Abstract
Genetic association studies can be used to identify factors that may contribute to disparities in disease evident across different racial and ethnic populations. However, such studies may not account for potential confounding if study populations are genetically heterogeneous. Racial and ethnic classifications have been used as proxies for genetic relatedness. We investigated genetic admixture and developed a questionnaire to explore variables used in constructing racial identity in two cohorts – 50 African Americans (AAs) and 40 Nigerians. Genetic ancestry was determined by genotyping 107 ancestry informative markers. Ancestry estimates calculated with maximum likelihood estimation were compared with population stratification detected with principal component analysis. Ancestry was approximately 95% west African, 4% European, and 1% Native American in the Nigerian cohort and 83% west African, 15% European, and 2% Native American in the AA cohort. Therefore, self-identification as AA agreed well with inferred west African ancestry. However, the cohorts differed significantly in mean percentage west African and European ancestries (P < 0.0001) and in the variance for individual ancestry (P ≤ 0.01). Among AAs, no set of questionnaire items effectively estimated degree of west African ancestry, and self-report of a high degree of African ancestry in a three-generation family tree did not accurately predict degree of African ancestry. Our findings suggest that self-reported race and ancestry can predict ancestral clusters, but do not reveal the extent of admixture. Genetic classifications of ancestry may provide a more objective and accurate method of defining homogenous populations for the investigation of specific population-disease associations.
Keywords: African American, race, ancestry, genetic admixture
Introduction
Genome-wide case-control association studies provide a powerful tool for investigating possible genetic factors that may contribute to the health disparities observed among different racial and ethnic populations. Populations with different ancestral backgrounds may carry different genetic variants, and these may contribute to the variations in disease incidence and outcomes seen in specific racial and ethnic groups(1). Association studies can most easily identify disease-associated alleles when study groups are genetically similar, sharing a similar ancestral background(2). However, individual ancestry is not an easily assayed, simple category, and consequently race continues to be used as a proxy for genetic relatedness in clinical and other biological studies(3-6). There is currently no consensus on how best to examine or characterize different racial or ethnic groups when designing and conducting such studies. Two main approaches have been used to approximate individual ancestry in biologic studies: (1) using self-identified race and ethnicity (SIRE) which may capture common environmental influences as well as ancestral background and (2) genotyping a panel of markers that show large frequency differentials between major geographic ancestral groupings(7, 8). Both approaches have limitations. Self-identified racial categories may not always consistently predict ancestral population clusters, and evidence suggests it may take large sample sizes and numerous markers to describe genetic clusters that correspond to SIRE groupings(9-11). Racial categories are also imprecise and inconsistent, since they may potentially vary within the same individual over time(12, 13). Furthermore, their use risks reinforcing racial divisions in society. On the other hand, more objective analyses that genotype markers which are highly informative for ancestry may not be economically practical and are limited by the requirement of serum or fresh tissue for DNA extraction. Genetically determined ancestry may not capture unmeasured social factors that may impact on differences in health outcomes. There are also unique ethical challenges when linking biological phenotypes with genetic markers for specific racial groups, and caution must always be used when attributing biological differences (e.g., disease risk, treatment response) to different populations.
Understanding the ancestral background of study subjects is most important in genetic studies of admixed populations, such as African Americans (AAs), who represent an admixture of Africans, Europeans, and Native Americans(14). Genetic studies have demonstrated that AAs form a diverse group with percent European admixture estimated to range between 7 and 23%(14-16). Genotyping of self-identified AAs participating in the Cardiovascular Health Study (CHS) revealed that among self-reported Africans there are differences in genetic ancestry that are correlated with some clinically important endpoints(15).
In this study, we compared the degree of genetic admixture in two cohorts – AAs and Nigerians – by genotyping a panel of 107 single nucleotide polymorphisms (SNPs) that are highly informative for ancestry (ancestry informative markers, AIMs). We developed a 26-item questionnaire to explore the variables used in constructing racial identity. We assessed how well self-reported race and ancestry matched genetic ancestry, as determined using our panel of AIMs. We also tested the association between questionnaire responses with degree of west African ancestry to identify questions and combinations of questions that may serve as proxies for estimating the proportion of west African ancestry. Specifically, we assessed whether selfreport of grandparents' ancestry among African Americans could be used to predict genetic ancestry.
Materials And Methods
Study Subjects
Study subjects were self-identified United States (US)-born AAs or Nigerian immigrants from either the Yoruba or Ibo cultures. The west African ancestral population that contributed to our panel of AIMs consisted of 37 people from West Africa, the majority of who were from Nigeria (please refer to Ancestral Populations section and unpublished correspondence). Furthermore, a significant number of Nigerians from the Yoruba and Ibo cultures were known to either work or reside in the communities where recruitment was planned (unpublished correspondence). Thus, we chose Nigerian immigrants as the comparison group. Subjects were recruited from the Washington Heights and Brooklyn communities of New York through postings, newspaper advertisements, and word of mouth, and through discussion with investigators at the Brooklyn campus of Long Island University (LIU). Study recruitment was conducted in collaboration with the Herbert Irving Comprehensive Cancer Center (HICCC) Research Recruitment and Minority Outreach Core, a shared facility for recruitment and retention of human subjects in clinical research which maintains a strong commitment to recruiting minority subjects to clinical trials. This study was approved by the Columbia University Medical Center (CUMC) Institutional Review Board (IRB; Protocol AAAA1500) and Long Island University IRB. Subjects were screened for participation using the following criteria: AAs – subjects identified themselves as AA, were born in the US, and identified both parents as AAs who were born in the US; Nigerians – subjects were from the Yoruba or Ibo cultures, and subjects were either born in Nigeria or both parents were born in Nigeria and immigrated to the US. These were the only entry criteria for study participation, and medical history information was not collected during screening. Subjects consented to participation in the study, donated blood specimens, and completed questionnaires at the General Clinical Research Center at the Irving Center for Clinical Research of Columbia University, CUMC (ICCR; Protocol 3324). Subjects received $40 compensation after completing all aspects of the study.
Ancestral Populations
Ancestral groups that were studied consisted of west African, European, and Native American populations. The west African ancestral population consisted of 37 people from West Africa, and DNA samples were provided by Paul McKeigue. We specifically chose to focus on West Africa since the history of AAs reflects the forced migration of slaves from mainly West Africa(14). The European population consisted of 42 European American samples from Coriell's North American Caucasian panel. The Native American population consisted of 15 people who were Mayan and 15 who were Nahua, with DNA samples provided by Mark Shriver.
Selection Of AIMs
The AIMs used in this study were bi-allelic SNPs that were selected from the Affymetrix 100K SNP chip (Affymetrix, Santa Clara, CA) based on “informativeness” for ancestry in the ancestral population samples genotyped. We used an iterative process for selecting our AIMs since the AA population is a mixture of three ancestral populations, west Africans, Europeans and Native Americans. For each of the three possible pairs of ancestral populations, we identified markers where the difference in allele frequency (δ) was at least 0.5 between any two ancestral populations. Once we identified such markers, we selected a group of 107 AIMs that were adequately distributed across the genome, with the markers being far enough apart that they were in linkage equilibrium in the ancestral populations. The average distance between markers was about 2.4 × 107 bp.
Table 1 lists the AIMs examined, their rs number, chromosomal location, calculated allele frequencies in each ancestral population, and delta values for each ancestral population pairing.
Table 1.
Table of 107 Ancestry Informative Markers
| Allele Frequencies | Delta Values | ||||||
|---|---|---|---|---|---|---|---|
| AIM rs# | Chromosome | African (AF) |
European (EU) |
Native American (NA) |
AF-EU | AF-NA | EU-NA |
| rs1004704 | 16 | 0.028 | 0.214 | 0.867 | 0.187 | 0.839 | 0.652 |
| rs10131076 | 14 | 0.694 | 0.071 | 0.000 | 0.623 | 0.694 | 0.071 |
| rs1013459 | 18 | 0.233 | 0.900 | 0.967 | 0.667 | 0.733 | 0.067 |
| rs10214949 | 7 | 0.281 | 0.845 | 0.967 | 0.564 | 0.685 | 0.121 |
| rs10248051 | 7 | 0.083 | 0.833 | 0.200 | 0.750 | 0.117 | 0.633 |
| rs1036543 | 2 | 0.750 | 0.024 | 0.767 | 0.726 | 0.017 | 0.743 |
| rs10484578 | 6 | 0.944 | 0.375 | 0.067 | 0.569 | 0.878 | 0.308 |
| rs10486576 | 7 | 0.944 | 0.900 | 0.200 | 0.044 | 0.744 | 0.700 |
| rs10488172 | 7 | 0.972 | 0.786 | 0.200 | 0.187 | 0.772 | 0.586 |
| rs10491097 | 17 | 0.056 | 0.647 | 0.133 | 0.592 | 0.078 | 0.514 |
| rs10491654 | 9 | 0.500 | 0.286 | 0.900 | 0.214 | 0.400 | 0.614 |
| rs10492585 | 13 | 0.100 | 0.940 | 1.000 | 0.840 | 0.900 | 0.060 |
| rs10497705 | 2 | 0.118 | 0.238 | 0.867 | 0.120 | 0.749 | 0.629 |
| rs10498255 | 2 | 0.250 | 0.810 | 0.967 | 0.560 | 0.717 | 0.157 |
| rs10498919 | 6 | 0.000 | 0.000 | 0.733 | 0.000 | 0.733 | 0.733 |
| rs10500505 | 16 | 0.056 | 0.214 | 0.800 | 0.159 | 0.744 | 0.586 |
| rs10501474 | 11 | 0.056 | 0.643 | 0.800 | 0.587 | 0.744 | 0.157 |
| rs10506816 | 12 | 0.861 | 0.000 | 0.100 | 0.861 | 0.761 | 0.100 |
| rs10507688 | 13 | 0.000 | 0.167 | 0.733 | 0.167 | 0.733 | 0.567 |
| rs10508349 | 10 | 0.056 | 0.012 | 0.767 | 0.044 | 0.711 | 0.755 |
| rs10510791 | 3 | 0.972 | 0.488 | 0.133 | 0.484 | 0.839 | 0.354 |
| rs10515535 | 5 | 1.000 | 0.286 | 0.133 | 0.714 | 0.867 | 0.152 |
| rs10515919 | 2 | 0.861 | 0.881 | 0.167 | 0.020 | 0.694 | 0.714 |
| rs10517518 | 4 | 0.944 | 0.857 | 0.133 | 0.087 | 0.811 | 0.724 |
| rs10519979 | 4 | 0.167 | 0.451 | 0.967 | 0.285 | 0.800 | 0.515 |
| rs10520440 | 4 | 1.000 | 0.786 | 0.167 | 0.214 | 0.833 | 0.619 |
| rs10520678 | 15 | 0.154 | 0.732 | 1.000 | 0.578 | 0.846 | 0.268 |
| rs1073319 | 2 | 0.917 | 0.810 | 0.167 | 0.107 | 0.750 | 0.643 |
| rs12953952 | 18 | 0.139 | 0.927 | 0.967 | 0.788 | 0.828 | 0.040 |
| rs1353251 | 5 | 0.972 | 0.798 | 0.267 | 0.175 | 0.706 | 0.531 |
| rs138022 | 22 | 0.833 | 0.262 | 0.033 | 0.571 | 0.800 | 0.229 |
| rs1395771 | 3 | 0.656 | 0.024 | 0.567 | 0.632 | 0.090 | 0.543 |
| rs1397618 | 10 | 0.294 | 0.952 | 1.000 | 0.658 | 0.706 | 0.048 |
| rs1398829 | 4 | 0.222 | 0.976 | 1.000 | 0.754 | 0.778 | 0.024 |
| rs1451928 | 14 | 0.861 | 0.857 | 0.200 | 0.004 | 0.661 | 0.657 |
| rs1470524 | 2 | 0.222 | 0.786 | 0.533 | 0.563 | 0.311 | 0.252 |
| rs1477277 | 5 | 1.000 | 0.345 | 0.300 | 0.655 | 0.700 | 0.045 |
| rs1498991 | 3 | 0.944 | 0.976 | 0.250 | 0.032 | 0.694 | 0.726 |
| rs1517634 | 2 | 0.833 | 0.833 | 0.000 | 0.000 | 0.833 | 0.833 |
| rs153898 | 5 | 0.083 | 0.738 | 0.033 | 0.655 | 0.050 | 0.705 |
| rs1898280 | 8 | 0.889 | 0.238 | 0.833 | 0.651 | 0.056 | 0.595 |
| rs1919550 | 3 | 0.028 | 0.024 | 0.833 | 0.004 | 0.806 | 0.810 |
| rs1934393 | 1 | 0.222 | 0.842 | 0.300 | 0.620 | 0.078 | 0.542 |
| rs1984473 | 3 | 0.056 | 0.631 | 0.967 | 0.575 | 0.911 | 0.336 |
| rs1990743 | 17 | 0.639 | 0.634 | 0.067 | 0.005 | 0.572 | 0.567 |
| rs1990745 | 5 | 1.000 | 0.881 | 0.233 | 0.119 | 0.767 | 0.648 |
| rs2035573 | 3 | 0.972 | 0.655 | 0.133 | 0.317 | 0.839 | 0.521 |
| rs2042762 | 18 | 0.000 | 0.000 | 0.733 | 0.000 | 0.733 | 0.733 |
| rs2208139 | 20 | 0.833 | 0.667 | 0.033 | 0.167 | 0.800 | 0.633 |
| rs2253624 | 17 | 0.176 | 1.000 | 1.000 | 0.824 | 0.824 | 0.000 |
| rs2296274 | 14 | 0.118 | 0.750 | 0.967 | 0.632 | 0.849 | 0.217 |
| rs249847 | 12 | 0.056 | 0.463 | 0.967 | 0.408 | 0.911 | 0.503 |
| rs2569029 | 5 | 0.889 | 0.667 | 0.133 | 0.222 | 0.756 | 0.533 |
| rs257748 | 5 | 0.806 | 0.381 | 0.967 | 0.425 | 0.161 | 0.586 |
| rs2585901 | 13 | 0.765 | 0.854 | 0.067 | 0.089 | 0.698 | 0.787 |
| rs2592888 | 1 | 0.088 | 0.762 | 1.000 | 0.674 | 0.912 | 0.238 |
| rs2595456 | 11 | 0.306 | 0.524 | 0.000 | 0.218 | 0.306 | 0.524 |
| rs2711070 | 2 | 0.139 | 0.464 | 0.967 | 0.325 | 0.828 | 0.502 |
| rs2785279 | 10 | 0.824 | 0.262 | 0.067 | 0.562 | 0.757 | 0.195 |
| rs2817611 | 1 | 0.278 | 0.952 | 0.967 | 0.675 | 0.689 | 0.014 |
| rs2829454 | 21 | 0.056 | 0.274 | 0.933 | 0.218 | 0.878 | 0.660 |
| rs2840290 | 9 | 0.861 | 0.268 | 0.900 | 0.593 | 0.039 | 0.632 |
| rs30125 | 16 | 0.639 | 0.049 | 0.100 | 0.590 | 0.539 | 0.051 |
| rs304051 | 3 | 0.750 | 0.548 | 0.000 | 0.202 | 0.750 | 0.548 |
| rs354747 | 20 | 0.083 | 0.643 | 0.767 | 0.560 | 0.683 | 0.124 |
| rs3768176 | 1 | 0.972 | 0.929 | 0.233 | 0.044 | 0.739 | 0.695 |
| rs3806218 | 1 | 0.059 | 0.667 | 0.821 | 0.608 | 0.763 | 0.155 |
| rs3828121 | 1 | 1.000 | 0.869 | 0.300 | 0.131 | 0.700 | 0.569 |
| rs3860446 | 2 | 0.972 | 0.357 | 1.000 | 0.615 | 0.028 | 0.643 |
| rs4013967 | 9 | 0.059 | 0.655 | 1.000 | 0.596 | 0.941 | 0.345 |
| rs4034627 | 12 | 0.781 | 0.048 | 0.033 | 0.734 | 0.748 | 0.014 |
| rs4076700 | 12 | 0.206 | 0.833 | 0.900 | 0.627 | 0.694 | 0.067 |
| rs4130405 | 8 | 0.000 | 0.190 | 0.800 | 0.190 | 0.800 | 0.610 |
| rs4130513 | 16 | 0.722 | 0.083 | 0.192 | 0.639 | 0.530 | 0.109 |
| rs4625554 | 12 | 0.167 | 0.298 | 0.867 | 0.131 | 0.700 | 0.569 |
| rs4657449 | 1 | 0.083 | 0.071 | 0.733 | 0.012 | 0.650 | 0.662 |
| rs4733652 | 8 | 0.917 | 0.762 | 0.100 | 0.155 | 0.817 | 0.662 |
| rs4762106 | 12 | 0.806 | 0.095 | 0.667 | 0.710 | 0.139 | 0.571 |
| rs4852696 | 2 | 0.250 | 0.905 | 0.700 | 0.655 | 0.450 | 0.205 |
| rs4934436 | 10 | 0.694 | 0.452 | 0.967 | 0.242 | 0.272 | 0.514 |
| rs5000507 | 13 | 0.139 | 0.713 | 1.000 | 0.574 | 0.861 | 0.288 |
| rs567992 | 11 | 1.000 | 0.845 | 0.333 | 0.155 | 0.667 | 0.512 |
| rs6569792 | 6 | 0.111 | 0.679 | 0.800 | 0.567 | 0.689 | 0.121 |
| rs6684063 | 1 | 0.222 | 0.833 | 0.167 | 0.611 | 0.056 | 0.667 |
| rs6804094 | 3 | 0.972 | 0.654 | 0.133 | 0.318 | 0.839 | 0.521 |
| rs6883095 | 5 | 0.861 | 0.548 | 0.033 | 0.313 | 0.828 | 0.514 |
| rs6911727 | 6 | 0.139 | 0.476 | 1.000 | 0.337 | 0.861 | 0.524 |
| rs708915 | 20 | 0.735 | 0.100 | 0.733 | 0.635 | 0.002 | 0.633 |
| rs719776 | 4 | 0.917 | 0.143 | 0.167 | 0.774 | 0.750 | 0.024 |
| rs7463344 | 8 | 0.639 | 0.000 | 0.000 | 0.639 | 0.639 | 0.000 |
| rs7535375 | 1 | 0.111 | 0.714 | 0.800 | 0.603 | 0.689 | 0.086 |
| rs798887 | 19 | 0.778 | 0.854 | 0.133 | 0.076 | 0.644 | 0.720 |
| rs802524 | 7 | 0.278 | 0.929 | 0.967 | 0.651 | 0.689 | 0.038 |
| rs842634 | 2 | 0.972 | 0.738 | 0.233 | 0.234 | 0.739 | 0.505 |
| rs868179 | 2 | 0.700 | 0.073 | 0.000 | 0.627 | 0.700 | 0.073 |
| rs879780 | 11 | 0.667 | 0.048 | 0.000 | 0.619 | 0.667 | 0.048 |
| rs888861 | 19 | 0.000 | 0.738 | 0.900 | 0.738 | 0.900 | 0.162 |
| rs9292118 | 5 | 0.176 | 0.833 | 0.967 | 0.657 | 0.790 | 0.133 |
| rs9295316 | 6 | 0.029 | 0.098 | 0.867 | 0.068 | 0.837 | 0.769 |
| rs9302185 | 15 | 0.875 | 0.190 | 0.033 | 0.685 | 0.842 | 0.157 |
| rs9307613 | 4 | 0.094 | 0.775 | 0.833 | 0.681 | 0.740 | 0.058 |
| rs9310888 | 3 | 0.667 | 0.075 | 0.000 | 0.592 | 0.667 | 0.075 |
| rs9320808 | 6 | 0.861 | 0.095 | 0.967 | 0.766 | 0.106 | 0.871 |
| rs9323178 | 14 | 0.176 | 0.452 | 0.967 | 0.276 | 0.790 | 0.514 |
| rs9325872 | 8 | 0.944 | 0.321 | 1.000 | 0.623 | 0.056 | 0.679 |
| rs948360 | 11 | 0.250 | 0.974 | 1.000 | 0.724 | 0.750 | 0.026 |
| rs993314 | 6 | 0.900 | 0.167 | 0.700 | 0.733 | 0.200 | 0.533 |
Genotyping approximately 100 AIMs was predicted to provide estimates of ancestry with correlation coefficients greater than 0.9 for the true individual ancestral proportions. Simulations with different numbers of AIMs using each of three major methods to estimate ancestry (maximum likelihood estimation, ADMIXMAP, and Structure) by Tsai et al. indicate that the Pearson's product moment correlation coefficient for agreement between individual ancestry estimates and true individual ancestry proportions is 0.79-0.81, 0.87-0.88, and 0.93 for 25, 50, and 100 AIMs, respectively(17). These simulations are based on a model where the markers being tested have a mean informativeness of 0.15. Furthermore, 100 markers were predicted to be an adequate number for identifying admixture proportions in a three-way population admixture as seen among AAs(17).
Genotype Analysis
All study participants signed an informed consent document for blood donation and DNA preparation and testing. Peripheral venous blood samples (12 mL) were collected by venipuncture from each participant in tubes containing EDTA. DNA extraction was conducted using the Gentra DNA isolation platform (Minneapolis, MN) as previously described (http://www.gentra.com/pdf/00191.pdf). Briefly, whole blood was combined with RBC Lysis Solution and then centrifuged to isolate the buffy coat. Peripheral blood leukocytes were then lysed with Cell Lysis Solution and mixed with Protein Precipitation Solution. This mixture was centrifuged and the protein pellet was discarded. DNA was precipitated from the supernatant using 100% isopropanol and cleaned with 70% ethanol. Final DNA concentrations were within the range of 220-660 μg/mL.
Genotyping of AIMs was performed using iPLEX reagents and protocols for multiplex polymerase chain reaction (PCR), single base primer extension (SBE), and generation of mass spectra, as per the manufacturer's instructions (for complete details see iPLEX Application Note, Sequenom, San Diego, CA). Genotyping was conducted at the Functional Genomics Core, Children's Hospital Oakland Research Institute (Oakland, CA). Four multiplexed assays containing 28, 27, 26, and 26 SNPs (total = 107 SNPs) were performed using each DNA sample. Briefly, initial multiplexed PCR was performed in 5-μL reactions on 384-well plates containing 5 ng of genomic DNA. Reactions contained 0.5 U HotStar Taq polymerase (Qiagen, Valencia, CA), 100 nM primers, 1.25X HotStar Taq buffer, 1.625 mM MgCl2, and 500 μM dNTPs. Following enzyme activation at 94 °C for 15 min, DNA was amplified with 45 cycles of 94 °C x 20 sec, 56 °C x 30 sec, 72 °C × 1 min, followed by a 3-min extension at 72 °C. Unincorporated dNTPs were removed using shrimp alkaline phosphatase (0.3 U, Sequenom, San Diego, CA). Single-base extension was carried out by addition of SBE primers at concentrations from 0.625 μM (low molecular weight primers) to 1.25 μM (high molecular weight primers) using iPLEX enzyme and buffers (Sequenom) in 9-μl reactions. Reactions were desalted, and SBE products were measured using the MassARRAY Compact system (Sequenom). Mass spectra were analyzed using TYPER software (Sequenom) to generate genotype calls and allele frequencies.
Development Of Study Questionnaire
Development of Study Questionnaire. We developed a 26-item questionnaire that explored beliefs about race, ethnicity, and nationality. This questionnaire asked participants standard demographic information, including gender, age, household income, and place of birth. The questionnaire included closed-ended questions that have been used previously to measure Racial-Ethnic Identity (REI) using the Racial-Ethnic Identity Subscales (http://www.sitemaker.umich.edu/culture.self/files/racial_identity_measures06.doc). This tool, based on the Oyserman, Gant, and Ager tripartite model of REI, assesses REI by measuring connectedness, embedded achievement, and awareness of racism(18). Using the 5-point Likert response scale (1 = strongly disagree, 2 = disagree, 3 = neither agree nor disagree, 4 = agree, 5 = strongly agree), participants indicated their agreement with 13 statements that tested all three aspects of REI. Alpha reliability for each of these aspects has been reported in the 0.6 to 0.7 range(19). The 26-item study questionnaire also used the Likert response scale to assess the importance of different physiognomic characteristics when estimating African ancestry. The questionnaire included a three-generation family tree in which participants filled in the race and birth-country of their grandparents and parents. To test response reliability, two separate questions that asked participants to write down their ethnicity were included in the questionnaire (questions 12 and 19).
Calculation Of Population Admixture And Estimates Of Ancestry
Population admixture proportions were calculated and compared using two methods, maximum likelihood estimates (MLE) and principal components analysis (PCA). The MLE approach was implemented in a JAVA program (available from S. Huntsman upon request). This program uses information on allele frequencies from each ancestral population and all the study participants(20, 21). (For a detailed description of the application of MLE to admixture proportion calculations, please refer to Tsai, 2005)(17). PCA was applied to the genotype data for the study participants using S-PLUS 7.0 for Windows (Insightful Corporation, Seattle, WA) to order participants by degree of genetic variation. The first principal component indicates a continuous axis of genetic variation which codes the greatest degree of variance among study participants.
Statistical Methods
Independent T-tests with equal variances not assumed were used to compare mean admixture proportions in the two cohorts. Levene's test for equality of variances was used to compare variances for each ancestral grouping in the two cohorts. Cohen's kappa measurement of reliability for questionnaire responses was calculated by comparing responses on two questions that asked for the same information with slightly different wording. Questionnaire responses were analyzed with univariate chi-square (X2) analyses to identify significant correlations between participants' responses and percentage west African ancestry. PCA and factor analysis were used to identify a set of questionnaire items that predicted degree of west African ancestry. PCA was applied to determine if the 26-item questionnaires could be reduced to fewer variables that describe the total variation in questionnaire responses. Factor analysis was then used to determine which questionnaire items contributed most to the variance in questionnaire responses. Logistic regression was applied to assess for an association between the identified factors and percentage west African ancestry. Self-reported grandparent race was used to assess the sensitivity, specificity, and positive predictive value of self-reported ancestry in a three-generation family tree for calculated degree of west African ancestry. Statistical significance was defined as P < 0.05. Statistical software used were SPSS 14.0 for Windows (SPSS Inc., Chicago, IL) (T-tests, Levene's test, Cohen's kappa, X2-analyses) and S-PLUS 7.0 for Windows (Insightful Corporation) (factor analysis, PCA).
Results
Participant Characteristics
50 AAs and 40 Nigerians from the New York metropolitan area participated in this study. Participant characteristics are shown in Table 2. Mean age, median household income bracket, and mean years of schooling all varied significantly between the two cohorts (P < 0.05). These demographic variables were therefore included in the factor analysis of the questionnaire items.
Table 2.
Demographic Characteristics of Study Participants
| Characteristic | African Americans (n=50) |
Nigerians (n=40) |
P-value |
|---|---|---|---|
| Mean age, years old | 43 | 30 | <0.0001 |
| Gender, % male | 41 | 48 | 0.527 |
| Median household income bracket | <$15,000 | $45,001-$60,000 | <0.0001 |
| Mean years of schooling | 13 | 16 | <0.0001 |
| Mean age immigrated to US, years old |
N/A | 20 |
Population Substructure and Admixture in the Two Cohorts
Population Substructure and Admixture in the Two Cohorts. Genetic scoring using AIMs and MLE revealed that ancestry was approximately 95% west African, 4% European, and 1% Native American in the Nigerian cohort and 83% west African, 15% European, and 2% Native American in the AA cohort (Table 3). The mean percentage of west African and European ancestries varied significantly between the two cohorts (P < 0.0001). The mean percentage of Native American ancestry did not vary significantly between the two cohorts (P = 0.087). Furthermore, the variance of individual ancestry within each cohort differed significantly between the two groups for all three geographic ancestries (P = 0.002, P < 0.0001, and P = 0.011 for the variance of west African, European, and Native-American ancestry, respectively; data not shown).
Table 3.
Ancestry Admixture Proportions of Study Participants
| Ancestry | N | Mean | Standard Deviation |
SEM* | P- value† |
|
|---|---|---|---|---|---|---|
| African | ||||||
| African-American | 50 | .8302 | .11515 | .01628 | <0.0001 | |
| Nigerian | 40 | .9510 | .06979 | .01103 | ||
| European | ||||||
| African-American | 50 | .1470 | .12407 | .01755 | <0.0001 | |
| Nigerian | 40 | .0365 | .06705 | .010606 | ||
| Native American | ||||||
| African-American | 50 | .0228 | .03301 | .00467 | 0.087 | |
| Nigerian | 40 | .0125 | .02329 | .00368 | ||
Standard Error of the Mean (SEM)
Independent sample t-test comparing mean ancestry, equal variances not assumed.
We analyzed AA study participants according to quartiles of increasing percentage of west African ancestry to determine how well self-reported race accorded with inferred genetic population cluster (Table 4). Of the 50 participants who self-identified as AA, only one was found to have a minority (i.e., < 50%) of west African ancestry upon genotype testing.
Table 4.
Quartiles of Percentage African Ancestry (African-American Subjects)
| Quartiles of Percentage African Ancestry | Number of Study Subjects |
|---|---|
| 0 – 25% | 0 |
| 26 – 50% | 1 |
| 51 – 75% | 10 |
| 76 – 100% | 39 |
Reliability of Questionnaire Responses
To assess the level of intra-participant reliability of the questionnaire responses, a pair of redundant questions was inserted into the questionnaire (questions 12 and 19). These items asked participants to indicate their ethnicity. The first question asked participants what they consider their ethnicity to be; the second question asked participants what they record when ethnicity information is requested on forms or surveys. The Cohen's kappa measurement of agreement for responses to these two questions was 0.658. This indicates a good reliability of questionnaire responses.
Correlation of Genetic Ancestry with Self-Reported Ancestry
For the AA cohort, we used a three-generation family tree to assess the correlation between self-reported grandparent race and genetic ancestry calculated from the AIMs. For this analysis, we dichotomized responses for race into those consistent with African ancestry and those consistent with non-African ancestry. Responses of “African,” “African-American,” and “black” were all keyed as consistent with African ancestry. Using this definition, all of the AAs described that the race of at least three of their four grandparents was consistent with African ancestry. Thus a self-report that three or more grandparents were of a race consistent with African ancestry had a sensitivity of 100%, specificity of 0%, and positive predictive value of 80% for determining a calculated west African ancestry of 76-100%.
We performed univariate X2-analyses to identify any questionnaire items that were significantly associated with percentage west African ancestry. West African ancestry was divided into high and low percentage ancestry at the mean value of 85% among AAs. When the entire dataset was analyzed, the following questionnaire items were found to significantly predict a high percentage of west African ancestry: birthplace, self-described nationality, language spoken at home, number of generations in one's family that have lived in the US, self-described ethnicity, and a high estimation of the importance for one's community that one succeed in school (P < 0.05). However, these effects were lost when the US-born AA cohort was examined alone (data not shown), and we therefore did not test these factors for an independent effect. Thus no single questionnaire item significantly predicted the degree of west African ancestry in the US-born AA cohort.
We next tried to identify a group of questions that may possibly serve as a proxy for estimating west African ancestry. Using PCA, we found that the two top components explain 42% of the total variation in the data. Factor analysis was then used to identify two factors, consisting of a group of questions, which explain this large portion of the variance in survey responses. The first factor included the REI subscales, participants' rating of the importance of different physiognomic characteristics when estimating African ancestry, their rating of agreement with the statement that “people of African ancestry share physical traits,” and their rating of agreement with the statement, “I have similar physical traits to other people in my racial/ethnic group.” The second factor consisted of an average of household income bracket and years of schooling. Scores from each factor were used in a logistic regression with percentage west African ancestry in the AA cohort. Unfortunately, the regression analysis revealed no significant association between responses on these two sets of questions (i.e., first two components) and degree of west African ancestry (P = 0.47).
Comparison of Different Methods to Estimate Ancestry
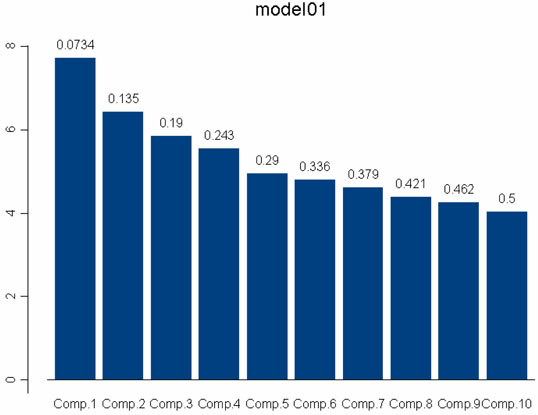
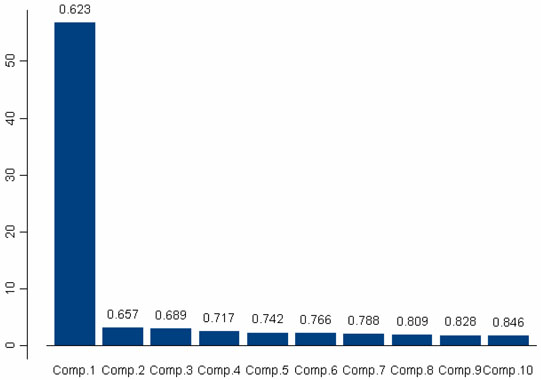
We applied PCA to the AIMs genotypes of our study population to investigate the variance in both the AIMs selected to determine ancestry and the actual study subjects. We attempted to identify a first principal component consisting of the AIMs that describe the largest part of the variance in genotype frequencies among participants. This could be used to order participants by degree of genetic variation. The relative level of genetic variation should approximate degree of African ancestry and would then be compared with the ancestry estimates calculated using MLE. Unfortunately, we were unable to identify a principal AIMs component using this method (Figure 1). As shown in Figure 1, all 107 AIMs appeared to be fairly independent and unrelated. This is consistent with the method by which the AIMs were selected to be widely spaced throughout the genome. (Please refer to Materials and Methods – Selection of AIMs.) Interestingly, when we performed PCA to investigate the variance in actual study subjects (Figure 2), we found that the first principal component separated out two groups of subjects that significantly varied in percentage west African ancestry as determined using the 107 AIMs and MLE (P < 0.0001). The first principal component explained 62% of the variability in west African ancestry and consisted of 62 of the study participants. For the subjects included in the first principal component, the average and median percentage west African ancestries were 93% and 95%, respectively. In other words, the majority of study subjects in both cohorts (AA and Nigerian) were found to be highly similar, and these “more similar” subjects comprised the principal component. For the remaining 28 subjects that were not included in the first principal component, the average and median percentage west African ancestries were 78% and 81%, respectively.
Figure 1. Principal Components Analysis of the Ancestry Informative Markers.
Principal components analysis was used to identify a principal component of AIMs that may explain the largest part of the variance in participant genotype frequencies and therefore be used to order participants by relative degree of genetic variation. Bars represent successive principal components; bar numbers indicate the cumulative proportion of variance explained by each component; vertical axis indicates the number of AIMs contributing to each component. The first principal component explains only 7% of the variation in AIMs, and no dominant principal component could be identified. All 107 AIMs thus appear to be independent and unrelated.
Figure 2. Principal Components Analysis of Participant Genotype Frequencies.
Bars represent successive principal components; bar numbers indicate the cumulative proportion of variance explained by each component; vertical axis indicates the number of participants comprising each component. The first principal component identifies a cluster of 62 participants with highly similar distributions of AIM genotypes.
Discussion
In this study, we genotyped a panel of 107 AIMs to investigate the degree of west African ancestry among AAs and Nigerians and administered a questionnaire to explore the variables used by each cohort in constructing racial identity. We found that while nearly all selfidentified AAs had a majority of west African ancestry, AAs had significantly more European admixture and greater admixture variability than the Nigerians. Self-report of a high degree of African ancestry in a three-generation family tree did not accurately predict the degree of west African ancestry calculated from our AIMs. Analysis of questionnaire responses revealed that no simple question proxy effectively estimates the degree of west African ancestry among US-born AAs. However, relative degree of west African ancestry could be effectively determined using both MLE and PCA. The results of our study thus suggest that while self-identified race could identify a cohort of individuals with a high degree (>80%) of west African ancestry, an admixture-matched case-control design may be more accurate and objective for conducting genetic association studies in admixed populations.
There are conflicting reports in the literature regarding the ability of self-identified race to serve as an accurate predictor of population clusters. In our study, self-reported race generally accorded well with inferred genetic population cluster. The mean west African ancestry among the AA participants was 83%, and only one of the participants in the US-born AA cohort did not have a majority of west African ancestry on genotype analysis. Thus, based on our genotyping data, if members of the AA cohort were assigned to one of the five major population genetic clusters (African, Caucasian, Pacific Islander, East Asian, and Native American, as defined by Risch, 2002)(7), all but one of the participants would be classified together in the African cluster. (Of note, our study is limited by the relatively small sample sizes of both cohorts, which may lead to skewing of both mean ancestry estimates and collected questionnaire information. Thus, the population level inferences should be taken with caution.) In contrast, in a previous study, Wilson et al. used microsatellite markers to analyze individuals from eight populations and observed that genetically inferred clusters corresponded poorly to commonly used racial labels(11). However, this study used far fewer markers and also classified Ethiopians as Blacks and New Guineans as Asians, while more recent population studies suggest that the genetic ancestries of these groups are European and Pacific Islander, respectively(7). Other studies have found that given sufficient numbers of markers and sample sizes, self-defined race may correspond well with inferred genetic clusters. Rosenberg et al. tested the correspondence of predefined population groups with those inferred from individual multilocus genotypes and found general agreement between the genetic and predefined populations(9). Tang et al. studied 3,636 subjects participating in the Family Blood Pressure Program who identified themselves as belonging to one of four racial groups (white, African American, East Asian, and Hispanic). Subsequent genetic cluster analysis using microsatellite markers produced four major clusters with near-perfect correspondence with the four racial categories(10).
The AA cohort in our study had a mean of 15% European admixture, which is consistenty with previous reports of a range of 7-23% European admixture among US AAs(14-16). Of note, the estimates of 4% European and 1% Native American ancestry in the Nigerian population is likely due to bias in ML estimates due to the limited number of markers. We found that among participants, there was a significantly higher proportion of admixture and higher variability in admixture proportions, in the US-born AA cohort compared to a population that emigrated from Africa (i.e., Nigerians) (Table 3). The significant variation in individual ancestry estimates among the AA cohort suggests that this group, like the CHS AA cohort(15), represents a diverse population consisting of several subpopulations. For participation in the AA cohort, subjects identified both parents as AAs who were born in the US. Although data regarding grandparental race were not used to screen study participation, these data were collected through a threegeneration family tree during administration of the questionnaire. In this study population, all AA subjects described that the race of at least three of their four grandparents was consistent with African ancestry. Individuals and society have historically classified children of mixed race ancestry as AA, even when one parent is Caucasian, Asian, or Native American. For AAs, this is a remnant of the “Jim Crow” laws and the “One Drop” rule or “Rule of Hypodescent”. Thus, identification as AA would still occur in cases where the parents and grandparents were of mixed race ancestry. This could also contribute to the greater European admixture and greater admixture variability seen in the AA cohort.
The two cohorts were found to differ significantly in income bracket and education level, raising the possibility of a confounding relationship between socioeconomic status (SES) and degree of west African ancestry. In fact, others have found significant interactions between SES, genetic ancestry, and disease outcome(22). In our study however, analyses of income and education within each cohort suggested that SES is unlikely to represent an important bias in our study population as neither income nor education significantly correlated with degree of west African ancestry within either group (data not shown). SES is a complex construct, operates on multiple levels, and may be time dependent(23). In our study, income and education within each cohort were not significant confounders. However, it is possible that there were unmeasured confounders for which no amount of correction would control. Therefore, generalizations cannot be made regarding the relationship between ancestry and SES, and the potential confounding effect of SES must be addressed specifically in individual studies.
The limited ability of self-reported race to effectively reveal population substructure was also seen in a recent study that compared population structure inferred from individual ancestry estimates with self-reported race(24). In a case-control study of early-onset lung cancer, Barnholtz-Sloan et al. reported that the frequency of the drug-metabolizing gene GSTM1 null “risk” genotype varied both by individual European ancestry and by case-control status within self-reported race, particularly among the AA study participants. Furthermore, they found that genetic risk models that adjusted for European ancestry provided a better fit for this relationship between GSTM1 genotype and lung cancer risk compared with the model that adjusted for selfreported race. The results of this and other studies suggest that the likelihood of identifying disease-susceptibility loci will be lower in studies that rely on less accurate measures of population stratification (e.g., self-reported race)(25). Thus, genetic classifications of ancestry may provide a more objective and accurate method of defining homogeneous populations which can be used to investigate specific population-disease associations.
Because of cost and feasibility issues that may discourage the incorporation of admixture testing in the design of both preclinical and clinical studies, we developed a questionnaire to search for questions or combinations of questions that may reliably serve as a proxy for west African ancestry. We are not aware of any previous reports that have investigated relationships between factors used by individuals in constructing racial identity and individual ancestry estimates as determined through genotyping. When the entire dataset (i.e., two cohorts) was examined as a whole, several questionnaire items were found to have a significant association with percentage west African ancestry. Many of these items appear to be related to characteristics of an immigrant population (e.g., Nigerians), such as birthplace, self-described nationality, language spoken at home, number of family generations living in the US, self described ethnicity, and estimation of importance of one's success in school for his/her community. However, when the AA cohort was examined separately, no question or set of questions significantly predicted degree of west African ancestry, as determined both by univariate analysis and factor analysis of survey items. Self-reported ancestry using a threegeneration family tree also could not accurately predict degree of west African ancestry. Although reported grandparent race was highly sensitive for ancestry, it was not specific. Since all participants in our study reported that at least three grandparents were of a race consistent with African ancestry, this information could not distinguish those who actually had a high degree of African ancestry. The lack of specificity of reported grandparent race likely is due to the imprecision of racial categories. Our family tree analysis was limited by the relatively similar background of our study participants; for example, all AA participants indicated that three or all of their grandparents were of a race consistent with African ancestry. Thus, studying a population with a greater degree of admixture may be more appropriate for investigating the utility of a three-generation family tree in predicting degree of African ancestry. A recent study by Burnett et al., however, suggests that self-reported ancestry may have poor reliability(26). In this study, Burnett et al. prospectively asked siblings to list the countries of origin of both parents. Participants in this study were recruited at the Mayo Clinic and were primarily Caucasian. Nevertheless, Burnett et al. found that only 49% of sibling pairs agreed completely on the countries of origin of both parents and this agreement only increased to 68% when named countries were postcoded into six population genetic clusters (Eurasia, East Asia, Oceania, America, Africa, and the Kalash group of Pakistan).
Applying PCA to the AIMs genotypes of our study population, we were unable to identify a principal component that could be used to order participants by relative degree of west African ancestry and compared with percentage west African ancestry calculated using MLE. However, PCA of the study participants' genotype frequencies was able to identify a cluster of subjects with highly similar SNP distributions. The majority of subjects (both AA and Nigerian) with a high percentage of MLE-calculated west African ancestry were included in the first principal component, indicating an overall concordance between individual ancestry estimates calculated with MLE and subjects groupings with PCA. A few subjects included in the first principal component had a lower percentage west African ancestry calculated with MLE. We suspect this difference may result from methodological differences and our use of point estimates rather than confidence intervals for estimated ancestry in our MLE calculations.
We began this study to investigate the degree of admixture among self-reported AAs following our previous studies of breast cancer tumor biology in AAs(3, 4, 27). Genetic heterogeneity in study subjects could impair the ability of a study to detect true biological differences between racially-defined, apparently uniform groups. We have found that genetic ancestry proportions can vary significantly within groups of individuals who would self-identify as the same racial group. Our work suggests that to maximize the predictive value of clinical inferences from genome-wide association studies, one must consider within-as well as between-population association. Thus, while self-identified race can identify a cohort of individuals with a high degree of African ancestry, admixture-matched case-control studies will be more effective in studying differences in disease incidence and outcomes in specific racial populations.
Acknowledgements
We wish to acknowledge Dr. I. Bernard Weinstein for invaluable advise during the preparation of this manuscript and Scott Huntsman for assistance in analyzing the AIM data. We also wish to thank Dr. Wendy K. Chung, Grace Ajuluchukwu, Marline Anderson-Slater, and Barbara Castro for assistance with this project.
Grant support: Long Island University/Herbert Irving Comprehensive Cancer Center Minority Institution/Cancer Center Partnership (CA91372, CA101388, CA101598) (AKJ, NSC, MGB); NIH fellowship T32CA09529 (RY); National Center for Minority Health Disparities Center of Excellence in Nutritional Genomics (5P60MD00022) (KBB); Tobacco-Related Disease Research Program New Investigator Award (15KT-0008) (SC); and National Institute of Heart, Lung and Blood (R01 HL078885), RWJ Amos Medical Faculty Development Award, NCMHD Health Disparities Scholar, Extramural Clinical Research Loan Repayment Program for Individuals from Disadvantaged Backgrounds (2001-2003), Sandler Center for Basic Research in Asthma and the Sandler Family Supporting Foundation (EGB).