Summary
Replicative DNA damage bypass, mediated by the ubiquitylation of the sliding clamp protein PCNA, facilitates the survival of a cell in the presence of genotoxic agents, but it can also promote genomic instability by damage-induced mutagenesis. We show here that PCNA ubiquitylation in budding yeast is activated independently of the replication-dependent S phase checkpoint but by similar conditions involving the accumulation of single-stranded DNA at stalled replication intermediates. The ssDNA-binding replication protein A (RPA), an essential complex involved in most DNA transactions, is required for damage-induced PCNA ubiquitylation. We found that RPA directly interacts with the ubiquitin ligase responsible for the modification of PCNA, Rad18, both in yeast and in mammalian cells. Association of the ligase with chromatin is detected where RPA is most abundant, and purified RPA can recruit Rad18 to ssDNA in vitro. Our results therefore implicate the RPA complex in the activation of DNA damage tolerance.
Keywords: DNA, PROTEINS
Introduction
DNA damage tolerance enables a cell to resolve replication problems such as damage-induced fork stalling (Lawrence, 1994; Lehmann, 2002; Ulrich, 2005). Replicative bypass of DNA lesions occurs via translesion synthesis (TLS) by specialized damage-tolerant DNA polymerases or by an error-free damage avoidance pathway that makes use of the information of the newly synthesized sister chromatid (Lawrence, 1994). Both pathways are activated by posttranslational modification of the replicative sliding clamp, proliferating cell nuclear antigen (PCNA) (Hoege et al., 2002; Kannouche et al., 2004; Stelter and Ulrich, 2003; Watanabe et al., 2004). In response to DNA damage, monoubiquitylation of PCNA at a conserved lysine, K164, activates the damage-tolerant polymerases for TLS, whereas polyubiquitylation is required for error-free damage avoidance. Independent of DNA damage, budding yeast PCNA is also modified during S phase by the ubiquitin-like protein SUMO (Papouli et al., 2005; Pfander et al., 2005). PCNA monoubiquitylation involves the ubiquitin-conjugating enzyme (E2) Rad6 and its cognate ubiquitin ligase (E3), Rad18. A second E2-E3 pair, the heterodimeric E2 Ubc13-Mms2 with the E3 Rad5, is responsible for polyubiquitylation. Although it promotes resistance to genotoxic agents, TLS is a potentially mutagenic process that endangers the cell's genomic stability. At the same time, failure to reactivate stalled replication forks may lead to genome instability by inducing DNA double-strand breaks (DSBs) and gross chromosomal aberrations (Branzei and Foiani, 2005). Stringent control over DNA damage bypass is therefore essential, but the upstream signals that activate PCNA modification in vivo are not well defined.
One of the important regulatory mechanisms responsible for the sensing of DNA damage and replication stress is the replication-dependent S phase checkpoint, a surveillance system that inhibits the firing of late-replication origins, prevents cell-cycle progression to mitosis, and is important for the stabilization of stalled replication forks (Branzei and Foiani, 2005; Nyberg et al., 2002). The ATR-ATRIP kinase complex and the PCNA-like 9-1-1 checkpoint clamp, responsible for initiating the checkpoint signaling cascade during S phase, are activated by replication fork stalling. Their recruitment to stretches of single-stranded (ss)DNA is mediated by interactions of ATRIP and the 9-1-1-specific clamp loader with the ssDNA-binding RPA complex (Zou and Elledge, 2003; Zou et al., 2003).
Our analysis of the mechanisms that control the activity of the RAD6 pathway in budding yeast now reveals striking parallels to the replication checkpoint response: although the two systems operate independently, they are activated in a similar fashion by stalled replication intermediates that involve an accumulation of ssDNA. We show that yeast and human Rad18, like ATRIP and the 9-1-1 clamp loader, directly interact with the RPA complex. In vivo, the abundance of Rad18 on DNA mirrors that of RPA even in the absence of its physiological target, PCNA, and depletion of RPA prevents damage-induced PCNA ubiquitylation in S phase. In vitro, the RPA complex can recruit the ubiquitin ligase to ssDNA. These results suggest an effective activation mechanism for ubiquitin-dependent damage bypass.
Results
Replication Forks Are Required for PCNA Ubiquitylation
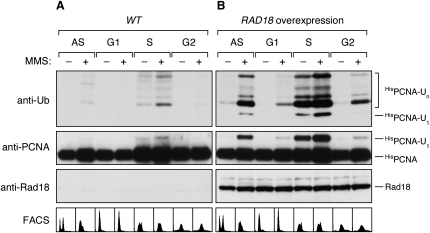
In order to characterize the conditions required for PCNA ubiquitylation in S. cerevisiae, we examined possible contributions of the DNA damage checkpoint and cell-cycle regulation. We found that, in budding yeast, as in X. laevis egg extracts and in S. pombe (Chang et al., 2006; Frampton et al., 2006), ubiquitin-dependent DNA damage tolerance and checkpoint signaling operate independently (see Figure S1 available online). Given the importance of ubiquitylated PCNA for replicative lesion bypass, the modification is expected to be most relevant during S phase. In fact, consistent with our previous findings (Papouli et al., 2005) and with the situation in mammalian cells (Kannouche et al., 2004), arrest in S phase with hydroxyurea (HU), which causes replication fork stalling by nucleotide depletion without directly damaging DNA, is sufficient to trigger PCNA modification (Figure 1A). In contrast, ubiquitylated PCNA was not detected in G1- or G2-arrested cells even after treatment with DNA-damaging agents. This indicates that, even in asynchronous populations, all detectable PCNA ubiquitylation arises from S phase cells.
Figure 1.
Effects of the Cell Cycle and RAD18 Overexpression on PCNA Ubiquitylation
(A) Cell-cycle dependence of PCNA modification. Cells arrested in G1, S, and G2 phase were treated with 0.02% MMS for 90 min where indicated, and modifications of HisPCNA, isolated under denaturing conditions, were detected by western blot. DNA contents were monitored by flow cytometry (FACS). Asynchronous cells (AS) were processed in parallel.
(B) Effects of RAD18 overexpression on PCNA modification throughout the cell cycle. Cells were treated and analyzed as in (A). Note that, in this panel, monoubiquitylated PCNA is abundant enough to be detected by the anti-ubiquitin antibody, which recognizes this form very poorly (Hoege et al., 2002). Rad18 was detected in total cell extracts.
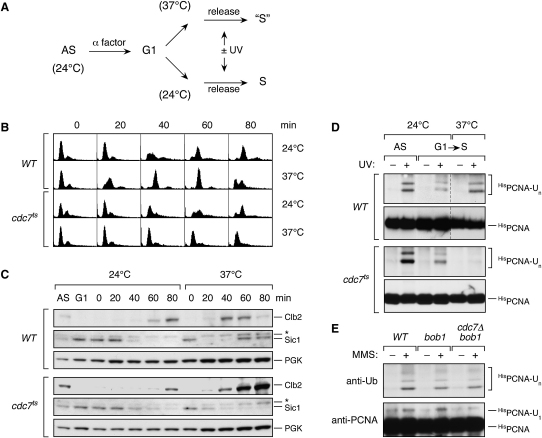
The absence of ubiquitylated PCNA outside of S phase could be due to the lack of replication forks. Alternatively, the physiological state of the cell, defined by the activities of cyclin-dependent kinases, could control PCNA modification. In order to directly examine the need for DNA replication, we made use of a temperature-sensitive mutant of an essential kinase gene responsible for DNA replication initiation, cdc7ts (Figure 2) (Hartwell, 1973). At the permissive temperature, cdc7ts cells undergo regular cycles of DNA replication and cell division, whereas upon release from G1 arrest at the restrictive temperature, the mutant enters the cell cycle without initiating DNA replication (Figure 2B). Degradation of the CDK inhibitor Sic1 at the beginning of S phase and later on the accumulation of the mitotic cyclin Clb2 proceeds normally in cdc7ts cells (Figure 2C), indicating that the physiological state under these conditions resembles a passage through the cell cycle. Following the scheme outlined in Figure 2A, we asked whether DNA damage would trigger PCNA modification in the mutant. We found that WT cells underwent PCNA ubiquitylation normally at 24°C and 37°C, but in cdc7ts the modification was visible only at the permissive temperature (Figure 2D). In order to exclude the possibility that the kinase activity of Cdc7 itself was needed for the modification, we examined a strain in which the requirement for CDC7 is bypassed by a mutation in MCM5, encoding a subunit of the replicative helicase (Hardy et al., 1997). In the context of this allele, bob1, deletion of CDC7 did not abolish PCNA ubiquitylation, implying that the Cdc7 kinase itself is dispensable for modification of the clamp (Figure 2E). This suggests that PCNA needs to be engaged in replication for efficient ubiquitylation in vivo.
Figure 2.
Active Replication Forks Are Required for PCNA Ubiquitylation
(A) Experimental strategy to prevent the formation of replication forks in the cdc7ts mutant.
(B) FACS profiles of WT and cdc7ts cells subjected to the treatment outlined in (A).
(C) Progression through the cell cycle in WT and cdc7ts cells as monitored by Clb2 and Sic1 levels. Note that at 37°C WT cells have completed mitosis and re-entered G1 phase at 80 min, whereas cdc7ts mutants accumulate in G2/M with abnormally high Clb2 and low Sic1 levels. The asterisk indicates a band crossreactive to the Sic1 antibody. Detection of phosphoglycerate kinase (PGK) served as a loading control.
(D) Damage-induced PCNA ubiquitylation in WT and cdc7ts cells after treatment as outlined in (A). S phase samples in cdc7ts cells and in WT at 24°C were taken 40 min after release, whereas the S phase sample in the WT at 37°C was taken after 25 min due to the faster cell-cycle progression at this temperature (see [B] and [C]).
(E) CDC7 is dispensable for PCNA ubiquitylation. Damage-induced PCNA modification was examined in asynchronous cultures of isogenic WT, bob1, and bob1 cdc7Δ cells.
Not All Types of DNA Damage Induce PCNA Ubiquitylation
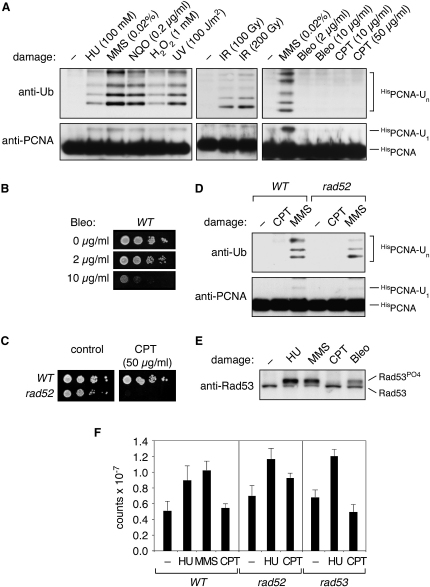
We next asked what types of lesions would induce PCNA modification during DNA replication. In addition to UV radiation and methyl methane sulfonate (MMS), which induce ubiquitylation in all species examined so far, agents that cause bulky adducts such as 4-nitroquinoline oxide (NQO) and also the oxidizing agent hydrogen peroxide (H2O2) were found to be active (Figure 3A). As observed in S. pombe (Frampton et al., 2006), PCNA was also ubiquitylated after treatment with ionizing radiation (IR) (Figure 3A), despite our previous finding that PCNA ubiquitylation site mutants are not particularly sensitive to IR, and DSBs induced by an endonuclease do not cause PCNA modification (Chen et al., 2005). Moreover, mammalian cells do not ubiquitylate PCNA after IR treatment (Kannouche et al., 2004). A possible explanation for the IR-induced modification of S. cerevisiae and S. pombe PCNA may therefore be base damage associated with the high doses of radiation used in the yeast system rather than DSBs per se.
Figure 3.
Influence of the Type of DNA Damage on PCNA Ubiquitylation
(A) PCNA ubiquitylation after treatment with HU, MMS, NQO, H2O2, UV, IR, Bleomycin (Bleo), and CPT at the indicated doses. Modified forms of PCNA were detected as described in the legend to Figure 1.
(B) Sensitivities of WT cells to Bleomycin as monitored by spot assays.
(C) Sensitivities of WT and rad52 cells to CPT.
(D) PCNA ubiquitylation in WT versus rad52 cells. Treatment with damaging agents (50 μg/ml CPT, 0.02% MMS) and detection of the modifications was performed as above.
(E) Induction of the DNA damage checkpoint after treatment with selected DNA-damaging agents. Rad53 phosphorylation, monitored by western blot analysis, was used as an indicator of checkpoint activation.
(F) Quantification of ssDNA after treatment with MMS, HU, or CPT by radioactive random-primed labeling of nondenatured genomic DNA. The graph shows the average labeling efficiencies with standard deviations (in cpm) from triplicate experiments, corrected for the amount of template DNA as determined by real-time PCR.
To further explore this hypothesis, we analyzed the response to chemicals that cause DSBs by different mechanisms. Bleomycin, which acts independently of the cell-cycle phase, did not trigger PCNA modification (Figure 3A), although it affected viability (Figure 3B). Camptothecin (CPT), a topoisomerase I inhibitor, induces covalent adducts of the enzyme to DNA, which are converted to DSBs in S phase through collisions with replication forks (Liu et al., 2000). Despite its S phase-specific action, CPT treatment did not result in PCNA ubiquitylation (Figure 3A). In order to exclude the possibility that the drug had simply no effect at the concentrations used in this experiment, we examined PCNA modifications in a rad52 mutant, which is highly sensitive to CPT (Figure 3C). Again, we did not observe PCNA ubiquitylation in response to CPT in rad52, whereas its reaction to MMS was comparable to that of WT cells (Figure 3D). Likewise, CPT induces PCNA modifications very modestly in S. pombe when compared to HU or UV (Frampton et al., 2006), and the drug is also a poor inducer of the replication checkpoint in budding yeast (Redon et al., 2003) (Figure 3E).
Replication fork stalling by treatment with HU or adduct-forming agents like MMS results in an inhibition of DNA polymerases and an accumulation of ssDNA due to the continuing movement of the replicative helicase (Branzei and Foiani, 2005). In contrast, collision of the replication machinery with CPT-induced topoisomerase adducts and DSB formation is expected to affect the entire replicon and should therefore not initially involve an accumulation of ssDNA. In support of this model, radioactive labeling of nondenatured total DNA by random priming was significantly enhanced after MMS or HU, but not after CPT treatment (Figure 3F). This even applied to the checkpoint-defective rad53 mutant, which is known to accumulate ssDNA in response to replication stress (Sogo et al., 2002). In rad52 cells, the difference was less pronounced, but we cannot rule out a replication-independent effect of the mutation—for example, on formation or resection of DSBs—that would have no consequences for PCNA modification. Thus, PCNA ubiquitylation correlates well with elevated levels of ssDNA during replication.
Overexpression of RAD18 Relaxes the Conditions for PCNA Modification
Given that the ubiquitin ligases, Rad18 and Rad5, are ssDNA-binding proteins, they might directly recognize a blocked fork by associating with the stretches of ssDNA adjacent to the stalled polymerase-PCNA complex. If these enzymes are limiting factors in transmitting the signal for modification, changes in their abundance should affect the extent of modification. We therefore analyzed the consequences of overexpressing RAD18, encoding the E3 responsible for PCNA monoubiquitylation. Figure 1B shows that overexpression of RAD18 caused significantly increased levels of ubiquitylated PCNA. In asynchronous populations, RAD18 overexpression resulted in constitutive modification of PCNA even in undamaged cells, suggesting that fork stalling was no longer required. Likewise, the modification was no longer restricted to S phase, as ubiquitylated PCNA now appeared in G1- and G2-arrested cells. Outside of S phase, however, DNA damage was still a prerequisite for the reaction. Taken together, elevated levels of Rad18 afforded PCNA ubiquitylation whenever the clamp was detectable in association with chromatin (Figure S2), strongly suggesting that the modification takes place on DNA. Consistent with this notion, loading of PCNA onto DNA was shown to be required for efficient modification in vitro (Garg and Burgers, 2005).
RPA Is Required for Ubiquitylation of PCNA
Our findings suggest that PCNA ubiquitylation is induced by the recruitment of Rad18 to regions of ssDNA exposed by the uncoupling of helicase movement from DNA synthesis, analogous to the way in which stalled forks are believed to activate the replication checkpoint (Branzei and Foiani, 2005). Considering that the factor responsible for recruitment of the ATR kinase complex and the 9-1-1 clamp loader is not naked ssDNA but rather ssDNA coated by the ssDNA-binding RPA complex (Zou and Elledge, 2003; Zou et al., 2003), we examined a possible contribution of RPA to PCNA ubiquitylation.
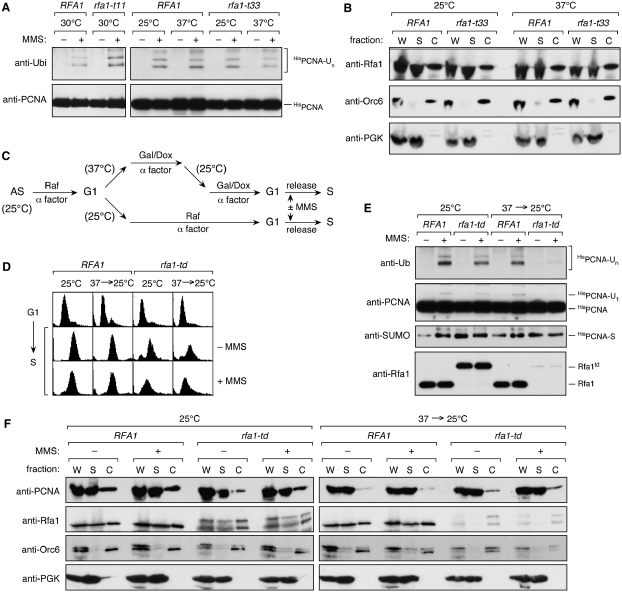
The rfa1-t11 mutant (K45E), defective in homologous recombination and recruitment of checkpoint factors (Umezu et al., 1998; Zou and Elledge, 2003; Zou et al., 2003), was competent in damage-induced PCNA ubiquitylation (Figure 4A). Likewise, a temperature-sensitive mutant, rfa1-t33 (S373P), which is unable to mediate DNA replication at the restrictive temperature (Umezu et al., 1998), ubiquitylated PCNA normally. This indicates that the replicative function of RPA is not required for PCNA modification. However, even at the restrictive temperature, the mutant protein was found in association with DNA (Figure 4B), which leaves the possibility that it was still able to promote the recruitment or activation of Rad18. A panel of rfa2 and rfa3 mutants defective in various aspects of DNA replication and repair (Maniar et al., 1997) had relatively minor effects on PCNA ubiquitylation (Figure S3). We therefore examined the consequences of depleting Rfa1 by means of a heat-inducible degradation signal (Dohmen et al., 1994). Following the procedure outlined in Figure 4C, we induced degradation of the Rfa1td protein in G1-arrested cells. The rfa1-td mutant engaged in DNA replication (Figure 4D) with less than 5% of the Rfa1td protein remaining, but damage-induced PCNA ubiquitylation was almost completely abolished (Figure 4E). In contrast, S phase-associated sumoylation of PCNA, which likewise depends on DNA replication (our unpublished results), was not impaired (Figure 4E), implying that the lack of ubiquitylation is not due to a general failure to modify PCNA. In addition, comparable amounts of PCNA were chromatin-associated in WT and rfa1-td (Figure 4F), suggesting that replication forks remain largely intact when Rfa1 levels are seriously reduced. These findings indicate that RPA is required for damage-dependent ubiquitylation of PCNA during S phase, independent of its function in DNA replication, recombination, and the recruitment of checkpoint factors.
Figure 4.
RPA Is Required for PCNA Ubiquitylation
(A) PCNA is ubiquitylated normally in the rfa1-t11 and rfa1-t33 mutants. MMS-induced PCNA ubiquitylation was detected as described in the legend to Figure 1 in WT (RFA1) and two rfa1 mutants.
(B) DNA association of the mutant protein encoded by rfa1-t33 is indistinguishable from the WT protein at 25°C and 37°C. Whole-cell extracts (W) were fractionated by centrifugation into soluble (S) and chromatin-associated (C) material. Proteins were detected by western blot. The distributions of the chromatin-associated Orc6 protein and soluble PGK served as controls for the quality of fractionation.
(C) Experimental strategy to deplete Rfa1td from yeast cells. In the rfa1-td strain, the construct encoding the heat-labile Rfa1td protein is controlled by a doxycycline (Dox)-repressible promoter, and UBR1, encoding the E3 responsible for degradation of Rfa1td, is induced by galactose (Gal). Rfa1td remains stable at 25°C in raffinose (Raf) medium. Temperature effects were excluded by returning the cells to 25°C without allowing resynthesis of Rfa1td before releasing them into S phase.
(D) DNA replication proceeds after depletion of the Rfa1td protein. FACS samples were taken before and 50 min after release from G1 arrest following the scheme shown in (C).
(E) Loss of damage-induced PCNA ubiquitylation after depletion of the Rfa1td protein. HisPCNA and its modified forms were detected as in Figure 1. Rfa1 was detected in total cell extracts.
(F) Chromatin association of PCNA in RFA1 and rfa1-td. Chromatin-binding assays were performed as described in (B). Note that the Rfa1td protein remaining after depletion is entirely associated with chromatin. The degron tag is partially cleaved from the protein during extract preparation.
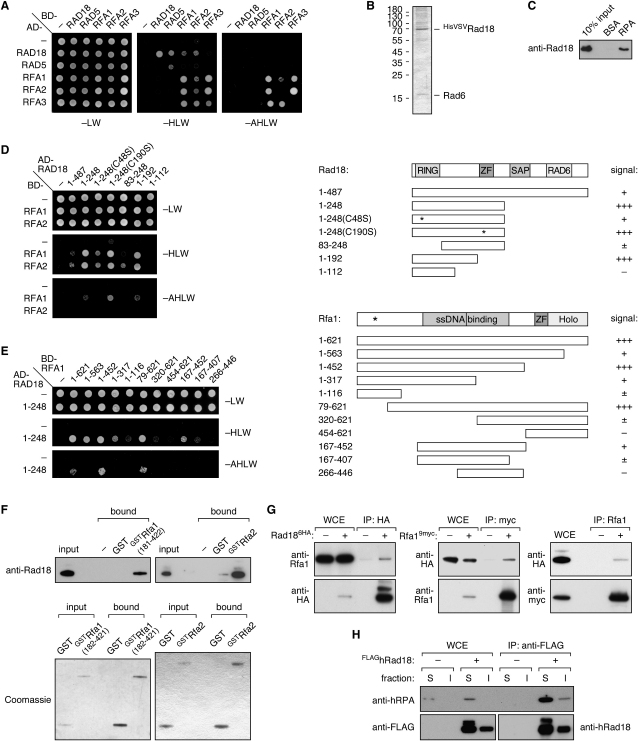
Rad18 Interacts with the RPA Complex
We asked whether RPA contributes directly to the recruitment of the ubiquitylation machinery to stalled replication intermediates by examining possible interactions between Rad18, Rad5, and the RPA subunits. In the two-hybrid system, we observed an interaction of Rad18, but not of Rad5, with the subunits Rfa1 and Rfa2 in addition to mutual interactions among the three RPA subunits (Figure 5A). In order to exclude indirect effects, we examined the interaction of recombinant yeast Rad6-Rad18 complex (Figure 5B) with RPA immobilized on Sepharose beads. Purified HisVSVRad18 was retained on the beads in the presence of a nuclease, indicating that the interaction was direct and not mediated by DNA or other cellular factors (Figure 5C).
Figure 5.
Interaction of Rad18 with the RPA Complex
(A) Protein-protein interactions between Rad18, Rad5, and the RPA subunits Rfa1, Rfa2, and Rfa3 in the yeast two-hybrid system. The open reading frames were expressed as fusions to the Gal4 activation (AD) and DNA-binding domains (BD), and the presence of the constructs was confirmed by growth on selective medium (-LW). Positive interactions were scored by growth on plates lacking histidine (-HLW) and stronger interactions on plates lacking histidine and adenine (-AHLW). Interactions between Rad18 and Rad5 (Ulrich and Jentsch, 2000) and between the individual RPA subunits are shown as internal controls.
(B) Coomassie-stained gel of recombinant HisVSVRad18 isolated in complex with untagged Rad6 from baculovirus-infected insect cells.
(C) Interaction between purified recombinant RPA, covalently coupled to Sepharose, and HisVSVRad18. HisVSVRad18 retained on the RPA beads was eluted in Laemmli buffer and detected by western blotting. BSA-coupled beads served as negative control.
(D) Two-hybrid analysis of the Rad18 domains interacting with Rfa1 and Rfa2. The scheme on the right indicates the RING domain, a Zinc finger (ZF), a SAP domain, and the region relevant for interaction with Rad6. Positions C48 and C190 are indicated by asterisks in the respective mutants.
(E) Two-hybrid analysis of the Rfa1 domains interacting with Rad18. The scheme on the right indicates the regions relevant for ssDNA binding and holocomplex formation as well as a Zinc finger domain. The position of the rfa1-t11 mutation (Umezu et al., 1998) is indicated by an asterisk.
(F) Interaction of Rad18 with the ssDNA-binding domain of Rfa1 and with Rfa2 in vitro. Purified recombinant GSTRfa1(182–421) or Rfa2, immobilized on glutathione Sepharose, was incubated with HisVSVRad18. Bound material was eluted and analyzed by western blot to detect Rad18 (upper panel) or by Coomassie staining to detect the GST constructs (lower panel).
(G) In vivo interaction of Rad18 with RPA was analyzed by coimmunoprecipitation of yeast Rad186HA and Rfa19myc by using the indicated antibodies. Strains expressing untagged RAD18 or RFA1 served as negative controls for precipitation of Rad186HA and Rfa19myc, respectively. In the anti-Rfa1 precipitation, antibody was omitted as a negative control. Bands of higher molecular weight in anti-HA precipitates represent ubiquitylated Rad186HA.
(H) Interaction between hRad18 and hRPA in cultured human cells. Immunoprecipitations (anti-FLAG) were performed from extracts derived from HEK293T cells transiently transfected with an expression construct for FLAGhRad18. Cells transfected with the empty vector served as negative control. Extracts were partitioned into detergent-soluble (S) and -insoluble (I) material. Note that FLAGhRad18 is monoubiquitylated in the soluble fraction, which includes the cytoplasmic material, but not in the chromatin-associated fraction.
Analysis of truncations and mutations of Rad18 in the two-hybrid system revealed that an N-terminal portion of Rad18, including the RING domain, is sufficient for interaction, whereas the central Zinc finger and a DNA-binding SAP domain (Notenboom et al., 2007) are irrelevant (Figure 5D). Within Rfa1, we mapped the interaction site to the central region spanning amino acids 167–452 (Figure 5E), which comprises the primary ssDNA-binding motifs of RPA. This interaction, as well as the association between Rad18 and Rfa2, was confirmed in vitro using recombinant constructs GSTRfa1(182–421), GSTRfa2, and HisVSVRad18 (Figure 5F). Thus, both subunits, Rfa1 and Rfa2, contribute independently to the interaction with the E3.
Coimmunoprecipitations from strains bearing epitope-tagged RAD186HA and RFA19myc in their native contexts confirmed the association of Rad18 with RPA in vivo (Figure 5G). The interaction did not vary measurably between damaged and undamaged cells (data not shown). Considering the conservation of PCNA modification in eukaryotes, we asked whether interaction of Rad18 with RPA was detectable in human cells. Upon transfection with an expression construct for FLAG-tagged human Rad18, isolation of FLAGhRad18 indeed resulted in copurification of endogenous hRPA both from the cytoplasmic and the chromatin-associated fraction (Figure 5H). Addition of nuclease did not abolish the interactions in yeast or mammalian extracts (data not shown), indicating that they are primarily mediated by protein-protein contacts. These observations raised the question of whether Rad18 is recruited to ssDNA by means of its interaction with RPA.
Association of Rad18 with ssDNA Correlates with that of RPA Even in the Absence of PCNA
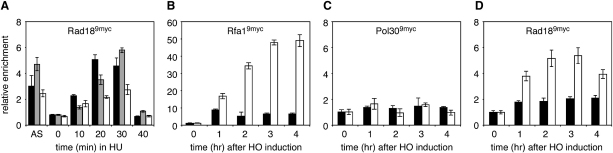
Chromatin immunoprecipitations (ChIPs) from yeast cells bearing a RAD189myc allele provided evidence for association of Rad18 with chromatin in vivo. In HU-treated cells, Rad189myc, like RPA and PCNA (Papouli et al., 2005), was detectable at replication forks in the vicinity of a replication origin (Figure 6A). In order to exclude the possibility that the presence of Rad18 at this site was merely due to its interaction with PCNA, we analyzed the chromatin association of Rad18 at stretches of ssDNA that result from the resection of a DSB induced by the HO endonuclease (Lee et al., 1998). In this context, RPA initially associates with sequences adjacent to the break but is displaced by the recombinogenic Rad51 filament in a Rad52-dependent manner (Wang and Haber, 2004) (Figure 6B). Although PCNA is easily detectable at replication forks by ChIP (Papouli et al., 2005), the protein was absent from the DSB (Figure 6C). We had previously failed to observe significant Rad189myc association by ChIP using multiplex PCR (Chen et al., 2005); however, with a more sensitive real-time PCR method, we were able to detect a 2-fold enrichment after induction of the HO endonuclease (Figure 6D). This small but reproducible signal is likely to be significant, as it was abolished in an mre11 mutant, in which ssDNA forms more slowly (Lee et al., 1998) (Figure S4). Importantly, inhibition of Rad51 filament formation by deletion of RAD52 resulted in strongly enhanced association of Rad189myc (Figure 6D), consistent with elevated RPA levels on the ssDNA in this mutant (Wang and Haber, 2004) (Figure 6B). Thus, the amount of Rad18 on ssDNA correlates with that of RPA even where PCNA is not present.
Figure 6.
In Vivo Association of Rad18 with ssDNA
(A) Association of Rad189myc with a replication fork in HU-treated cells detected by ChIP after release from G1 arrest. Rad189myc was precipitated (anti-myc) from formaldehyde-crosslinked cells at the indicated times, and associated DNA was quantified by real-time PCNA using primers specific for an early-firing origin, ARS607 (black); a sequence 4 kbp removed from ARS607 (gray); and a late-firing origin, ARS501 (white). AS, asynchronous cells. Error bars in all panels represent combined standard deviations from three independent amplifications.
(B) Rfa19myc is detectable by ChIP at sequences adjacent to an HO-induced DSB. Enrichment of sequences adjacent to the HO site (MAT) relative to those at an unrelated locus (ACT1) in immunoprecipitates (anti-myc) from crosslinked cells was followed over the indicated time using real-time PCR as in (A). ChIP assays were performed in WT (black) and rad52 mutants (white).
(C) PCNA (Pol309myc) is not detectable by ChIP at the site of an HO-induced DSB in WT (black) or rad52 (white) cells.
(D) Association of Rad189myc next to a DSB, analyzed as above in WT (black) and rad52 (white) cells.
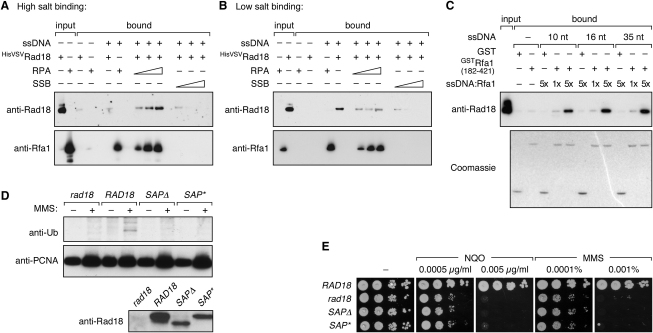
RPA Can Recruit Rad18 to ssDNA In Vitro
To gain insight into how RPA might associate with Rad18 on ssDNA, we examined the effect of RPA on the DNA binding of HisVSVRad18 to an immobilized 75 nt oligonucleotide in vitro. At 250 mM sodium chloride, binding of HisVSVRad18 by itself was undetectable, but increasing levels of RPA afforded a robust and RPA-specific enrichment of the E3 in the bound material, indicating that RPA recruits HisVSVRad18 to ssDNA (Figure 7A). Similar results were obtained at 150 mM NaCl (data not shown). Surprisingly, under low-salt conditions, HisVSVRad18 alone associated with the oligonucleotide, and RPA now caused a reduction in the amount of bound HisVSVRad18 (Figure 7B). However, comparable quantities of the bacterial ssDNA-binding protein, SSB, competed much more effectively with HisVSVRad18 than RPA, indicating that, even at low ionic strength, RPA and the E3 can occupy a common stretch of DNA.
Figure 7.
Effects of DNA on the Interaction between Rad18 and RPA
(A) Recruitment of HisVSVRad18 to RPA-coated ssDNA under high-salt conditions. A 75 nt 5′-biotinylated oligonucleotide was immobilized on streptavidin agarose and preincubated with increasing amounts of purified yeast RPA, and retention of HisVSVRad18 was analyzed by western blotting. RPA and HisVSVRad18 were allowed to bind in a buffer containing 15 mM KCl and 250 mM NaCl.
(B) DNA binding of HisVSVRad18 under low-salt conditions. The experiment was performed exactly as in (A), except that the proteins were allowed to bind to the ssDNA at 15 mM KCl.
(C) Preincubation of short oligonucleotides with GSTRfa1(182–421) at a ratio of 1:1 or 5:1 enhances interaction with Rad18. Pull-down assays were performed as described in the legend to Figure 5F.
(D) Deletion (Δ) or mutation (∗) of the Rad18 SAP domain impairs PCNA ubiquitylation in vivo. The rad18 deletion mutant was complemented by the indicated alleles, and PCNA modifications were analyzed as in Figure 1. Presence of the mutant proteins was verified by immunoprecipitation and western blot (lower panel).
(E) The Rad18 SAP domain is required for protection from DNA damage in vivo. Spot assays for NQO and MMS sensitivity were performed on a rad18 deletion mutant complemented by the indicated RAD18 constructs.
In order to assess in a simplified system whether RPA can simultaneously interact with ssDNA and Rad18, we examined the interaction of the E3 with GSTRfa1(182–421) in the presence of short oligonucleotides with no measurable affinity for Rad18 (data not shown) and varying affinities for Rfa1 (Fanning et al., 2006) (Figure S5). We found that ssDNA prebound to Rfa1 enhanced retention of Rad18, suggesting that Rfa1 in its DNA-bound conformation interacts with Rad18 better than free in solution (Figure 7C). An interaction of RPA with Rad18 not involving DNA binding of Rad18 itself thus appears possible. Yet, the Rad18 SAP domain, responsible for DNA binding of the human protein (Notenboom et al., 2007), was previously shown to be involved in the assembly of the E3 at replication foci (Nakajima et al., 2006). Inactivation of this domain in yeast Rad18 abolished its function in vivo, as judged by PCNA modification and DNA damage sensitivity (Figures 7C and 7D), despite the ability of the mutants to autoubiquitylate in vitro (data not shown). Hence, DNA binding by Rad18 itself also appears to be important for its activity toward PCNA.
Discussion
RPA has been implicated in virtually all eukaryotic DNA transactions (Fanning et al., 2006; Zou et al., 2006). Its stabilizing effect on ssDNA is essential for replication, recombination, and nucleotide excision repair, but the complex engages in DNA metabolism even more directly by means of protein-protein interactions, for example with DNA polymerase α/primase or the recombination protein Rad52. During S phase, RPA-coated ssDNA signals replication stress by recruiting the PCNA-like 9-1-1 clamp and the ATR/ATRIP checkpoint kinase complex to sites where polymerase stalling has resulted in exposure of extended stretches of ssDNA (Zou and Elledge, 2003; Zou et al., 2003). Our findings now point to an even more general role of RPA in dealing with replication problems that appears to be conserved from yeast to mammals.
RPA Contributes Independently to Damage Signaling and Damage Bypass
Loss of PCNA ubiquitylation upon depletion of Rfa1 implicates RPA in the activation of the RAD6 pathway, suggesting that the complex is responsible not only for the sensing of damaged DNA and the stabilization of stalled forks but also for initiating the steps necessary to overcome the damage that caused the arrest. Our results indicate that the requirements of RPA for replication and recombination are genetically and physically separable from its role in DNA damage bypass.
Identification of RPA-covered ssDNA as an upstream signal for the RAD6 pathway explains a large body of circumstantial evidence defining the conditions that induce PCNA ubiquitylation. Although experiments with synchronized cells suggest that DNA damage causes PCNA ubiquitylation primarily during S phase, the cell-cycle stage itself is irrelevant for the modification: on one hand, a replication initiation defect abolishes PCNA ubiquitylation under conditions that physiologically resemble S phase. On the other hand, the reaction can be triggered in G1 by stalled replication intermediates resulting from the processing of DNA interstrand crosslinks (Sarkar et al., 2006). Hence, stalled replication intermediates appear to be both necessary and sufficient for activation of PCNA ubiquitylation.
A need for ssDNA at the sites of stalled replication intermediates is implied by our inability to detect PCNA modification in response to DSB-inducing agents that do not cause base damage. Even CPT, which induces strand breaks in a replication-dependent manner and stalls replication forks, did not result in PCNA ubiquitylation, consistent with our finding that this drug does not cause accumulation of ssDNA. By similar reasoning, uncoupling between polymerase and helicase has been suggested to activate damage bypass in X. laevis egg extracts (Chang et al., 2006).
Elevated levels of Rad18 can overcome the requirement for fork stalling and promote ubiquitylation of PCNA whenever the clamp is associated with DNA (Figure 1B). This suggests that the E3 is rate limiting for the reaction, and higher concentrations will drive ubiquitylation at shorter or more transient stretches of ssDNA during undisturbed replication. Constitutive modification of PCNA, as observed in S. pombe and in X. laevis egg extracts (Frampton et al., 2006; Leach and Michael, 2005), might therefore be caused by a higher expression level or a lower activation threshold of Rad18 in these systems. A higher incidence of spontaneous fork stalling or intrinsic damage might have the same effect.
Finally, a placement of RPA-covered ssDNA upstream of PCNA ubiquitylation explains the similarities between the signals for modification of the clamp and those that elicit the replication-dependent S phase checkpoint (Branzei and Foiani, 2005). Given that the domains required for the recruitment of the checkpoint factors and for interaction with Rad18 are distinct, the two pathways—damage sensing and damage bypass—appear to originate independently from a common structure. Intriguingly, a reduction in PCNA ubiquitylation has been reported in mammalian cells upon siRNA-mediated downregulation of ATR (Bi et al., 2006). Hence, while damage tolerance is clearly independent of checkpoint function in S. cerevisiae (Figure S1), S. pombe, and X. laevis (Chang et al., 2006; Frampton et al., 2006), an indirect contribution of the kinase in the mammalian system remains possible.
Interactions of RPA and Rad18 on ssDNA
RPA is well known for a complicated DNA-binding behavior that involves several distinct binding modes and varies considerably with ionic strength (Fanning et al., 2006). Interactions with other cellular factors may involve either one or multiple of its subunits, and these interactions often induce conformational changes of RPA. Recruitment of other proteins by RPA can involve cooperative binding but also a “trading of places” whereby a replication or repair factor would replace the RPA domains in a stepwise manner (Fanning et al., 2006). Our in vitro experiments reveal that both protein-protein and protein-DNA interactions are likely to contribute to the recruitment of Rad18 to ssDNA. In particular, we found that the affinity of Rad18 for ssDNA and RPA strongly depends on ionic strength. While modulation of the reaction buffer imposes rather artificial conditions on the RPA-Rad18-ssDNA interaction, our experiments have nevertheless revealed two patterns that might explain in principle how the two proteins can interact on DNA: at low ionic strength, Rad18 exhibits detectable affinity for ssDNA by itself but does not engage in cooperative interactions with RPA. In physiological salt conditions, however, RPA is clearly necessary for recruitment of Rad18 to ssDNA, and interaction of Rad18 with the DNA-binding domain of Rfa1 is enhanced when the latter is bound to ssDNA (Figure 7). Whether the association in vivo relies solely on protein-protein interactions or whether RPA and Rad18 bind cooperatively to the DNA needs to be determined. A change between the two interaction modes during recruitment of Rad18 would explain the requirement of both RPA and the Rad18 SAP domain for PCNA ubiquitylation. However, the SAP domain may also have other functions in addition to DNA binding.
Implications of the Rad18-RPA Interaction for Rad18 Function
The physical interactions between RPA and Rad18, detectable in yeast and mammalian cells, suggest that RPA recruits Rad18 to ssDNA by direct association. In the context of a stalled replication fork, it is likely that additional cellular factors will also influence the behavior of Rad18. For example, interaction of the E3 with its target, PCNA, might affect its DNA-binding properties. Alternatively, Rad18 might undergo a conformational change upon DNA binding that would in turn modulate its interactions with PCNA, RPA, or Rad5. Notably, C-terminal truncations of Rad18 excluding the DNA-binding SAP domain consistently exhibit stronger interaction signals than the full-length protein in two-hybrid assays (Figure 5D) (Ulrich and Jentsch, 2000), possibly indicating an inhibitory function of the C-terminal domain that might be relieved by DNA binding. Further biochemical studies and isolation of selectively interaction-deficient mutants of both Rad18 and RPA will be necessary to define the molecular mechanism by which RPA targets Rad18 to its sites of action. However, the presence of Rad18 at ssDNA in vivo correlates with that of RPA even in the absence of its ubiquitylation target, PCNA. Association of the E3 with HO-induced DSBs might therefore suggest additional, PCNA-independent functions. Consistent with this notion, yeast rad18, but not ubiquitylation-deficient PCNA mutants, are highly sensitive to IR (Chen et al., 2005; Friedl et al., 2001), and the phenotypes of vertebrate rad18−/− cells have implicated the E3 in the repair of DNA-strand breaks and recombination (Shiomi et al., 2007; Szuts et al., 2006). However, the relevant ubiquitylation targets as well as possible contributions of the RPA complex to these alternative functions remain to be identified.
Experimental Procedures
Yeast Strains and Plasmids
Standard procedures were followed for the cultivation of yeast. Mutants in rad18, rad53, and rad52 as well as HisPOL30 and RAD186HA have been described previously (Stelter and Ulrich, 2003; Ulrich and Jentsch, 2000), and other epitope-marked alleles were created in the same background. The cdc7ts allele was transferred into HisPOL30 by cloning of the open reading frame with flanking regions into a URA3 plasmid, integration into the CDC7 locus, selection for loss of the marker, and screening for temperature-sensitive growth. Mutants bob1, bob1 cdc7Δ, and an isogenic WT (Hardy et al., 1997) were modified by introduction of HisPOL30. ChIP experiments at HO-induced DSBs were performed in the background of JKM179 (Lee et al., 1998). The rfa1-t11 strain was generated in HisPOL30 by direct replacement of the WT allele with a URA3-marked construct. Experiments involving rfa1-t33 were performed in the S288c background by introduction of HisPOL30 into isogenic RFA1 and rfa1-t33 strains (Umezu et al., 1998). The rfa1-td strain carries the degron allele (Dohmen et al., 1994) under control of a dual tetracycline-repressible promoter system (Belli et al., 1998). Both rfa1-td and its isogenic WT carry an integrative plasmid expressing mycUBR1 under control of the GAL1 promoter and the HisPOL30 allele in addition to endogenous POL30. Overexpression of RAD18 was accomplished using an integrative vector with the GAL1 promoter. For complementation of rad18 with WT, SAPΔ (amino acids 279–312), or SAP∗ (G299A, R301A, M304A), we used integrative vectors carrying the respective alleles under control of the RAD18 promoter. Strains with a bar1 deletion were used for experiments involving G1 synchronization, except for those used in Figure 4. Details of strains and plasmids are available on request.
Antibodies
Rabbit polyclonal antibodies were obtained from S. Brill (Rfa1), J. Diffley (Sic1, Rad53, Orc6), or S. West (hRPA); purchased from Santa Cruz Biotechnology (Clb2, myc, HA) or Abcam (hRad18); or generated for our lab (PCNA, Rad18); monoclonals were from Molecular Probes (PGK), Sigma (FLAG), Cell Signaling Technology (ubiquitin, P4D1) or Cancer Research UK (myc, 9E10; HA, 12CA5).
Detection of PCNA Modifications
Strains bearing the HisPOL30 allele were subjected to the desired treatment and processed for isolation of HisPCNA by Ni-NTA pull-downs under denaturing conditions followed by detection of PCNA and ubiquitin as described previously (Hoege et al., 2002; Stelter and Ulrich, 2003). Responses to DNA damage were analyzed by treatment with HU, MMS, NQO, Bleo, CPT, or H2O2 in liquid YPD medium for 90 min. For UV irradiation, exponentially growing cultures were plated onto solid YPD medium, irradiated at 254 nm (UV Stratalinker 2400, Stratagene), resuspended in fresh medium, and incubated for 40 min prior to analysis. Treatment with IR was performed with a 137Cs γ source in liquid YPD. Cell-cycle arrests in G1, S, and G2 were achieved by treatment with α factor (10 ng/ml), HU (100 mM), or nocodazole (15 μg/ml), respectively, and monitored by propidium iodide staining and flow cytometry.
Induction of Rfa1td Degradation
Cells were pregrown in raffinose medium at 25°C and blocked in G1 with 5 μg/ml α factor for 3 hr. After exchange into galactose medium with 100 μg/ml doxycyclin and α factor and incubation for 1 hr, the temperature was raised to 37°C, and incubation was continued for 2 hr. After returning the cultures to 25°C and addition of more α factor (2 μg/ml), cells were incubated for another 2 hr before being released into S phase by washing in galactose/doxycyclin medium. Control cultures were blocked in G1 and kept in raffinose medium at 25°C for 3 hr before release in raffinose medium. Damage was induced after release by addition of 0.02% MMS for 90 min.
Analysis of DNA Damage Sensitivities and Checkpoint Activation
Sensitivities to Bleo, CPT, NQO, and MMS were analyzed by spot assays on YPD plates containing the indicated concentrations of damaging agents as described previously (Stelter and Ulrich, 2003). Strains were spotted onto the plates in 10-fold serial dilutions. Phosphorylation of Rad53 was followed by preparation of total cell extracts under denaturing conditions and western blotting.
Analysis of Protein-Protein Interactions in the Two-Hybrid System
Protein-protein interactions were monitored in the reporter strain PJ69-4A as described previously (Ulrich and Jentsch, 2000) by using constructs based on fusions to the Gal4 activation and DNA-binding domains. Constructs for full-length RAD18 and RAD5 have been described (Ulrich and Jentsch, 2000). Mutations C48S and C190S were introduced by PCR. The RFA1, RFA2, and RFA3 open reading frames were amplified by PCR from genomic DNA. Truncations were generated using appropriate internal restriction sites.
Immunoprecipitations in Yeast
Total cell extracts were prepared from yeast strains expressing RFA19myc and/or RAD186HA by lysis with zirconium/silica beads in 50 mM Tris-HCl (pH 7.5), 250 mM KCl, 20% glycerol, 1% NP40, 2 mM DTT, and protease inhibitors. Immunoprecipitations were performed from ∼6 × 108 cells (∼4 mg total protein) by incubation with 3–5 μg antibody for 2 hr, addition of 20 μl Protein G agarose (Roche), and further incubation for 1 hr followed by washing five times with lysis buffer and once with the same buffer containing no detergent.
Immunoprecipitations in Mammalian Cell Culture
HEK293T cells were transiently transfected with a construct encoding FLAG-tagged human Rad18 (Mulder et al., 2002). After 3 days, cell extracts were prepared from one 90 mm plate per immunoprecipitation in 50 mM Tris-HCl (pH 7.5), 25 mM NaCl, 1.05 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% NP40, and protease inhibitors. Soluble material was recovered by centrifugation at 2000 rpm for 5 min. The pellet was washed twice with lysis buffer and extracted with 50 mM Tris-HCl (pH 7.5), 125 mM NaCl, 1 mM EDTA, 0.5% NP40, and protease inhibitors for 30 min on ice to yield the detergent-insoluble chromatin fraction. Both extracts were centrifuged at 13,000 rpm for 15 min, and the lysates were precleared with Protein G agarose for 1 hr. Immunoprecipitations were performed with 20 μl anti-FLAG agarose (Sigma) for 3 hr. The beads were washed four times with 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1% NP40, and protease inhibitors and twice with the same buffer without NP40.
Chromatin Immunoprecipitations
ChIP assays at replication forks were performed as described previously (Papouli et al., 2005). A full description of the assay conditions for ChIP at HO-induced DSBs (Chen et al., 2005) is available in the Supplemental Data.
Chromatin-Binding Assays
Fractionation of total cell extracts prepared by spheroplast lysis into soluble and chromatin-bound fractions was achieved by centrifugation through a sucrose cushion essentially as described (Liang and Stillman, 1997).
Quantification of ssDNA
After the desired treatment of exponential cultures, genomic DNA was isolated from ∼5 × 107 cells with the DNeasy Blood & Tissue Kit (QIAGEN) according to the manufacturer's instructions. Klenow enzyme (0.5 U) with 0.5 μl hexanucleotide mix (Roche), 7.5 μCi 32P-dCTP, and 5 mM each of cold dATP, dGTP, and dTTP were used in 5 μl random-primed labeling reactions on 100 ng of nondenatured DNA for 16 hr at room temperature (RT). Unincorporated nucleotides were removed by two steps of ammonium acetate precipitation. Aliquots of 2 μl spotted in duplicate on nitrocellulose were quantified by phosphorimaging. A correction factor for the exact amount of input DNA was determined by real-time PCR quantification of the template with primers specific for the PAC2 gene (5′-AATAACGAATTGAGCTATGACACCAA-3′ and 5′-AGCTTACTCATATCGATTTCATACGACTT-3′). Standard deviations were calculated from triplicate measurements.
Protein Preparations
Bacterial SSB was from Promega. Yeast RPA was purified from E. coli as described (Sibenaller et al., 1998). The expression constructs for GSTRfa1(181–422) and GSTRfa2 were generated in pGEX-4T-3 (GE Healthcare); the proteins were produced in E. coli and purified by affinity chromatography on glutathione Sepharose. Expression constructs for yeast HisVSVRad18 and untagged Rad6 were generated in the vector pFastBAC (Invitrogen). The proteins were produced in Sf9 insect cells and purified as a complex from cytoplasmic extracts on Ni-NTA agarose. Thus, Rad6 was present in all experiments involving HisVSVRad18. Protein concentrations were determined by comparison to a BSA standard.
Analysis of Protein-Protein Interactions In Vitro
Interaction of HisVSVRad18 with RPA in the absence of DNA was examined by covalent coupling of yeast RPA to activated CH Sepharose 4B (at 2.5 μg/ml) and monitoring retention of HisVSVRad18 in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 0.05% Triton X-100, and 2.5 U benzonase (Novagen). Beads (20 μl) were incubated with ∼2 pmol HisVSVRad18 for 2 hr at 4°C and washed five times with the same buffer containing 250 mM NaCl. BSA-derivatized beads served as negative control. Bound material was eluted with Laemmli buffer and analyzed by western blotting.
Interaction of Rad18 with GSTRfa1(182–421) or GSTRfa2 was assayed by immobilizing 0.5 nmol fusion protein on glutathione Sepharose for 30 min at 4°C, followed by a brief wash in a buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, and 0.05% Triton X-100. HisVSVRad18 (10 pmol) was added in the same buffer containing 100 μg/ml BSA and incubated for 60 min at 4°C. Beads were washed five times in buffer without BSA, and bound material was eluted as above and analyzed by western blotting and Coomassie blue staining. Where indicated, GST or GSTRfa1(182–421) was incubated with a 1× or 5× molar excess of oligo-dT (10, 16, or 35 nt) for 45 min before binding to the beads.
Binding of HisVSVRad18 to RPA-covered ssDNA was examined by immobilizing a 75 nt, 5′-biotinylated oligonucleotide of mixed sequence on streptavidin agarose (Pierce) in phosphate-buffered saline (PBS) with 1 mM EDTA for 1 hr at RT. After washing three times in PBS with 1 mM EDTA and three times in binding buffer (25 mM HEPES-NaOH [pH 7.5], 15 mM KCl, 1 mM EDTA, 0.05% Triton X-100, 0.5 mM DTT, 100 μg/ml BSA, supplemented with 150 or 250 mM NaCl for high-salt conditions), 15 μl beads per reaction (∼5 pmol of the oligonucleotide) were incubated with increasing amounts of purified yeast RPA or bacterial SSB (2.4, 8, and 24 pmol) for 30 min at 4°C. Beads were washed once, and ∼5 pmol HisVSVRad18 was added. Incubation was continued for 1 hr at 4°C, and the beads were washed three times with high-salt binding buffer and twice with the same buffer containing no BSA. Bound material was eluted as above and detected by western blotting. For RPA blots, equal fractions of input and bound material were loaded. For Rad18 blots, 1% of the input and 35% of the bound material were loaded.
Acknowledgments
We thank T. Sixma for helpful discussions and communication of unpublished results; C. Zierhut and J. Diffley for the gift of and advice on the rfa1-td strain; and S. Brill, J. Diffley, R. Kolodner, L. Mulder, R. Sclafani, S. West, and M. Wold for generous gifts of reagents. This work was funded by Cancer Research UK, the German Ministry for Education and Research (BMBF), the EMBO Young Investigator Programme and a predoctoral fellowship from the Boehringer Ingelheim Foundation (to D.H.).
Published: March 13, 2008
Footnotes
Supplemental Data include Supplemental Experimental Procedures and five figures and can be found with this article online at http://www.molecule.org/cgi/content/full/29/5/625/DC1/.
Supplemental Data
References
- Belli G., Gari E., Piedrafita L., Aldea M., Herrero E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–947. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Barkley L.R., Slater D.M., Tateishi S., Yamaizumi M., Ohmori H., Vaziri C. Rad18 regulates DNA polymerase κ and is required for recovery from S-phase checkpoint-mediated arrest. Mol. Cell. Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005;17:568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Chang D.J., Lupardus P.J., Cimprich K.A. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J. Biol. Chem. 2006;281:32081–32088. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- Chen S., Davies A.A., Sagan D., Ulrich H.D. The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res. 2005;33:5878–5886. doi: 10.1093/nar/gki902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R.J., Wu P., Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Fanning E., Klimovich V., Nager A.R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J., Irmisch A., Green C.M., Neiss A., Trickey M., Ulrich H.D., Furuya K., Watts F.Z., Carr A.M., Lehmann A.R. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol. Biol. Cell. 2006;17:2976–2985. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl A.A., Liefshitz B., Steinlauf R., Kupiec M. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res. 2001;486:137–146. doi: 10.1016/s0921-8777(01)00086-6. [DOI] [PubMed] [Google Scholar]
- Garg P., Burgers P.M. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl. Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F., Dryga O., Seematter S., Pahl P.M., Sclafani R.A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J. Bacteriol. 1973;115:966–974. doi: 10.1128/jb.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Kannouche P.L., Wing J., Lehmann A.R. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- Lawrence C. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays. 1994;16:253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- Leach C.A., Michael W.M. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J. Cell Biol. 2005;171:947–954. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lehmann A.R. Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat. Res. 2002;509:23–34. doi: 10.1016/s0027-5107(02)00227-0. [DOI] [PubMed] [Google Scholar]
- Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F., Desai S.D., Li T.K., Mao Y., Sun M., Sim S.P. Mechanism of action of camptothecin. Ann. N Y Acad. Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- Maniar H.S., Wilson R., Brill S.J. Roles of replication protein-A subunits 2 and 3 in DNA replication fork movement in Saccharomyces cerevisiae. Genetics. 1997;145:891–902. doi: 10.1093/genetics/145.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L.C., Chakrabarti L.A., Muesing M.A. Interaction of HIV-1 integrase with DNA repair protein hRad18. J. Biol. Chem. 2002;277:27489–27493. doi: 10.1074/jbc.M203061200. [DOI] [PubMed] [Google Scholar]
- Nakajima S., Lan L., Kanno S., Usami N., Kobayashi K., Mori M., Shiomi T., Yasui A. Replication-dependent and -independent responses of RAD18 to DNA damage in human cells. J. Biol. Chem. 2006;281:34687–34695. doi: 10.1074/jbc.M605545200. [DOI] [PubMed] [Google Scholar]
- Notenboom V., Hibbert R.G., van Rossum-Fikkert S.E., Olsen J.V., Mann M., Sixma T.K. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 2007;35:5819–5830. doi: 10.1093/nar/gkm615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- Papouli E., Chen S., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Redon C., Pilch D.R., Rogakou E.P., Orr A.H., Lowndes N.F., Bonner W.M. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 2003;4:678–684. doi: 10.1038/sj.embor.embor871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Davies A.A., Ulrich H.D., McHugh P.J. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase ζ. EMBO J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi N., Mori M., Tsuji H., Imai T., Inoue H., Tateishi S., Yamaizumi M., Shiomi T. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibenaller Z.A., Sorensen B.R., Wold M.S. The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- Sogo J.M., Lopes M., Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- Stelter P., Ulrich H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Szuts D., Simpson L.J., Kabani S., Yamazoe M., Sale J.E. Role for RAD18 in homologous recombination in DT40 cells. Mol. Cell. Biol. 2006;26:8032–8041. doi: 10.1128/MCB.01291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H.D. The RAD6 pathway: control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. ChemBioChem. 2005;6:1735–1743. doi: 10.1002/cbic.200500139. [DOI] [PubMed] [Google Scholar]
- Ulrich H.D., Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K., Sugawara N., Chen C., Haber J.E., Kolodner R.D. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Haber J.E. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2004;2:E21. doi: 10.1371/journal.pbio.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L., Liu D., Elledge S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Liu Y., Wu X., Shell S.M. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.