), respectively, caused strong activity of independent reporter gene systems.
(G) Domain structure of murine Cobl (gi32251014) and of clone Y2H 337. Domain abbreviations, PRD, proline-rich domain, NLS, putative nuclear localization signal, WH2, WASP-homology 2.
Since our analyses have not revealed a direct influence of syndapins or Abp1 on actin polymerization (our unpublished observations; Kessels et al., 2000), this observation led to the exciting hypothesis that filament formation is promoted through an interaction with yet another actin nucleator present in brain extracts. Toward identification of this factor, we conducted yeast-2-hybrid screens with brain cDNA libraries and syndapin I as a bait. 5.3 million clones were screened. 742 candidates were obtained, subjected to plasmid isolation and retransformation into yeast for verification by directed yeast-2-hybrid analyses (Figure 1E and S1G). Preys verified for syndapin interaction were sequenced and finally crosschecked for interaction with Abp1 (Figure 1F and S1H). Only one candidate fulfilled the required criteria, i.e. contained actin-interaction modules, had no known functional connections to the Arp2/3 complex or formins and interacted strongly with both Abp1 and syndapin. The corresponding cDNA of this ORF was found five times and represented a large portion of the mRNA of Cobl, a protein of unknown function that contains three C-terminal WASP homology 2 (WH2) domains and is present in all vertebrates but absent from lower eukaryotes (Figure 1G).
Individual Cobl WH2 domains bind and sequester actin
The three predicted WH2 domains of Cobl are highly similar to each other (Figure 2A), show high sequence conservation from fish to man (Figure S2) and form a distinct subfamily of WH2 domains (Figure 2B and S3).
Figure 2. The individual Cobl WH2 domains are highly related and sequester G-actin.
(A) Sequence alignment of different WH2 motifs by Clustal W 1.83 with depicted position of the amphiphatic α-helix. Identical residues are shaded in dark grey; those with conservation or similarities in at least two of the Cobl WH2 domains are shaded in light grey. (B) Phylogenetic analysis of WH2 domains by Clustal W (1.83) (alignment see Figure S3).
(C) Affinity purifications of actin from rat brain extracts with equal amounts of GST-Cobl WH2 domains.
(D-G) Actin bead assay with Cobl GST-WH2 domains. Images after 5 minutes (WH2 (1) and WH2 (2) (D, E)) and 30 minutes of incubation (WH2 (3) (F)) show actin on the surface. Beads coated with GST alone did not accumulate any actin (G). Bar in F for D, E and F, 15 μm; bar in G, 10 μm. (H-J) In vitro reconstitutions of actin dynamics (pyrene-actin assays). Actin (actin, 4 μM; pyrene-actin, 0.4 μM) was polymerized in the presence of the indicated concentrations of GST-WH2 (1) (H), GST-WH2 (2) (I) and GST-WH2 (3) (J). WH2 (1) and (2) quench G-actin very effectively and thus prevent spontaneous actin filament formation. Insets show Coomassie stainings of the WH2 domains.
(K-M) Determination of G-actin dissociation constants of WH2 (1) (K), WH2 (2) (L), and WH2 (3) (M) by measuring the fluorescence of NBD-labeled G-actin (0.4 μM) incubated with different concentrations of the WH2 domains.
We first addressed whether all three predicted WH2 domains are functional, i.e. bind to G-actin. Affinity purifications demonstrated that both the first and the second WH2 domain interact with actin albeit the first did so with slightly lower efficiency. The third WH2 domain showed no binding in this assay (Figure 2C). In parallel, we visualized the association of each individual WH2 domain immobilized on beads with actin from rat brain extracts endowed with fluorescently labeled actin. Brightly fluorescent beads showed that monomeric actin was bound effectively by both the first and the second WH2 domain. The assay furthermore revealed that also the third WH2 domain is able to recruit actin, albeit in a less efficient manner (Figure 2D-G).
We next analyzed putative actin sequestering capabilities of the individual WH2 domains by testing their inhibitory effects on spontaneous actin polymerization. In fluorimetric analysis, we indeed observed a strong and dose-dependent sequestering effect of both the first and the second WH2 domain of Cobl, while the third had a comparatively modest effect (Figure 2H-J). Similar results were obtained with GFP-fusion proteins immunoisolated from mammalian cells (Figure S4).
Determination of the G-actin dissociation constants for the three WH2 domains by measuring the fluorescence of NBD-labeled actin incubated with different concentrations of the GST-WH2 domain fusion proteins according to Mattila et al. (2003) quantitatively demonstrated their distinct affinities for G-actin, 39 nM for the first, 40 nM for the second, and 432 nM for the third WH2 domain of Cobl (Figure 2K-M).
Cobl associates with actin in vivo and promotes cell ruffling
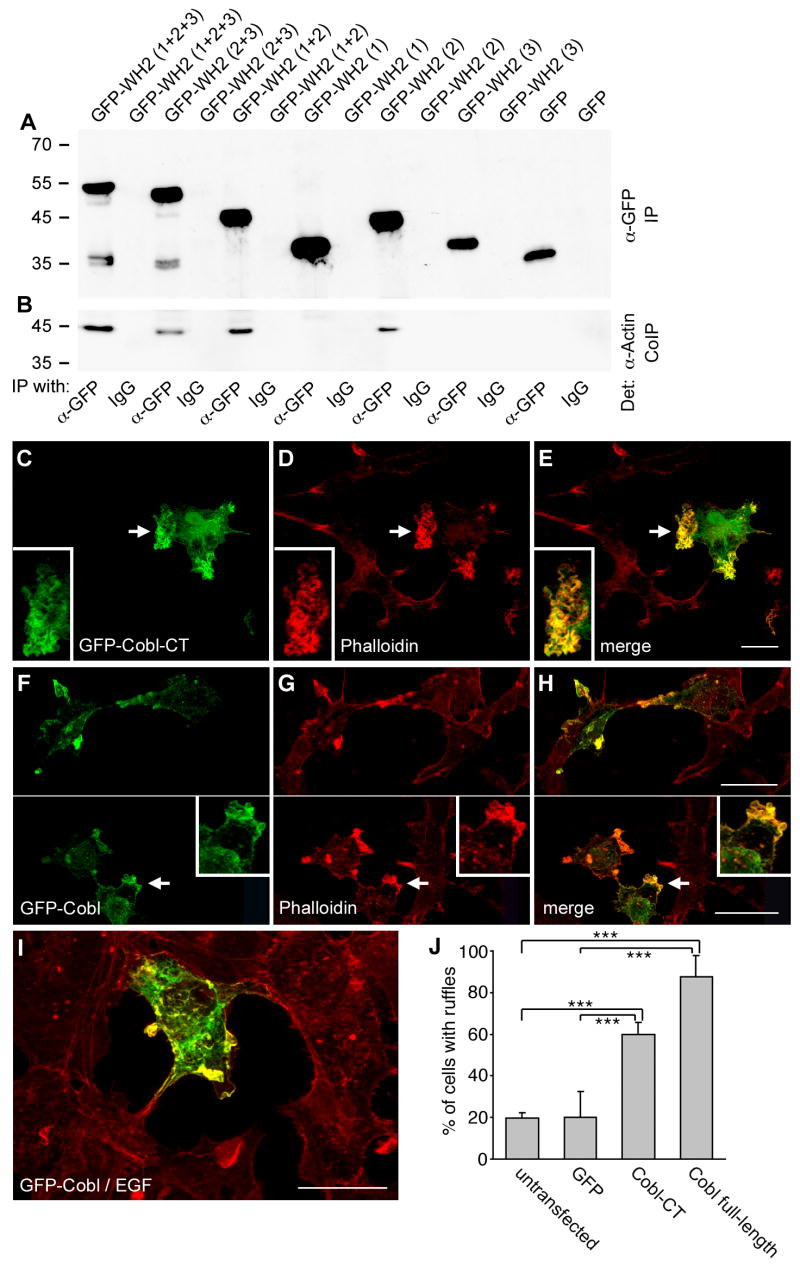
Next we asked whether the actin associations are of relevance in vivo. Immunoblot analyses showed that GFP fusion proteins containing the second Cobl WH2 domain (Figure 3A) all specifically coimmunoprecipitated actin, while the individual first and third WH2 domains did not. The amount of tightly associated actin was higher for Cobl fusion proteins containing all three WH2 domains or WH2 domain 1 and 2 than for those encompassing a combination of the second and the third WH2 domain or the second WH2 domain alone (Figure 3B).
Figure 3. Cobl WH2 domains bind actin in vivo and Cobl induces the formation of actin-rich ruffles in COS-7 cells.
(A and B) Coimmunoprecipitation of endogenous actin from HEK293 cells with different GFP-Cobl constructs. Immunoblot analysis of immunoprecipitated GFP-fusion proteins (A) and of coimmunoprecipitated endogenous actin (B).
(C-H) COS-7 cells transiently transfected with GFP-Cobl-CT (C-E) and GFP-Cobl full-length (F-H) exhibit ruffles enriched for the Cobl proteins (C and F; arrows) and F-actin detected by phalloidin staining and confocal microscopy (D and G; arrows). Insets are enlargements of the ruffles marked by arrows.
(I) EGF-treated cells (15 min, final concentration 100 ng/ml) show a colocalization of Cobl full-length (green) and F-actin (red) in ruffles, too.
(J) Quantification of the percentage of cells with ruffle induction (n > 300 each; 3-6 independent assays). Data are represented as mean ± SEM (p < 0.001, ***). Bar, 12 μm.
A GFP-tagged C-terminal segment of Cobl that contained all three WH2 domains (Cobl-CT) as well as full-length Cobl accumulated in areas of strong three-dimensional ruffling in COS-7 cells where they colocalized extensively with F-actin (Figure 3C-H). Cells treated with EGF also exhibited a strong accumulation of Cobl in all ruffles induced (Figure 3I). Since the F-actin-rich ruffles to which Cobl and Cobl-CT preferentially localized were rarely seen in untransfected or GFP-transfected cells, we asked whether these cortical actin structures were specifically induced by the expression of Cobl and Cobl-CT. A quantitative comparison of the abundance of the cortical ruffles highlighted by phalloidin staining for filamentous actin show that only 20% of the untransfected and GFP-transfected cells showed ruffles, while 60% of the GFP-Cobl-CT-transfected cells and 88% of the GFP-Cobl-transfected cells showed strong ruffling (Figure 3J).
Cobl nucleates actin filaments
Cell ruffling depends on de novo generation of actin filaments. We therefore hypothesized that Cobl in toto is not merely a G-actin sequestering protein but promotes filament formation. To test this, we reconstituted actin polymerization in vitro using purified components and followed the kinetics of filament assembly via the fluorescence increase of pyrene-labeled actin upon polymerization. Cobl-CT was expressed as a Flag- and GFP-tagged fusion protein in COS-7 cells and immunoisolated with anti-Flag antibodies. As a control, Flag-GFP was immunoisolated. As already expected from our coimmunoprecipitation analyses (Figure 3), Cobl-CT was complexed in a one-to-one ratio with actin. Further protein bands were not detected demonstrating the high purity of the purified protein material; importantly immunoisolated Cobl-CT was free of potential Arp2/3 complex contaminations (Figure S5).
At 2 μM, actin showed a long lag period preceding a phase of minor actin assembly. Cobl-CT addition induced rapid actin polymerization and caused a dramatic shortening of the lag phase, i.e. of the actin nucleation step. This effect was specific for Cobl-CT because we observed no increase of actin polymerization upon Flag-GFP control addition (Figure 4A).
Figure 4. Cobl-CT promotes the assembly of actin filaments.

(A, B and D) In vitro reconstitutions of actin nucleation and polymerization. (A) Cobl-CT nucleates actin filament assembly in a dose-dependent manner (2 μM actin) and shortens the lag phase when compared to actin alone and to addition of Flag-GFP (Cont.), respectively. Actin polymerization induced by 20 nM Arp2/3 complex and 60 nM N-WASP WA is shown for comparison. (B) At higher Cobl-CT concentrations, a dose-dependent decrease of Cobl-induced actin filament formation is observed. (C) Relative rate of actin assembly (maximal slope measured during polymerization) for different concentrations of Cobl-CT plotted on a logarithmic (black) and a linear scale (green). (D) Comparison of Cobl-CT to WH2 (1+2) and WH2 (2+3) shows that all three WH2 domains are required.
(E-H) Visual evaluation of actin structures produced by Cobl-CT and Arp2/3. Filaments produced by 40 nM of Cobl-CT are long and mostly unbranched (E), whereas those produced by 25 nM of Arp2/3 complex and 100 nM N-WASP WA are often branched (F). Bar, 10 μm. (G) Quantitative analysis of the percentage of filaments appearing branched. (H) Quantitative analysis of filament lengths produced by Arp2/3 plus WA and Cobl-CT, respectively (n= 150 filaments each).
The effect of Cobl-CT showed a clear dose-dependence. Addition of as little as 4 nM Cobl-CT caused a significant increase. At 20 and 40 nM Cobl-CT, actin polymerization was comparable to that induced by N-WASP WA-activated Arp2/3 complex (20 nM), which leads to a massive nucleation of actin filaments. With Cobl-CT, the lag phase was even shorter than that observed with Arp2/3 complex and N-WASP WA. The maximum rate of assembly, however, remained slightly below that of Arp2/3 complex/N-WASP-mediated filament formation (Figure 4A).
In contrast to Arp2/3-mediated actin nucleation (data not shown), addition of higher amounts of Cobl-CT did not promote the speed of actin assembly further but instead led to a dose-dependent decrease in the polymerization rate and the steady state level of actin filaments (Figure 4B). The maximum was reached between 20 and 40 nM Cobl-CT (Figure 4C). When the concentration of Cobl-CT was raised to 750 nM there was no promotion of filament formation by Cobl anymore but filaments formed as slowly as with actin alone (Figure 4B). This biphasic dose-response (Figure 4C) is likely to reflect the G-actin binding and sequestering properties of the individual Cobl WH2 domains (Figure 2). While interesting and useful for distinguishing the nucleation mechanism of Cobl from those of formins and the Arp2/3 complex, the observed effects of Cobl-CT on cellular morphology (Figures 3, 5) suggest that promotion of filament formation rather than sequestration of actin monomers represents the physiologically relevant effect of Cobl on actin dynamics in vivo.
Figure 5. Cobl affects the morphology of primary hippocampal neurons.
(A) Characterization of rabbit anti-Cobl antibodies in immunoblot experiments. (B) Western blot analysis of postnuclear supernatants of various rat tissues (100 μg protein each) probed with antibodies against Cobl.
(C-E) Colocalization (E, merge) of endogenous Cobl detected by anti-Cobl antibodies (C) with phalloidin-stained F-actin (D) in growth cones of primary hippocampal neurons at DIV2 analyzed by confocal microscopy. Insets represent high magnifications of the axonal growth cone (arrowheads). Lower panels show a cell with more F-actin-rich dendritic growth cones (arrows). Bar in E, 20 μm.
(F-N) Morphological analyses of Cobl functions in developing primary hippocampal neurons transfected at DIV1. 48 h later, neurons expressing GFP-Cobl-CT (F), GFP (H), GFP-WH2 (2+3) and GFP-WH2 (1+2), respectively, were costained for MAP2 (G and I) and evaluated for length of axon (J; n= 80 cells each), branching points per axon (K; n= 55), branching points of the dendritic arbor (L; n= 55), dendrites per cell (M; n= 55) and dendrite length (N; n= 55). Bar in I, 20 μm.
(O-T) Morphological analyses of Cobl functions in hippocampal neurons transfected at DIV5 with GFP-Cobl full-length (O), GFP control (Q), GFP-WH2 (1+2) and GFP-WH2 (2+3) (S, T). Neuronal morphology was analyzed based on MAP2 immunostainings (P, R) 48 later. Quantitative analyses (n= 60) confirmed that Cobl-CT and especially Cobl full-length expression led to an increase of branching points (S) as well of neurites (T) compared to WH2 (2+3), WH2 (1+2) and GFP expressing neurons. Bar in O, 15 μm.
(U) RNAi-mediated knock down of Cobl shown by anti-GFP immunoblotting of extracts from COS-7 cells transfected with bicistronic derivatives of the RNAi tools additionally encoding for GFP-Cobl(1-408).
(V-Y) Morphology of Cobl-depleted neurons (V) transfected at DIV5 versus control (X), as detected by cell fillers (GFP, RFP; V, X) and MAP2 labeling (W, Y). Bar in Y, 20 μm. Quantitative analyses show that RNAi-mediated knock down of Cobl leads to fewer neurites (Ä) and a loss of branching (Z) when compared to controls, such as a non-functional and an unrelated RNAi construct, empty RNAi vector pRNAT, GFP, RFP (n=80 each) as well as rescue experiments with Cobl mutants insensitive for RNAi#1 and #2 (n=50 each).
Data are represented as mean ± SEM (p < 0.05, *; p < 0.01, **; p < 0.001, ***).
We next determined the contribution of the individual WH2 domains to the observed actin filament formation by Cobl-CT. Removing the first WH2 domain completely abolished the capability of Cobl-CT to promote actin polymerization; Cobl WH2 (2+3) caused even an inhibition of spontaneous actin polymerization (Figure 4D) comparable to the effects of the individual WH2 domains (Figure 2). Higher amounts of Cobl WH2 (2+3) showed inhibition as well (data not shown).
Deletion of the third WH2 domain resulted in some remaining polymerization activity at 40 nM (Figure 4D). Also here, increasing concentrations of Cobl WH2 (1+2) led to an inhibition of actin nucleation (data not shown). These results demonstrate that a combination of all three Cobl WH2 domains is required for actin filament assembly.
In order to prove that the Cobl-mediated increase of fluorescence in the pyrene-actin assays indeed represents the formation of filaments, we next visualized the actin structures produced. Our analyses proved that under conditions where actin alone fails to form filaments (not shown) Cobl strongly promotes formation of unbranched filaments (Figure 4E). Under identical conditions a significant portion of Arp2/3 complex-induced filaments were branched, such as those shown in Figure 4F. Quantitative analysis of filament branching highlights this difference (Figure 4G). The lengths of filaments produced by both nucleators were equal (Figure 4H).
Cobl is a brain-enriched protein controlling neuronal morphology
To study the presence and the localization of endogenous Cobl, we used polyclonal antibodies raised against the amino-terminal third of Cobl. Immunoblot and immunofluorescence analyses demonstrated the high specificity and affinity of the antibodies (Figures 5A and S6). Screening different tissue homogenates revealed that expression of Cobl is strongest in brain. At low levels, Cobl was furthermore detected as a 180 kD band in testis, lung and spleen (Figure 5B).
Since Cobl is most highly expressed in the brain, we next asked whether it is present in neurons and whether its localization would provide any hints on its role in actin dynamics. In embryonic, primary hippocampal neurons at DIV2, Cobl was observed throughout the cell body, the axon and the dendrites. It showed accumulations in both axonal and dendritic growth cones, which are also enriched for F-actin (Figure 5C-E).
Since Cobl promotes actin filament assembly in vitro and elicited cell ruffling in COS-7 cells, we tested whether overexpression of Cobl or Cobl-CT altered the morphology of developing neurons. Neurons transfected with Cobl-CT after DIV1 (Figure 5F) indeed exhibited morphologies very different from control cells, as highlighted by immunostaining for the endogenous components MAP2 (Figure 5G). Tubulin beta3 immunostaining gave similar results (Figure S7). Transfected cells showed a strong branching of both the axon and the dendrites and additionally had more dendrites protruding from the cell body (Figure 5F). Quantitative evaluations showed that the length of the axon was not significantly altered (Figure 5J) but expression of Cobl-CT caused a strong increase in branching of the axon (Figure 5K), more than doubled the overall branch points of the dendritic arbor (Figure 5L) and furthermore significantly increased the number of dendrites (Figure 5M). Since the length of dendrites was decreased (Figure 5N), the excessive neurite induction and branching induced by Cobl-CT obviously occurred at the expense of longitudinal growth of dendrites.
None of the effects on neuronal morphology was observed with WH2 (1+2) or WH2 (2+3) (Figure 5J-N). This confirms our hypothesis that promotion of nucleation, a process requiring all three Cobl WH2 domains in combination - rather than monomer binding, which can be observed for the individual WH2 domains and combinations thereof - represents the physiologically relevant function of Cobl.
Since we were unable to express full-length Cobl in neurons just taken into culture, we studied the effects of full-length Cobl and fragments thereof on morphology control in neurons kept in culture for about five days. These cells are less vulnerable and showed higher transfection efficiencies. Due to the higher morphological variability at this stage of development, however, these cells are not suitable for determinations of axon and dendrite length differences. As in younger neurons, Cobl-CT induced significantly more neurites and branching points (Figure 5S and T). Cells expressing GFP-Cobl full-length were also marked by a much more excessive arborization than control cells (Figure 5O-R). Quantitative evaluations confirmed these observations (Figure 5S and T). Effects of full-length Cobl were even more pronounced than those induced by Cobl-CT suggesting that interactions of the more N-terminal parts of Cobl contribute to its function in neuronal morphogenesis.
In order to address for which morphogenesis processes Cobl is necessary, we designed and tested several RNAi constructs. Knock down efficiencies were analyzed in extracts of cells transfected with bicistronic derivatives of RNAi vectors additionally encoding for GFP-Cobl 1-408. Two siRNAs resulted in a significant Cobl knock down, one did not (Figure 5U). In neurons, RNAi-mediated Cobl knock down resulted in a loss of branching and fewer dendrites (Figure 5V-Y). Quantitative analyses showed that the effects of the two functional siRNAs were consistent and significantly different from the various controls (cells transfected with an empty pRNAT vector, pEGFP, pRFP, a non-functional (RNAi#3) and an unrelated RNAi plasmid) (Figure 5Z and Ä). To show that these effects are due to Cobl depletion specifically, the functional RNAi tools were combined with plasmids encoding for full-length Cobl carrying silent mutations rendering it resistant to RNAi#1 and RNAi#2, respectively. In such rescue experiments, we observed a complete abolishment of the RNAi-mediated decrease in neurite number and branching (Figure 5Z and Ä; Figure S8). Thus, Cobl plays a critical role in neuritogenesis and initiation of new branches.
Cobl nucleates barbed-end actin polymerization independently of the Arp2/3 complex
As a powerful nucleator and multiple G-actin binding protein, Cobl is likely to reside on the newly formed filament. Indeed, in vitro cosedimentation assays showed that Cobl-CT efficiently coprecipitated with actin filaments. In contrast, removal of the first or third WH2 domain, respectively, completely abolished the F-actin binding of Cobl-CT (Figure 6A). We thus observed a tight correlation between the abilities to efficiently induce actin filament polymerization (Figure 4) and to associate with actin filaments (Figure 6A).
Figure 6. Cobl-CT nucleates barbed-end actin polymerization in a mechanism distinct from that of the Arp2/3 complex.
(A) GFP-Cobl-CT but not WH2 (1+2), WH2 (2+3) and the linker L2 specifically associates with F-actin, as shown by cosedimentation assays with 1 μM protein and 20 μM actin. Supernatant (S) and pellet (P) fractions were immunoblotted with anti-GFP (upper panel) and detected by Coomassie staining (actin), respectively (middle panel). The lower panel shows control reactions without actin.
(B; C) Cytochalasin D (CD) completely inhibits Cobl-CT-induced actin assembly. Actin (2 μM; 0.2 μM pyrene-actin) was assembled in the presence and absence of 20 nM CD and Cobl-CT (10 nM). 100 nM Cytochalasin B (CB) exhibited similar effects.
(D) Cobl and the Arp2/3 complex (40 nM each) promote filament assembly independently from each other.
(E-G) Cobl-CT does not interfere with actin filament disassembly from the pointed end but slows disassembly at concentrations below the critical concentration of barbed ends. Preformed pyrene-labeled actin filaments (2 μM actin; 0.2 μM pyrene-actin) were diluted to a final concentration of 0.1 μM (E) and 0.05 μM (F), respectively. (G) Quantitative determinations of depolymerization rates, i.e. of slopes of dilution-induced depolymerization - normalized to corresponding experiments with actin alone.
In vivo, actin filaments assemble primarily from their fast-growing barbed end. To address whether Cobl-mediated actin filaments elongate from their barbed ends, we compared the kinetics of Cobl-mediated actin filament growth in the absence and presence of suitable concentrations of the barbed end cappers cytochalasin D (Sampath and Pollard, 1991) and cytochalasin B (MacLean-Fletcher and Pollard, 1980), respectively. 20 nM cytochalasin D inhibited not only spontaneous actin polymerization but also completely blocked actin assembly induced by Cobl-CT (Figure 6B). Consistently, we observed that actin assembly promoted by Cobl-CT was also inhibited by cytochalasin B and rates of actin filament formation were indistinguishable from control (Figure 6C). Thus, Cobl-induced actin filaments grow by barbed end assembly.
Cobl has multiple WH2 domains, and is in this sense reminiscent of N-WASP, which has two WH2 domains and strongly activates the Arp2/3 complex. In order to analyze whether Cobl and the Arp2/3 complex might functionally interact or interfere with each other, we combined Cobl and the Arp2/3 complex in actin polymerization assays. We observed that the initial rates of nucleation are strictly additive for Cobl-CT and the Arp2/3 complex (Figure 6D). This suggests that these two nucleators act independently.
Since the Arp2/3 complex is furthermore known to bind to the pointed ends of actin filaments with nanomolar affinity (Mullins et al., 1998), the increased actin polymerization in the presence of the Arp2/3 complex is consistent with the conclusion that Cobl-promoted actin filaments grow at their barbed ends.
Additionally, we elucidated Cobl's positioning on filaments by subjecting actin filaments nucleated by Cobl-CT to dilutions (0.5 and 0.1 μM) below the critical concentration of the pointed end of 0.6 μM (Pollard et al., 2000). If Cobl caps the pointed end similar to the Arp2/3 complex (Welch and Mullins, 2002), then depolymerization from this end should be inhibited. Depolymerization rates in the presence of Cobl-CT and with actin alone, however revealed no obvious differences (Figure 6E and G). In contrast, when the reactions were diluted below the critical concentration of the barbed end of 0.1 μM (Pollard et al., 2000), depolymerization rates were reduced to about 50% in the presence of Cobl-CT (Figure 6F and G). These results suggest that Cobl is unable to protect the pointed ends but is able to stabilize the barbed ends of filaments.
Actin nuclei formed by Cobl are likely to be in cross-filament orientation
Actin filament nucleation by the three Cobl WH2 domains could in principle be catalyzed by generation/stabilization of a trimer along one strand of the filament helix or by assembling actin monomers belonging to both strands. Our working hypothesis was that first a linear dimer is formed by the first two WH2 domains and that actin-binding of WH2 domain 3 then adds the third monomer in cross-filament orientation and thereby completes the actin nucleus required for spontaneous polymerization (Figure 7A and B). The linker between WH2 domain 1 and 2 (L1) is short (17 amino acids) and would indeed only allow for linear dimer formation. Formation of a nucleus via adding the third monomer in cross-filament orientation demands that the WH2 domain 3 is spaced away from the others by a longer linker that can go around the formed nucleus, i.e. can span a distance of at least 7.5 nm. Indeed, the linker between WH2 domain 2 and 3 (L2) is 68 amino acids in length (Figure S2), i.e. could theoretically bridge 23.8 nm, if fully extended.
Figure 7. Working model of Cobl-induced actin nucleation by cross-filament assembly of an actin trimer.
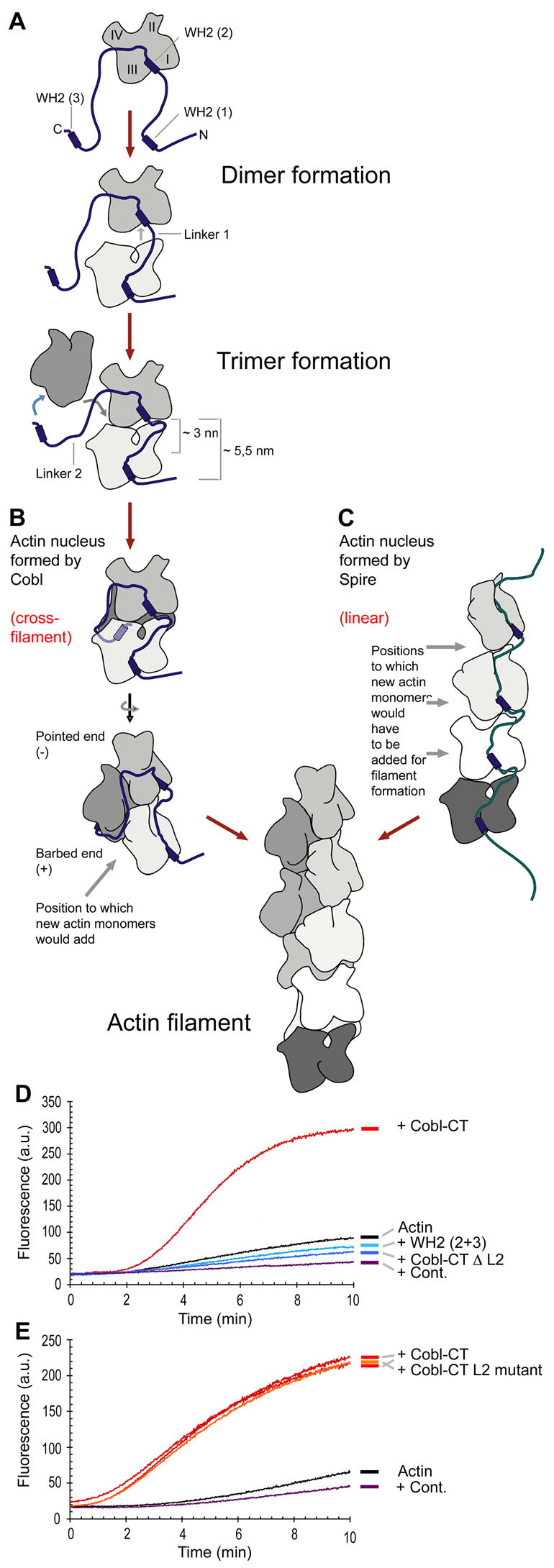
(A) Model of Cobl/actin complex formation. Throughout the figure, the N-terminal, amphiphatic α-helix of the Cobl WH2 domains was placed into the shear zone between actin subdomains 1 and 3, which are oriented towards the barbed end, based on crystal structures of other WH2-proteins (Hertzog et al., 2004; Irobi et al., 2004).
(B) Proposed structure of a Cobl-induced actin nucleus (front view and rotated by 90°) and mechanism of filament formation (elongation).
(C) Filament formation and elongation by addition of actin monomers to linear actin trimers proposed to be assembled by Spire (Quinlan et al., 2005).
(D) Experimental test of the cross-filament nucleation hypothesis by deleting the extended linker L2, which allows for the third WH2 domain to reach the backside of a forming nucleus. Cobl-CT ΔL2 (40 nM) failed to induce actin nucleation. Results were indistinguishable from WH2 deletions and GFP control.
(E) Cobl-CT with a mutated L2 sequence of similar length (orange) nucleates as effectively as wild-type Cobl-CT (two examples in red).
We tested the cross-filament hypothesis by shortening the linker L2 to 17 residues (compare Figure S9). If Cobl nucleates actin filaments via linear trimer formation, as proposed for Spire, a four WH2 domain-containing protein that can elicit some actin nucleation (Quinlan et al., 2005; Figure 7C), Cobl-mediated actin nucleation should also be induced by the mutant (Cobl-CT ΔL2), whereas cross-filament monomer addition would be disrupted. Our analyses revealed that the exclusive presence of short linkers, which can only accommodate binding of actin monomers in a linear fashion, completely abolished the capability of Cobl-CT to promote actin filament formation (Figure 7D).
Although the reconstitutions are done with purified actin and Cobl proteins and the linker does not bind to F-actin (Figure 6A), it could theoretically still be that not shortening the linker L2 but disrupting other L2 functions and/or interactions was the reason for the lack of actin nucleation capability of Cobl-CT ΔL2. We therefore replaced the linker L2 by inserting a stretch of similar length but of different amino acid composition into Cobl-CT ΔL2 (Figure S9). The resulting Cobl-CT L2 mutant triggered actin nucleation as effectively as wild-type Cobl-CT (Figure 7E). This result clearly shows that the length but not the sequence of linker L2 is critical for actin nucleation by Cobl. This finding is well in line with the observation that although the sequence conservation among Cobl proteins in L2 is low, its length is relatively well conserved from fish to man (Figure S2).
Discussion
Unraveling the molecular mechanisms of cell morphogenesis is indispensable for an understanding of functions of both individual cells and superstructures, such as organs and tissues, which these cells form. The formation and reorganization of neuronal networks is particularly dependent on extensive morphological changes of the individual, specialized cells. Despite the wealth of different actin structures and of cellular processes involving actin dynamics and organization, currently merely two actin nucleators are well established in vertebrates, the Arp2/3 complex and formins (Welch and Mullins, 2002; Higgs, 2005; Kovar, 2006). Here, we describe the discovery and characterization of a novel actin nucleator, Cobl, a brain-enriched vertebrate protein that uses three C-terminal WH2 domains to associate with actin. Cobl gives rise to long, non-bundled and unbranched filaments, which elongate by barbed end growth.
How does Cobl nucleate actin filaments? In principle, actin filament formation can be promoted by different means that all generate free barbed ends for rapid elongation, i) severing of existing actin filaments, ii) uncapping of filaments and iii) de novo formation of actin nuclei. Since the kinetics of actin polymerization induced by Cobl-CT are not characteristic of those obtained with known severing proteins (Du and Frieden, 1998) and Cobl addition to preformed filaments did not cause an increase in the rate of disassembly when filaments were subjected to dilution to an actin concentration below that of the critical concentration of the pointed end, we exclude that Cobl promotes actin polymerization by severing. The Cobl-CT-induced actin polymerization is also unlikely to reflect an uncapping activity of Cobl-CT, because we reconstituted filament formation with actin and Cobl as the sole components. Instead, our analyses clearly demonstrate that Cobl nucleates filaments. Besides the observed dose-dependent increase in the rate of assembly, the reduction of the lag phase suggests a Cobl-mediated formation of actin nuclei. At 20 and 40 nM Cobl-CT, the lag phase was even shorter than that observed upon addition of 20 nM Arp2/3 complex activated by an excess of the N-WASP C-terminus. This power of Cobl most likely reflects its ability to directly associate with at least one actin molecule extremely tightly, as seen in our immunoprecipitation experiments. Further monomers can then be added rapidly by associations with the WH2 domains and with bound actin monomers.
Extensive mechanistic studies revealed fundamental differences in the molecular mechanisms of filament nucleation by Cobl compared to the Arp2/3 complex. First, Cobl shows very strong G-actin binding, whereas the Arp2/3 complex does not. Second, the actin structures formed by both types of nucleators were different; the side-binding activity of the Arp2/3 complex gave rise to branched filaments, while Cobl-induced filaments were unbranched. Third, while the Arp2/3 complex resides at the pointed end and thereby stabilizes this end under depolymerization conditions (Mullins et al., 1998), Cobl's interaction with actin does not protect the pointed end. Instead, Cobl slows depolymerization only when filaments are diluted to concentrations below the critical concentrations of both the barbed and the pointed end. In line with these mechanistic differences, we did not observe any competition or inhibitory or stimulatory cross-talk between Cobl and the Arp2/3 complex but their effects on actin nucleation were additive. This suggests that both nucleators act independently from each other and that Cobl is associated with barbed ends. This conclusion is in line with the fact that superimposition of a WH2 domain/actin structure and an actin filament suggested that the barbed end could accommodate a WH2 domain without any steric clashes (Chereau et al., 2005), that WH2 domains favor ATP actin (Paavilainen et al., 2004) as occurring at the barbed end and with the very recent finding that WH2 domains mediate dynamic attachments of actin filament barbed ends to membranes (Co et al., 2007).
Some of the properties of Cobl resemble those of formins. In both cases, actin filaments induced growth via barbed end elongation but form with slightly slower rates when compared to unhindered growth of barbed ends, such as provided by Arp2/3 complex-mediated actin nucleation (Pruyne et al., 2002; Sagot et al., 2002). In the case of formins, this reflects processive associations with barbed end (for review see Higgs, 2005; Kovar, 2006).
Compared to formins, Cobl is more powerful. Cobl-induced actin nucleation occurs with actin concentrations as low as 1 μM (our unpublished data), while formins fail to induce filaments at such low actin concentrations (Pruyne et al., 2002). Furthermore, only 20 nM Cobl are required for actin polymerization similar to activated Arp2/3 complex. Formins, however, require concentrations of about 200 nM to reach this performance (Sagot et al., 2002).
Interestingly, Cobl-induced nucleation shows a biphasic dose-response. The most likely explanation for the observed decrease in Cobl-induced acceleration of filament formation at high Cobl concentrations is that the actin concentration is not sufficient anymore to ensure full loading of each Cobl molecule. This results in incomplete and thus inactive Cobl/actin complexes and in G-actin sequestering. One nucleator has recently been reported to act in a biphasic way similar to Cobl, the D. melanogaster protein Spire. Spire is an actin-binding, WH2 domain-containing protein that nucleates actin filaments when employed in high concentrations in in vitro reconstitutions (Quinlan et al., 2005). Since Spire function seems based on a combination of several G-actin binding modules, here too G-actin sequestering may explain the biphasic behavior observed.
The molecular mechanism of actin nucleation by Cobl, however, seems to be very different from that of Spire. While Cobl and the Arp2/3 complex work independently, Spire failed to show any additive performance when combined with the Arp2/3 complex (Quinlan et al., 2005). Also in contrast to Cobl, Spire somewhat slowed depolymerization from the pointed end. Similar to the Arp2/3 complex, Spire was therefore proposed to associate with the pointed end (Quinlan et al., 2005). Furthermore, the type of actin nuclei we propose for Cobl-induced filament formation are completely different from those proposed for Spire. The affinities of the individual G-actin interactions of the Cobl WH2 domains, the topology of Cobl and its properties revealed in kinetic analyses suggest that Cobl assembles a nucleus by first forming a complex composed of WH2 domain 2 and G-actin, to which a second actin monomer can then rapidly be added by interactions with the WH2 domain 1, as reflected by the relatively strong actin association of this domain in our experiments. Cobl WH2 domain 3 binding then helps to add the third monomer to the two actin molecules already assembled and thereby completes the actin nucleus (Figure 7A). Importantly, the linker L2 between WH2 domains 2 and 3 is long enough to accommodate the addition of the third actin monomer in cross-filament orientation (Figure 7A and B). Consistently, Cobl-induced actin nucleation i) critically depends on all three Cobl WH2 domains, ii) the length (but not the sequence) of the linker L2 and iii) is very efficient when compared to other nucleators. In contrast, the linkers between the four Spire WH2 domains are so short that they can only allow for linear assembly of actin monomers. Cross-filament monomers would then have to be added by spontaneous actin/actin interactions (Figure 7C). Biophysical analyses show that such a mechanism is not favored; instead, cross-filament orientations of trimeric actin nuclei, such as those proposed to be assembled by Cobl-mediated mechanisms, are highly favored in actin nucleation and were suggested to account for the vast majority of all actin nuclei formed (Sept and McCammon, 2001). Consistently, high concentrations of Spire (0.5 to 1 μM) were needed to cause actin nucleation in in vitro reconstitutions but still did not lead to activities comparable to the performance of 20 nM activated Arp2/3 complex (Quinlan et al., 2005).
Cobl-induced actin nucleation is likely required for structural remodeling in a variety of cell types. In fibroblastic cells, increased cell ruffling was observed. The molecular process underlying ruffle formation is massive generation of new actin filaments at the leading edges (for reviews see Pollard et al., 2000; Welch and Mullins, 2002). In the gastrulating embryo cobl mRNA (Carroll et al., 2003) shows a remarkable restriction to a small group of cells, the organizer, giving rise to the axial midline - a critical source of patterning and morphogenesis cues in vertebrate embryos. Later, cobl mRNA and protein are also found in other tissue domains known to involve active morphogenesis (Carroll et al., manuscript submitted). In mature organisms, we observed highest Cobl expression levels in the brain. We suggest that one molecular function behind this distribution of Cobl is its ability to modulate the actin cytoskeleton and to control neuronal cellular architecture. Neuronal networks are undergoing functional and structural remodeling also in the adult and Cobl has dramatic influences on cellular morphology. Cobl expression led to drastic increases in the number of neurites protruding from the cell body and of the branching of both axon and of dendrites. These effects can be clearly linked to actin nucleation caused by Cobl, because they did not occur when nucleation-deficient Cobl mutants were employed. RNAi-mediated Cobl knock down revealed that Cobl functions are critical for neuritogenesis and initiation of new branches.
Interestingly, the formation of neurites and their arborization critically involving Cobl has been revealed not to rely on Arp2/3 complex-mediated cytoskeletal functions. The Arp2/3 complex selectively influences axon length (Strasser et al., 2004; Pinyol et al., 2007). Thus, cells use different actin nucleators to steer the complex remodeling processes underlying cell morphogenesis, the formation of cellular networks and the development of complex body plans.
Experimental Procedures
DNA Constructs, Recombinant Proteins and Antibodies
For a detailed description of the DNA constructs used, of the purifications of recombinant proteins performed and of the antibodies used in this study please see the Supplementary Material.
Yeast two-Hybrid
Yeast-2-hybrid screening was carried out as described in Braun et al. (2005).
Cell Culture and Immunofluorescence Microscopy
HEK293 and COS-7 cells and primary hippocampal cultures were cultured and transfected as described (Kessels and Qualmann, 2006; Pinyol et al., 2007). Quantification of ruffles was performed for 300-760 cells from 3-6 independent assays.
Primary neurons were transfected at day 1 and 5 in vitro, respectively, grown for 48 more hours, fixed and subjected to immunofluorescence experiments as described (Pinyol et al. 2007). Morphometric analyses of randomly sampled neurons are based on highlighting cell morphologies by anti-MAP2 immunostaining using NIH Image Software (ImageJ). The axon length was measured by defining the longest neurite of the not yet fully polarized cells as developing axon, the remaining MAP2-positive neurites as dendrites. Statistical analysis was performed using the two-tailed Student test. Further details are available as supplementary information.
Homogenate Preparations, Coprecipitation and Coimmunoprecipitation Analyses
Preparation of postnuclear supernatants from different rat tissues and from cultured cells and coprecipitation and coimmunoprecipitation assays were essentially performed as described (Qualmann and Kelly, 2000; Kessels and Qualmann, 2006).
Biochemical examinations of cytoskeletal properties
The dissociation constants of the three Cobl WH2 domains and G-actin were determined according to Mattila et al. (2003).
The actin polymerization bead assay was designed according to Pinyol et al. (2007). To deplete rat brain extracts from Arp2/3 complex, homogenate was repeatedly incubated overnight at 4°C with covalently immobilized GST-N-WASP WA.
The pyrene-actin polymerization assay was designed according to Pinyol et al. (2007). Polymerization studies were conducted with 2 μM actin. Sequestering experiments were additionally done with 4 μM G-actin to visualize quench effects better.
Fluorescence microscopy of actin filaments was done according to Blanchoin et al., 2000. Briefly, 4 μM of actin was polymerized for 20 min in presence of 4 μM of Alexa 568-phalloidin, diluted, mounted onto coverslips and viewed. Further technical details of the above assays are provided in the Supplementary Material.
For actin disassembly assays, 2 μM actin was assembled in presence of 20 nM Cobl-CT for 15 min. Disassembly was then induced by diluting these pre-assembled actin filaments (in assay buffer) to final concentrations below the critical concentration of the barbed end (20 and 50 nM) and to concentrations just below the critical concentration of the pointed end (100 and 500 nM) (Pollard et al., 2000).
Actin filaments cosedimentation assays were done according to Kessels et al. (2000).
Supplementary Material
Acknowledgments
We thank K. Hartung and K. Keller for technical assistance, O. Kobler for help with confocal microscopy, M.D. Welch for providing anti-Arp2/3 complex antibodies and E.D. Gundelfinger for support. This work was supported by grants from the NIH (to JK), the Kultusministerium Land Sachsen-Anhalt (XN3571A to BQ) and the Deutsche Forschungsgemeinschaft (to BQ and MMK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–11. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Braun A, Pinyol R, Koch D, Dahlhaus R, Fonarev P, Grant BD, Kessels MM, Qualmann B. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll EA, Gerrelli D, Gasca S, Berg E, Beier DR, Copp AJ, Klingensmith J. Cordon-bleu is a conserved gene involved in neural tube formation. Dev Biol. 2003;262:16–31. doi: 10.1016/s0012-1606(03)00323-3. [DOI] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of WASP-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- Du J, Frieden C. Kinetic studies on the effect of yeast cofilin on yeast actin polymerization. Biochemistry. 1998;37:13276–13284. doi: 10.1021/bi981117r. [DOI] [PubMed] [Google Scholar]
- Gasca S, Hill DP, Klingensmith J, Rossant J. Characterization of a gene trap insertion into a novel gene, cordon-bleu, expressed in axial structures of the gastrulating mouse embryo. Dev Genet. 1995;17:141–154. doi: 10.1002/dvg.1020170206. [DOI] [PubMed] [Google Scholar]
- Hertzog M, Van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Irobi E, Aguda AH, Larsson M, Guerin C, Yin HL, Burtnick LD, Blanchoin L, Robinson RC. Structural basis of actin sequestration by thymosin-β4: implications for WH2 proteins. EMBO J. 2004;23:3599–3608. doi: 10.1038/sj.emboj.7600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Engqvist-Goldstein ÅEY, Drubin DG. Association of Mouse Actin-binding Protein 1 (mAbp1/SH3P7), an Src Kinase Target, with Dynamic Regions of the Cortical Actin Cytoskeleton in Response to Rac1 Activation. Mol Biol Cell. 2000;11:393–412. doi: 10.1091/mbc.11.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B. Syndapins integrate N-WASP in receptor-mediated endocytosis. EMBO J. 2002;21:6083–6094. doi: 10.1093/emboj/cdf604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B. Syndapin oligomers interconnect the machineries for endocytic vesicle formation and actin polymerization. J Biol Chem. 2006;281:13285–13299. doi: 10.1074/jbc.M510226200. [DOI] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- MacLean-Fletcher S, Pollard TD. Mechanism of action of cytochalasin B on actin. Cell. 1980;20:329–341. doi: 10.1016/0092-8674(80)90619-4. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Salminen M, Yamashiro T, Lappalainen P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J Biol Chem. 2003;278:8452–8459. doi: 10.1074/jbc.M212113200. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavilainen VO, Bertling E, Falck S, Lappalainen P. Regulation of cytoskeletal dynamics by actin-monomer-binding proteins. Trends Cell Biol. 2004;14:386–394. doi: 10.1016/j.tcb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Paunola E, Mattila PK, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS ONE. 2007;2:e400. doi: 10.1371/journal.pone.0000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kelly RB. Syndapin Isoforms Participate in Receptor-mediated Endocytosis and Actin Organization. J Cell Biol. 2000;148:1047–1061. doi: 10.1083/jcb.148.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM. Endocytosis and the cytoskeleton. Int Rev Cytol. 2002;220:93–144. doi: 10.1016/s0074-7696(02)20004-2. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Sampath P, Pollard TD. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry. 1991;30:1973–1980. doi: 10.1021/bi00221a034. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal TE, Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Curr Opin Cell Biol. 2006;18:4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Strasser GA, Rahim NA, VanderWaal KE, Gertler FB, Lanier LM. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 2004;43:81–94. doi: 10.1016/j.neuron.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Machesky LM. The WASP-Arp2/3 pathway: genetic insights. Curr Opin Cell Biol. 2004;16:174–181. doi: 10.1016/j.ceb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:47–88. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.