Abstract
The neurotrophins nerve growth factor (NGF) and neurotrophin-3 (NT3) support the survival of subpopulations of primary sensory neurons with defined and distinct physiological characteristics. Only a few genes have been identified as being differentially expressed in these subpopulations, and not much is known about the nature of the molecules involved in the processing of sensory information in NGF-dependent nociceptive neurons or NT3-dependent proprioceptive neurons. We devised a simple dorsal root ganglion (DRG) explant culture system, allowing the selection of neuronal populations preferentially responsive to NGF or NT3. The reliability of this assay was first monitored by the differential expression of the NGF and NT3 receptors trkA and trkC, as well as that of neuropeptides and calcium-binding proteins. We then identified four differentially expressed sodium channels, two enriched in the NGF population and two others in the NT3 population. Finally, using an optimized RNA fingerprinting protocol, we identified 20 additional genes, all differentially expressed in DRG explants cultured with NGF or NT3. This approach thus allows the identification of large number of genes expressed in subpopulations of primary sensory neurons and opens the possibility of studying the molecular mechanisms of nociception and proprioception.
The dorsal root ganglia (DRGs) of higher vertebrates comprise more than 20 different subtypes of sensory neurons (1). These neurons convey information about mechanical, thermal, chemical, and noxious stimuli both from the outside world and from within the body. Much is known about the development and the morphological and physiological properties of DRG neurons. For example, neurons involved in the perception of noxious stimuli are small, and their unmyelinated axons conduct action potentials with slow velocities, whereas neurons relaying mechanical information about limb posture are large with thickly myelinated fibers and high conduction velocities (1). However, the molecular mechanisms underlying the detection and processing of sensory information by these neurons are largely unknown.
As fundamental biological mechanisms are often conserved through evolution, studying genetically accessible invertebrates such as Drosophila or Caenorhabditis elegans could eventually help us to understand sensory processing in vertebrates also. Indeed, recent results suggest that it might be possible to elucidate the molecular apparatus of mechanosensation in C. elegans (2). However, there is little evidence so far that molecules homologous to those found in the nematode play similar roles in vertebrates. Thus, a more direct approach using vertebrate tissue is desirable.
The rationale of the present study is based on the findings that during development, different subpopulations of sensory neurons with distinct physiological properties selectively depend for survival on the activation of trkA by its ligand nerve growth factor (NGF) or the activation of trkC by neurotrophin-3 (NT3) (3, 4). Thus, NGF deprivation (using gene targeting or antibody treatment), or deletion of the trkA receptor gene caused dramatic losses of small-diameter neurons projecting to the superficial laminae of the dorsal horn and of thin, unmyelinated fibers in peripheral nerves (5–9). In such animals, the behavioral response to potentially noxious stimuli was profoundly impaired. By contrast, after targeted deletion of the trkC or NT3 gene, the large neurons and their myelinated axons were affected, including the Ia muscle afferents contacting motoneurons in the ventral spinal cord, as well as skin afferents terminating on Merkel cells. These animals had grossly abnormal movements and impaired coordination. Proprioceptive neurons were essentially absent from such animals, whereas innervation of the superficial layers of the spinal cord appeared to be largely normal (10–13). Work performed with avian embryos after administration of neutralizing antibodies against NGF, NT3, or trkC led to very similar conclusions, indicating that the involvement of NGF and NT3 in the development of specific subtypes of sensory afferents has been strongly conserved in widely separated vertebrate species (14–17).
In the present study, we used DRG explants from chicken embryos and the selective survival properties of NGF and NT3 as a means to separate distinct subsets of DRG neurons and to screen for genes that are relevant for their specific physiological properties.
MATERIALS AND METHODS
Cultures of Dissociated DRG Neurons.
Lumbosacral DRGs from embryonic day 8 (E8) chicken embryos were dissected, incubated in 0.1% trypsin/PBS, and gently triturated with a fire-polished, silanized Pasteur pipette. E8 was selected because the death of isolated neurons cultured in the absence of neurotrophins is more rapid than at E10. To enrich neuronal cells, the resulting cell suspension was preplated for 2 hr at 37°C in Nunclon dishes (Nunc). Nonadhering cells were plated onto 35-mm wells of six-well tissue culture plates (Falcon) coated with polyornithine (Sigma) and mouse laminin (Life Technologies) according to ref. 18. Densities were adjusted to 20,000 or 40,000 cells per well in the NGF or NT3 experiments, respectively. Culture medium consisted of Ham’s F-14 (Life Technologies) with 10% horse serum, 5% fetal calf serum, 100 μg/ml penicillin G (Sigma), 60 μg/ml streptomycin sulfate (Sigma), and 20 ng/ml either NGF (recombinant human NGF, Genentech) or NT3 (recombinant human NT3, Genentech). After 3 days of incubation at 37°C in a 3% CO2 atmosphere at 95% relative humidity, total RNA was isolated by using an RNeasy kit (Qiagen, Chatsworth, CA).
DRG Explant Cultures.
Lumbosacral ganglia 4, 5, and 6 of E10 chicken embryos were carefully dissected, washed in PBS, and transferred to 48-well tissue culture plates (Falcon) coated with polyornithine (Sigma) and mouse laminin (Life Technologies) according to ref. 18. Each well contained 0.5 ml of culture medium consisting of Ham’s F-14 (Life Technologies) supplemented with 0.1 mg/ml transferrin, 16 μg/ml putrescine, 6 ng/ml progesterone, 8 ng/ml sodium selenite, 100 μg/ml penicillin G, 60 μg/ml streptomycin sulfate (all reagents from Sigma), and 2 ng/ml NGF or NT3. After 4 days of incubation at 37°C in a 3% CO2 atmosphere at 95% relative humidity, ganglia were detached from the wells with a small spatula and total RNA was isolated by using an RNeasy kit (Qiagen).
RNA Fingerprinting.
cDNAs were synthesized from total RNA with oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (Superscript II; Life Technologies) at 45°C for 1 hr. Reaction products were purified from excess oligonucleotides by using a QIAquick kit from Qiagen. cDNA derived from 60 ng of total RNA was used as template for a PCR. Amplification was performed in a volume of 25 μl containing 1× PCR buffer (Perkin–Elmer), 20 μM dNTPs (Pharmacia), 50 nM each primer, and 5 μCi (1 μCi = 37 kBq) of [α-33P]dATP (Amersham). For each reaction, two 18-mer oligonucleotides of arbitrary sequence and a G+C content of 61% were used (see list below). All PCRs were run on thermal cyclers 2400/9600 from Perkin–Elmer. After 3 min of denaturation at 95°C, 1.5 units of AmpliTaq DNA polymerase (Perkin–Elmer) was added (manual “hot start”). After 2 min at 95°C the profile was started with two low-stringency cycles (95°C for 30 sec, 50°C for 2 min, and 72°C for 2 min) followed by 25–30 high-stringency cycles (95°C for 30 sec, 65°C for 1 min, and 72°C for 1 min), and a final 5-min extension at 72°C. Reaction products were separated by electrophoresis on nondenaturing 4% polyacrylamide gels for 3 hr at 40 W. Gels were vacuum dried on chromatography paper (3MM; Whatman), fixed in a cassette, and overlaid with x-ray film. Exposure times varied from overnight to 3 days. Bands of interest were marked on the gel by piercing through the developed film. Gel pieces were excised with a scalpel and DNA was eluted by boiling in 50 μl of 10 mM Tris⋅HCl/1 mM EDTA buffer, pH 7.5, for 5 min. One microliter of the eluted DNA was used as template in a second PCR (reamplification). The reaction volume was 50 μl containing 1× PCR buffer, 0.2 mM dNTPs, and 0.1 μM primers (the same as used for first PCR). Then 1.5 units of AmpliTaq were added by manual “hot start.” The temperature profile consisted of 30 cycles (95°C for 15 sec, 65°C for 30 sec, and 72°C for 30 sec) and a final 5-min extension at 72°C. A 10-μl aliquot of the reaction was separated on an ethidium bromide-stained 1% agarose gel. Occasionally, reamplification products contained by-products of lower molecular weight. These were excluded by cutting out only the band of the highest molecular weight. The DNA was purified with a QIAquick kit and cloned in the pCRII vector of the TA-Cloning kit (Invitrogen). DNA sequencing was done with an automated sequencer (ABI 373A; Applied Biosystems). Primers used for RNA fingerprinting: P1, AGTCGCAGCACAGGTGAG; P2, CGTCCGTAGGTCCAGGT; P3, CCCAGGTCGTCGTGTTCA; P4, GTCAGGGATACCGAGAGC; P5, CCAGCCGCCAGAGTTTTG; P6, GAGAGTTAGCCGCAGCG; P7, GTTACAGCAACCGCCAGG; P8, GGGGTCCTTCAATGGGAG; P9, TCCGAGGGAGATTCCGTC; and P10, GCCACAGGGCAGAGTCTT.
Reverse transcriptase–PCR (RT-PCR).
Relative levels of gene expression in NGF- and NT3-treated explants were determined by RT-PCR. Primer pairs were designed to allow specific amplification of cDNA fragments. cDNA from total RNA was synthesized with oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (Superscript II; Life Technologies) at 45°C for 1 hr. For each experiment two independent RNA preparations of NGF/NT3-treated ganglia were used. cDNA derived from 25–60 ng of RNA was used as template for PCR amplification in a 50-μl reaction volume containing 1× PCR buffer, 0.2 mM dNTPs, and 0.1 μM each primer. “Hot start” was performed by adding 1.5 units of AmpliTaq. The temperature profile consisted of 15 to 30 cycles (95°C for 15 sec, 65°C for 30 sec, and 72°C for 30 sec) and a final 5-min extension at 72°C. To achieve accurate quantification, 10-μl aliquots were collected during the PCR run at various cycle numbers. PCR products were separated by electrophoresis on 1% agarose gels stained with SYBR Green I (Molecular Probes), and their fluorescence intensities were measured by using the EASY digital imaging system (Herolab). In all experiments, amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA fragments was run in parallel to normalize different cDNA samples. Quantitative differences stronger than 1:20 could not be accurately measured. These values are therefore indicated as “>20.”
cDNA Cloning.
cDNA fragments for chicken GAPDH, trkA, trkC, calretinin, parvalbumin/avian thymic hormone, calbindin, and calcitonin gene-related peptide (CGRP) were obtained by PCR using primers derived from published sequences. Fragments were cloned into pCRII vectors (Invitrogen). For preprotachykinin (PPT)A, two degenerate oligonucleotides were designed on the basis of the protein sequence of chicken substance P (GICCICARCARTTYTTYGG) and substance K (TTICCCATIARICCIACRAA). With these primers a 102-bp cDNA fragment was amplified, using E8 chicken DRG cDNA as template. The 102-bp fragment was used as a probe to screen a λgt10 cDNA library from E16 chicken DRGs. A cDNA fragment of 673 bp was isolated, which comprised the coding region of γPPT. This fragment was cloned into pBluescript (Stratagene). For voltage-gated sodium channels, two degenerate primers were designed on the the basis of the conserved protein sequences of the S3 and S6 segments of domain I in mammalian proteins (AYYYITGGAAYTGGYTIGAYTT, CRTAIGYCATIGYIAIIAYISC) and used in a PCR with a cDNA template from E16 chicken DRGs. The amplification products were cloned directly or after digestion by ScaI or PvuII in the pCRII vector (Invitrogen). Four different sodium channels were identified (see Table 1). Twenty-five clones were sequenced, and the relative abundances of sodium channels II, SNS, I, and H were 8, 3, 1, and 1, respectively.
Table 1.
Expression ratios of neuronal markers in neurotrophin-treated DRG explants
| Marker | Relative expression
|
|
|---|---|---|
| +NGF | +NT3 | |
| CGRP | >20 | 1 |
| PPTA | >20 | 1 |
| Calretinin | 1 | 20 |
| Parvalbumin | 1 | 15 |
| Calbindin | 1 | 1 |
| Sodium channel SNS | >20 | 1 |
| Sodium channel I | 8 | 1 |
| Sodium channel II | 1 | 15 |
| Sodium channel H | 1 | 3 |
In Situ Hybridization.
Preparation of tissue sections and hybridization with single-stranded RNA probes were performed as described (19).
RESULTS
DRG Explants Differentially Express Neurotrophin Receptors and Subpopulation Markers.
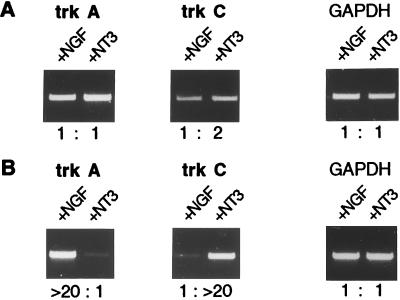
In an initial set of experiments, neuron-enriched preparations from dissociated lumbosacral DRGs were incubated with NGF or NT3. The number of surviving cells with either neurotrophin was in agreement with published data (20), and the morphologies of the neurons selected with NGF or NT3 were clearly different. We anticipated that NGF treatment would rescue trkA-expressing neurons and NT3 trkC-expressing neurons, but RT-PCR analysis revealed that trkA and trkC expressions were almost equal in NGF-treated and NT3-treated cultures (Fig. 1A).
Figure 1.
Expression of trkA and trkC in dissociated neuronal cultures (A) and in DRG explants (B). Expression levels of both trks were determined by RT-PCR in NGF-treated (+NGF) and in NT3-treated (+NT3) cultures or explants. The intensities of the bands were determined with a digital imaging system (Herolab) and the ratios (e.g., >20:1) are indicated. The absolute differences may be even higher, as the imaging system did not allow reliable quantifications beyond a 20:1 ratio. GAPDH expression served as a control. Dissociated neurons from E8 embryos were cultured for 3 days. DRG explants from E10 embryos were cultured for 4 days. Cycle numbers: 30 for trkA/C and 20 for GAPDH (A); 30 for trkA, 25 for trkC, and 20 for GAPDH (B).
We then turned to whole explants cultured in the presence of NGF or NT3, and they exhibited strong differences in the relative expression levels of trkA and trkC (Fig. 1B). In the presence of NGF, trkA expression was more than 20 times higher than in the presence of NT3, and conversely ganglia that had been incubated with NT3 expressed at least 20 times more trkC message than did NGF-treated ganglia. Strong differential expression of the trk receptors critically depended on the culture conditions, such as low neurotrophin concentrations, a serum-free medium, and omission of insulin (data not shown). In the absence of either NGF or NT3, the explants disintegrated within the 4-day culture period. Culturing the DRG explants for longer than 4 days did not increase the differences in trk expression levels. We took this differential expression of the trk receptors as an indication for a successful, though not complete, separation of two different subpopulations of neurons.
We next asked whether genes other than the trks were also differentially expressed in the two types of explants, and we used the neuropeptides and calcium-binding proteins as markers. The PPTA gene encodes three small neuropeptides. One of them, substance P, has been associated with small sensory neurons and is thought to play a role in pain modulation (21). PPTA transcript levels were about 20 times higher in NGF-treated ganglia than in NT3-treated ganglia (Table 1). CGRP immunoreactivity has been found mainly in small neurons and was reported to overlap with the trkA-positive population in rat DRGs (22). Similar to PPTA, CGRP mRNA was strongly enriched in NGF-treated ganglia. Calcium-binding proteins are thought to function as modulators of intracellular calcium levels and are often found in distinct subsets of neurons (23). We determined the levels of mRNA encoding calretinin, calbindin, and parvalbumin in the explants. When NGF-treated ganglia were compared with NT3-treated ganglia, we found a 1:>20 expression ratio for calretinin and a 1:15 ratio for parvalbumin. No differential expression was observed for calbindin.
DRG Explants Differentially Express Voltage-Gated Sodium Channels.
To test the possibility that differences in electrophysiological properties of subsets of sensory neurons might be accounted for, in part, by the differential expression of voltage-gated sodium channels, we designed degenerate oligonucleotides based on conserved sequences in mammalian channels. Four different sodium channels were identified as chicken homologues of mammalian sodium channel α subunits, designated sodium channel I, SNS, II, and H, respectively.
At the amino acid level, channel I has a 82% identity to the corresponding region of type I channel from rat brain and was 8 times more strongly expressed in ganglia that had been incubated with NGF (Table 1). With 70% amino acid identity, channel SNS represents a homologue of the recently described rat SNS channel, which was found to be expressed exclusively in small-diameter neurons of rat DRGs (24). Sodium channel SNS was more than 20 times more abundant in NGF-treated ganglia (Table 1). Channel H constitutes a homologue of a tetrodotoxin (TTX)-resistant channel originally described in rat heart muscle (87% amino acid identity), and its mRNA levels were 3 times higher in NT3-treated ganglia (Table 1). Channel II shares 90% sequence identity with the type II channel from rat brain. A 15-fold enrichment in NT3-treated ganglia was measured for channel II mRNA (Table 1).
Differential Expression in Vivo of trks, Subpopulation Markers, and Sodium Channels.
We then performed in situ hybridization on adjacent sections with probes for differentially expressed markers (see above) and for the sodium channels with the aim of correlating patterns of gene expression with those of trkA and trkC. Lumbosacral DRGs from E14 embryos were used, corresponding to the end of the 4-day culture period.
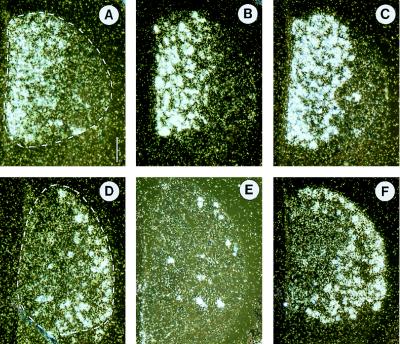
As reported in previous studies, trkA message was detectable mainly in the medial part of the DRG [(25); Fig. 2A]. This pattern was nearly identical to that of CGRP and sodium channel SNS, suggesting coexpression of the three genes (Fig. 2 A–C). Neurons marked by the trkC probe were concentrated on the lateral aspect of the ganglia, revealing a topography that is largely complementary to trkA (Fig. 2D). Calretinin showed the most restricted pattern within the DRGs. Only a few isolated neurons distributed in a lateral zone were strongly labeled. Obviously, calretinin expression overlaps with that of trkC, and it seems that most, if not all, calretinin-positive cells also express trkC mRNA (Fig. 2E). As with calretinin, the sodium channel II expression strongly colocalizes with that of trkC (Fig. 2F). However, cells labeled by the sodium channel II probe seemed to be more numerous than those labeled by the trkC probe.
Figure 2.
Micrographs of in situ hybridizations for trkA (A), CGRP (B), sodium channel SNS (C), trkC (D), calretinin (E), and sodium channel II (F). Consecutive transverse sections containing a lumbosacral DRG from an E14 embryo are shown. Boundaries of DRGs are marked with broken white lines. Signals for trkA, CGRP, and sodium channel SNS are found medially; those for trkC, calretinin, and sodium channel II are concentrated laterally. The patterns of expression for trkA and trkC are almost complementary. Dorsal is up and medial is left. (Bar represents 110 μm.)
In conclusion, ganglia cultured in the presence of NGF markedly differed from those incubated with NT3. These differences were reflected by characteristic expression levels of relevant neuronal markers and sodium channels. Most important, genes that are strongly expressed in one type of explant could be detected in a subpopulation of DRG neurons in vivo, and the expression pattern overlapped with the pattern of the receptor whose ligand was used during the culture period. We then went on to use this system to identify differentially expressed genes with an RNA fingerprinting method.
Differentially Expressed Genes Identified by RNA Fingerprinting.
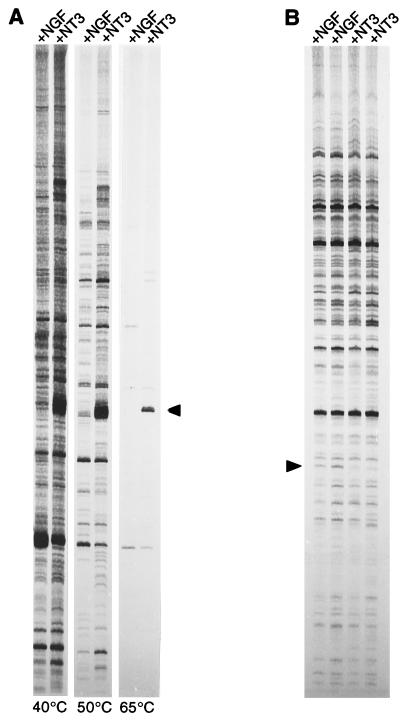
As published protocols for differential display gave unsatisfactory results (26), we developed an RNA fingerprinting method that was optimized on the basis of the differential expression of calretinin. This method involves two initial low-stringency cycles during which arbitrary 18-mers can anneal in a way such that mismatches are tolerated, thus increasing the number of possible target sequences. The subsequent high-stringency cycles lead to the efficient enrichment of the initially formed products. Fig. 3A shows the optimization of the annealing temperature for the first two cycles by using the calretinin signal as an internal control, and Fig. 3B represents an example of a RNA fingerprint obtained by using the final protocol. Two independent cDNA preparations per type of explant were always compared with two cDNAs from the other type.
Figure 3.
The RNA fingerprinting method. (A) Effect of the annealing temperature during the first two PCR cycles on the band pattern. cDNAs from NGF-treated (+NGF) and NT3-treated (+NT3) explants were compared. Changes were monitored on the basis of calretinin, known to be enriched in NT3-treated ganglia (see Table 1). Calretinin-specific primers were designed with the same length and G+C content as standard fingerprinting primers. The calretinin band (arrowhead, size marker was generated by amplifying a calretinin plasmid) was the predominant product at 65°C. Lowering the annealing temperature of the first two cycles to 40°C or 50°C allowed the primers to anneal less specifically, thus increasing the number of bands. In the final protocol 50°C was used. (B) Representative example of a fingerprint prepared with the final protocol. Routinely, two independent RNAs from NGF-treated explants were compared with two independent RNAs from NT3-treated explants. The arrowhead indicates a cDNA that is more abundant in NGF-treated explants.
We designed 10 primers and used them in all possible combinations, resulting in 45 sets of PCRs. A total of 43 candidate bands were excised. DNA fragments were eluted, reamplified, cloned, and sequenced. Twelve fragments could not be amplified or they contained more than one DNA species and were discarded. The remaining 31 cDNA fragments were analyzed by RT-PCR with fragment-specific primers. Twenty-two showed at least 2-fold differences in expression between NGF- and NT3-treated ganglia (Table 2).
Table 2.
cDNA fragments identified by RNA fingerprinting
| Fragment | Identity | Relative expression*
|
In situ† | |
|---|---|---|---|---|
| +NGF | +NT3 | |||
| Known avian sequences | ||||
| R2 | Delta EF1 | 1 | 4 | ND |
| R16 | Axonin-1 | 4 | 1 | + |
| R18 | Visinin-like protein (VILIP) | 1 | 10 | + |
| R48 | ret | 10 | 1 | + |
| High similarities to vertebrate sequences | ||||
| R1 | Fibronectin | 1 | 4 | − |
| R24‡ | Olfactomedin-related glycoprotein | 4 | 1 | − |
| R33 | Lipoprotein lipase-like | 6 | 1 | ? |
| R37 | Calcium-dependent actin-binding protein | 3 | 1 | − |
| R41 | Heparansulfate sulfotransferase | 1 | 3 | ND |
| R44 | Diacylglycerol kinase-γ | 5 | 1 | + |
| R45 | Cannabinoid receptor CB1 | 7 | 1 | + |
| R49 | LAMP | 16 | 1 | + |
| ORFs with low sequence similarities | ||||
| R3 | Zinc finger motif | 1 | 3 | ? |
| R29 | Proline-rich sequence | 2 | 1 | ND |
| R39‡ | Motif of IGF-binding proteins | 3 | 1 | − |
| Unknown sequences, no ORF | ||||
| R23 | 10 | 1 | + | |
| R30 | 3 | 1 | ND | |
| R31 | 1 | 3 | ND | |
| R34 | 1 | 3 | ND | |
| R35 | 8 | 1 | + | |
EF, enhancer binding factor; LAMP, limbic system-associated membrane protein; IGF, insulin-like growth factor.
Expression ratios of NGF- and NT3-treated DRG explants.
In situ mRNA hybridization signals in DRGs were assessed as follows: +, neuronal subpopulation labeled; −, no subpopulation labeled; ?, signal too weak for assessment; and ND, not determined.
Fragments of these genes were isolated twice independently with different primer combinations.
The cDNA sequences were placed into four groups (see Table 2). The first group comprises fragments of already known chicken genes. The second group includes probable chicken homologues of known vertebrate sequences. Fragments with an open reading frame but with similarities only to conserved structural protein motifs form the third group. The fourth group contains fragments with no open reading frame and with no similarities to known sequences.
A typical fingerprint PCR led to the amplification of approximately 100 bands, reflecting different mRNA species. On average, one candidate band could be identified per fingerprint reaction, indicating that about 1% of the transcribed genes may be expressed differentially in the DRG explant system. If we assume 15,000 different mRNA species per cell, roughly 30% of the expressed genes would have been covered with 45 fingerprint PCRs as used in this study. Clearly, the same cDNAs may be amplified more than once, and in two cases (see Table 2) the same cDNAs were identified with different primer combinations.
In Vivo Expression of Genes Identified by RNA Fingerprinting.
The in vivo expression patterns of the genes identified with the fingerprinting method were analyzed by in situ hybridization (Fig. 4; see also Table 2, right column).
Figure 4.
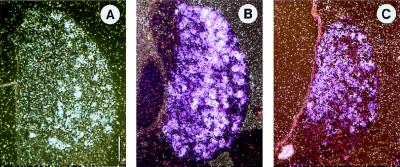
Micrographs of in situ hybridizations for R18 (A), R48 (B), and R45 (C). Consecutive transverse sections containing a lumbosacral DRG from an E14 embryo are shown. R18-positive cells are concentrated laterally. Maximal R48 expression is medial. R45 expression is concentrated in mediodorsal cells. Dorsal is up and medial is left. (Bar represents 110 μm.)
R18 encodes part of VILIP (27) and was strongly enriched in NT3-treated ganglia (Table 2). Few DRG neurons in the ventrolateral part were labeled (Fig. 4A). No signals were seen in the rest of the DRGs. The subset of neurons in which R18/VILIP could be detected was nearly identical with the trkC-positive population.
R48 encodes part of the receptor tyrosine kinase ret, which is the catalytic receptor for glial cell line-derived neurotrophic factor (GDNF) (28, 29) and was highly expressed in NGF-treated ganglia (Table 2). The most heavily labeled neurons lay more toward the medial part of the ganglion (Fig. 4B). Thus, the R48 pattern resembles that of trkA, whereby the number of R48-positive neurons exceeds the number of trkA-expressing neurons.
R45, a cDNA fragment with high similarities to mammalian cannabinoid receptor CB1 gene, was strongly expressed in NGF-treated ganglia (Table 2). R45-positive neurons were concentrated in the mediodorsal half of the ganglion, a topography that is similar to the trkA expression pattern (Fig. 4C).
DISCUSSION
Differential Expression of trk Receptors, Peptide Markers, and Sodium Channels in DRG Explants.
Our approach to study molecular constituents of sensory neurons involved two steps: first, a simple enrichment of two populations, and second, a comparison of their patterns of gene expression by RNA fingerprinting. Although it would have been conceivable to dissociate ganglia and then separate neurons by, for example, gradient centrifugation or immunopanning, such methods involve extensive manipulations, making subsequent comparisons difficult. A strategy based on the selective survival properties of neurotrophins seemed to be more straightforward, since previous experiments have demonstrated that peripheral sensory neurons with similar physiological properties depend on a single neurotrophin for survival. This is in contrast to the central nervous system, where selection of homogeneous neuronal populations by using single survival factors may not be possible. A culture system with peripheral sensory neurons from embryos thus offers a unique opportunity to enrich distinct subpopulations by simply supplementing the medium with the appropriate neurotrophin. The most obvious criterion for a successful selection of neurons by NGF or by NT3 is the differential expression of their receptors trkA and trkC. Strong differential expression of the trk genes was observed with DRG explants incubated in an optimized defined medium. This was not seen when dissociated neurons were cultured either under standard conditions or with the defined medium used for the explants, indicating that dissociated DRG neurons change patterns of gene expression in vitro. This surprising result was of critical relevance with regard to the goal of this study, and it suggests that isolated neurons start to express genes that are repressed under physiological conditions. This may result from loss of cell–cell interactions, including those involving satellite cells. In a previous study with isolated rat nodose neurons, which are normally not NGF responsive, NGF has been shown to induce the formation of dendrites, which is repressed by the presence of satellite cells (30). In explants, the physiological gene expression patterns were much better preserved, as confirmed by demonstrating that CGRP and calretinin, two markers for distinct subpopulations frequently used in rodents (21), were coexpressed in vitro and in vivo with trkA and trkC, respectively. Likewise, PPTA and parvalbumin revealed the expected differential levels of expression. Calbindin, a marker for a heterogeneous group of neurons (31), was not preferentially expressed.
In the course of our studies, we also found that voltage-gated sodium channels were differentially expressed in the DRG explants. In situ hybridization experiments revealed that sodium channel II was enriched in NT3-treated ganglia and exhibited an expression pattern very similar to that of trkC. The fact that sodium channel II is expressed in a subpopulation of DRG neurons is novel, and its association with trkC has not, to our knowledge, been reported before. In rat DRG neurons, at least three different types of sodium currents were identified on the basis of kinetic and pharmacological criteria (32). Large-diameter neurons show a single TTX-sensitive current with a inactivation constant described as intermediate (32). As trkC is expressed in large-diameter neurons of avian DRGs as in mammalian DRGs (refs. 33–36; see also Fig. 2), it is likely that sodium channel II contributes to the electrical properties of proprioceptive neurons.
The sodium channel SNS was strongly enriched in NGF-treated ganglia and revealed an expression pattern nearly identical to that of trkA. Sodium channel SNS is a chicken homologue of a voltage-gated sodium channel recently cloned from rat DRG (24, 37). This channel, which was named SNS or PN3, was found to be expressed specifically in sensory neurons. Within DRGs, SNS/PN3 mRNA was localized to small-diameter neurons. This expression pattern and the pharmacological properties, such as resistance to TTX, suggested that SNS/PN3 might be involved in the transmission of noxious stimuli. Our data provide further evidence for its exclusive expression in a defined subset of DRG neurons. Moreover, the in situ hybridization data indicate an almost complete overlap of the trkA- and SNS/PN3-expressing populations, suggesting coexpression of the two genes in DRG neurons. On the basis of gene disruption experiments in mice, NGF signaling through trkA has been implicated in nociception. The colocalization of trkA and SNS/PN3 strengthens the idea that this sodium channel is used by neurons involved in pain perception. It should be noted that the residue serine, which is thought to confer TTX resistence in the rat, is substituted for the aromatic residue tyrosine in chicken SNS/PN3 (38). This finding is in agreement with the observation that in chicken DRG neurons, only TTX-sensitive sodium currents are observed (39).
RNA Fingerprinting with DRG Explants.
Because our results indicated a strong correlation between differential gene expression in the explants and subset-specific expression patterns in vivo, we used these explants as a source for the identification of additional subset-specific genes by simply screening for expression differences. To this end, we established an RNA fingerprinting protocol that reliably displayed even low differences in expression levels when NGF- and NT3-treated explants were compared. The method uses oligo(dT)-primed cDNA in combination with two different fingerprint primers. This approach makes the method more versatile than other protocols (40) because one pool of cDNA can be used for many PCRs with different primer combinations. Another improvement is the possibility to design fingerprint primers that detect a known differentially expressed marker gene (calretinin in our case) and to adjust the reaction parameters with the help of the corresponding signal. We could thus identify a number of genes that have not been localized in a subpopulation of DRG neurons so far (see Table 2). The cannabinoid receptor CB1 (R45), the glial cell line-derived neurotrophic factor receptor ret (R48), and the calcium-binding protein VILIP (R18) are three examples of such genes (see Fig. 4). Our data demonstrate that strong quantitative difference anticipated a subset-specific expression pattern with high probability. In the case of low to moderate differences, this was less predictable. In one instance, a 4: 1 ratio corresponded with differential expression (R16), while in others it did not (R1, R24). Presumably, the small quantitative differences in expression levels may not always be revealed by in situ hybridization.
The genes identified in our screen relate to several aspects of DRG biology. Thus, cell surface proteins such as limbic system-associated membrane protein (LAMP) (R49) and axonin-1 (R16) provide hints as to how subpopulations of DRG neurons may form distinct topographic projections into the spinal cord and find specific peripheral target fields. Also, the identification of differentially expressed intracellular signaling molecules (diacylglycerol kinase-γ, R44) as well as known (delta EF1, R2) or putative transcription factors (R3) will help us to understand how the heterogeneity of DRG neurons is established and maintained. The discovery that trkA-positive neurons express CB1 (R45) indicates that cannabinoid receptors might be involved in modulating pain sensitivity not only in the central nervous system (41) but also in the peripheral nervous system.
In conclusion, the DRG explant system used here in combination with the optimized RNA fingerprinting method provides rapid and efficient means to uncover large numbers of genes correlated with the ability of sensory neurons to detect and transduce mechanical as well as noxious stimuli.
Acknowledgments
We thank Claudia Cap for oligonucleotide synthesis and sequencing.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CGRP, calcitonin gene-related peptide; DRG, dorsal root ganglion; En, embryonic day n; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NGF, nerve growth factor; NT3, neurotrophin-3; PPT, preprotachykinin; RT-PCR, reverse transcriptase–PCR; TTX, tetrodotoxin; VILIP, visinin-like protein.
References
- 1.Perl E R. In: Sensory Neurons: Diversity, Development, and Plasticity. Scott S A, editor. Oxford: Oxford Univ. Press; 1992. pp. 3–23. [Google Scholar]
- 2.Corey D P, Garcia-Anoveros J. Science. 1996;273:323–324. doi: 10.1126/science.273.5273.323. [DOI] [PubMed] [Google Scholar]
- 3.Lewin G R, Barde Y A. Annu Rev Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 4.Snider W D. Cell. 1994;77:627–638. doi: 10.1016/0092-8674(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 5.Goedert M, Otten U, Hunt S P, Bond A, Chapman D, Schlumpf M, Lichtensteiger W. Proc Natl Acad Sci USA. 1984;81:1580–1584. doi: 10.1073/pnas.81.5.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urschel B A, Brown P N, Hulsebosch C E. Exp Neurol. 1991;114:44–52. doi: 10.1016/0014-4886(91)90083-o. [DOI] [PubMed] [Google Scholar]
- 7.Ruit K G, Elliott J L, Osborne P A, Yan Q, Snider W D. Neuron. 1992;8:573–587. doi: 10.1016/0896-6273(92)90284-k. [DOI] [PubMed] [Google Scholar]
- 8.Smeyne R J, Klein R, Schnapp A, Long L K, Bryant S, Lewin A, Lira S A, Barbacid M. Nature (London) 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- 9.Crowley C, Spencer S D, Nishimura M C, Chen K S, Pitts-Meek S, Armanini M P, Ling L H, MacMahon S B, Shelton D L, Levinson A D, Phillips H S. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 10.Ernfors P, Lee K F, Kucera J, Jaenisch R. Cell. 1994;77:503–512. doi: 10.1016/0092-8674(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 11.Klein R, Silos-Santiago I, Smeyne R J, Lira S A, Brambilla R, Bryant S, Zhang L, Snider W D, Barbacid M. Nature (London) 1994;368:249–251. doi: 10.1038/368249a0. [DOI] [PubMed] [Google Scholar]
- 12.Fariñas I, Jones K R, Backus C, Wang X Y, Reichardt L F. Nature (London) 1994;369:658–661. doi: 10.1038/369658a0. [DOI] [PubMed] [Google Scholar]
- 13.Airaksinen M S, Koltzenburg M, Lewin G R, Masu Y, Helbig C, Wolf E, Brem G, Toyka K V, Thoenen H, Meyer M. Neuron. 1996;16:287–295. doi: 10.1016/s0896-6273(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 14.Gaese F, Kolbeck R, Barde Y-A. Development. 1994;120:1613–1619. doi: 10.1242/dev.120.6.1613. [DOI] [PubMed] [Google Scholar]
- 15.Oakley R A, Garner A S, Large T H, Frank E. Development. 1995;121:1341–1350. doi: 10.1242/dev.121.5.1341. [DOI] [PubMed] [Google Scholar]
- 16.Lefcort F, Clary D O, Rusoff A C, Reichardt L F. J Neurosci. 1996;16:3704–3713. doi: 10.1523/JNEUROSCI.16-11-03704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eide A L, Lewin G R, Barde Y-A. Soc Neurosci Abstr. 1994;20:1094. [Google Scholar]
- 18.Ernsberger U, Rohrer H. Dev Biol. 1988;126:420–432. doi: 10.1016/0012-1606(88)90151-0. [DOI] [PubMed] [Google Scholar]
- 19.Schnürch H, Risau W. Development. 1991;111:1143–1154. doi: 10.1242/dev.111.4.1143. [DOI] [PubMed] [Google Scholar]
- 20.LoPresti P, Scott S A. J Neurobiol. 1994;25:1613–1624. doi: 10.1002/neu.480251212. [DOI] [PubMed] [Google Scholar]
- 21.Lawson S R. In: Sensory Neurons: Diversity, Development, and Plasticity. Scott S A, editor. Oxford: Oxford Univ. Press; 1992. pp. 27–59. [Google Scholar]
- 22.Averill S, McMahon S B, Clary D O, Reichardt L F, Priestley J V. Eur J Neurosci. 1995;7:1484–1494. doi: 10.1111/j.1460-9568.1995.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baimbridge K G, Celio M R, Rogers J H. Trends Neurosci. 1992;15:303–308. doi: 10.1016/0166-2236(92)90081-i. [DOI] [PubMed] [Google Scholar]
- 24.Akopian A N, Sivilotti L, Wood J N. Nature (London) 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- 25.Schröpel A, von Schack D, Dechant G, Barde Y A. Mol Cell Neurosci. 1995;6:544–556. doi: 10.1006/mcne.1995.0006. [DOI] [PubMed] [Google Scholar]
- 26.Debouck C. Curr Opin Biotechnol. 1995;6:597–599. [Google Scholar]
- 27.Lenz S E, Henschel Y, Zopf D, Voss B, Gundelfinger E D. Mol Brain Res. 1992;15:133–140. doi: 10.1016/0169-328x(92)90160-d. [DOI] [PubMed] [Google Scholar]
- 28.Buj-Bello A, Buchman V L, Horton A, Rosenthal A, Davies A M. Neuron. 1995;15:821–828. doi: 10.1016/0896-6273(95)90173-6. [DOI] [PubMed] [Google Scholar]
- 29.Trupp M, Rydén M, Jörnvall H, Funakoshi H, Timmusk T, Arenas E, Ibáñez C F. J Cell Biol. 1995;130:137–148. doi: 10.1083/jcb.130.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Koninck P, Carbonetto S, Cooper E. J Neurosci. 1993;13:577–585. doi: 10.1523/JNEUROSCI.13-02-00577.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carr P A, Yamamoto T, Karmy G, Baimbridge K G, Nagy J I. Brain Res. 1989;497:163–170. doi: 10.1016/0006-8993(89)90983-9. [DOI] [PubMed] [Google Scholar]
- 32.Caffrey J M, Eng D L, Black J A, Waxman S G, Kocsis J D. Brain Res. 1992;592:283–297. doi: 10.1016/0006-8993(92)91687-a. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Zhou X F, Rush R A. Neurochem Res. 1996;21:809–814. doi: 10.1007/BF02532304. [DOI] [PubMed] [Google Scholar]
- 34.Williams R, Ebendal T. Neuroreport. 1995;6:2277–2282. doi: 10.1097/00001756-199511270-00003. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Yao L, Bernd P. J Neurobiol. 1994;25:1517–1532. doi: 10.1002/neu.480251205. [DOI] [PubMed] [Google Scholar]
- 36.Mu X, Silos-Santiago I, Carroll S L, Snider W D. J Neurosci. 1993;13:4029–4041. doi: 10.1523/JNEUROSCI.13-09-04029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sangameswaran L, Delgado S G, Fish L M, Koch B D, Jakeman L B, Stewart G R, Sze P, Hunter J C, Eglen R M, Herman R C. J Biol Chem. 1996;271:5953–5956. doi: 10.1074/jbc.271.11.5953. [DOI] [PubMed] [Google Scholar]
- 38.Satin J, Kyle J W, Chen M, Bell P, Cribbs L L, Fozzard H A, Rogart R B. Science. 1992;256:1202–1205. doi: 10.1126/science.256.5060.1202. [DOI] [PubMed] [Google Scholar]
- 39.Petersen M, Pierau F K, Weyrich M. Pflügers Arch. 1987;409:403–410. doi: 10.1007/BF00583794. [DOI] [PubMed] [Google Scholar]
- 40.Welsh J, Chada K, Dalal S S, Cheng R, Ralph D, McClelland M. Nucleic Acids Res. 1992;20:4965–4970. doi: 10.1093/nar/20.19.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin W J, Hohmann A G, Walker J M. J Neurosci. 1996;16:6601–6611. doi: 10.1523/JNEUROSCI.16-20-06601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]