Abstract
The peptides bound to class II major histocompatibility complex (MHC) molecules extend out both ends of the peptide binding groove. This structural feature provided the opportunity to design multivalent polypeptide chains that cross-link class II MHC molecules through multiple, repetitive MHC binding sites. By using recombinant techniques, polypeptide oligomers were constructed that consist of up to 32 copies of an HLA-DR1-restricted T cell epitope. The epitope HA306–318, derived from influenza virus hemagglutinin, was connected by 12- to 36-aa long spacer sequences. These oligomers were found to cross-link soluble HLA-DR1 molecules efficiently and, upon binding to the MHC molecules of a monocyte line, to trigger signal transduction indicated by the enhanced expression of some cell surface molecules. A particularly strong effect was evident in the T cell response. A hemagglutinin-specific T cell clone recognized these antigens at concentrations up to three to four orders of magnitude lower than that of the peptide or the hemagglutinin protein. Both signal transduction in the monocyte and the proliferative response of the T cell were affected greatly by the length of the oligomer (i.e., the number of repetitive units) and the distance of the epitopes within the oligomer (spacing). Thus, the formation of defined clusters of T cell receptor/MHC/peptide antigen complexes appears to be crucial for triggering the immune response and can be used to enhance the antigenicity of a peptide antigen by oligomerizing the epitope.
Dimerization or oligomerization of cell surface receptors upon binding of ligand is a common mechanism that initiates signal transduction (1). The T cell-mediated immune response results from association of the surface T cell receptor (TCR) with the major histocompatibility complex (MHC)/peptide antigen complex, and it is very likely that TCR-mediated signal transduction also is triggered by this mechanism. Although crystallographic studies of both class II MHC/peptide molecules (2, 3) and TCR Vα-chains (4) revealed structures corresponding to a putative dimeric complex, so far the co-crystallization of TCR and class I MHC/peptide complexes reveal only monomeric forms (5, 6). Nevertheless, evidence in support of the aggregation model arises from the simple fact that a T cell can be activated by adding divalent TCR-specific antibodies but not by adding monovalent Fab fragments (7). In a recent study (8) (using a quasi-elastic light scattering technique), the antigen-induced formation of TCR/MHC aggregates even was demonstrated directly. Previous experiments with hapten-linked polymers already had suggested that the extent and qualitative nature of the T cell response is effected by the size of these clusters (9).
Experiments with MHC-specific antibodies have shown that class II MHC molecules also represent receptor molecules sensitive to cross-linking (10). Because the clustering of TCR/MHC/peptide complexes apparently plays such a crucial role for both T cells and antigen-presenting cells (APC), we attempted to influence the immune response by promoting the artificial formation of such clusters. Class II MHC molecules, in contrast to class I MHC molecules, do not impose length restrictions on their ligands (2, 11). The peptide is attached to the binding site like a string of rope to a clamp, with both ends extruding from the MHC molecule. It was therefore assumed that the formation of antigen-loaded MHC clusters can be achieved simply by allowing multiple class II MHC molecules to bind to a single multivalent chain of peptide antigens. For this purpose, polypeptides were produced that contained multiple copies of a T cell epitope derived from the influenza virus hemagglutinin protein (HA306–318; ref. 12). This peptide was known to bind to the class II MHC molecule HLA-DR1 with high affinity (13) and a high-resolution crystal structure of the respective MHC/peptide complex already was available (3).
MATERIALS AND METHODS
Construction and Production of T Cell Epitope Oligomers.
Double-stranded oligonucleotide units were generated by annealing two complementary strands of synthetic oligonucleotides (see Fig. 1A; HA306–318 T cell epitope, + strand: 5′-TCCGAAATACGTTAAACAGAACACCCTGAAACTGGCTACCGG, − strand: 5′-GGTAGCCAGTTTCAGGGTGTTCTGTTTAACGTATTTCGGACC; HA 306–318 (Y308D) T cell epitope, + strand: 5′-TCCGAAAGACGTTAAACAGAACACCCTGAAACTGGCTACCGG, − strand: 5′-GGTAGCCAGTTTCAGGGTGTTCTGTTTAACGTCTTTCGGACC; S3 spacer, + strand: 5′-CGGTCCGGGTGGCGGTCCGGGTGGCGGTCCGGGTGG, − strand: 5′-ACCCGGACCGCCACCCGGACCGCCACCCGGACCGCC). The codon usage was adapted to the preferences of the host bacterium. For the construction of the full length DNA, an interface oligonucleotide (see Fig. 1B; + strand: 5′-TATGGCAATGGGATTC, − strand: 5′-TCGAGAATGCCCATTGCCA) was introduced into the pCITE 3a and pET 22b vectors (Novagen; one internal BsmI and two internal BsrDI sites of the pCITE 3a vector had been removed previously by site-directed mutagenesis). For cloning, the bacterial host TOP 10 (Invitrogen) was used, the production of the polypeptide oligomers was carried out in TOP 10 λDE3 pLysS [generated by using a lysogenation kit (Novagen)]. Endotoxin and other impurities were removed from the polypeptide oligomers by separation on a reversed phase C4 HPLC column (Vydac, Hesperia, CA).
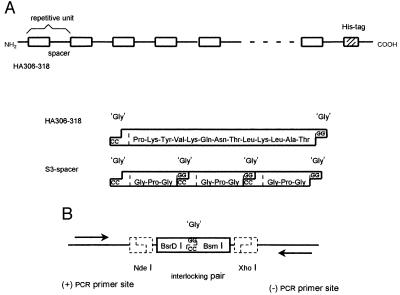
Figure 1.
Design and construction of T cell epitope oligomers. (A) Structure of HA306–318 T cell epitope oligomers and of the repetitive elements. The full length construct and the units representing the HA306–318 epitope and the S3 spacer are shown (amino acid sequence is indicated in 3-letter code). The oligonucleotide units are connected through short 3′-2-bp overhangs, GG on the coding and CC on the noncoding strand. They constitute a glycine codon (GGX) at the connection and ensure orientation-specific ligation. (B) Schematic presentation of the vector interface. For the generation of compatible GG/CC overhangs, an interface oligonucleotide was introduced into the cloning and the expression vector. Central to this interface are two nonpalindromic restriction sites, BsrDI and BsmI (the boxed symbol represents the interlocking pair of their recognition sites). They flank a cutting site, which can be used independently with either one of the two enzymes (the location of GG/CC overhangs, created after opening of the cutting site, is indicated). The interface is integrated into the vector by the restriction sites NdeI and XhoI (the NdeI site also provides the ATG start codon). The two primer sites, actually part of the vector and located outside the interface, are used to amplify cloned inserts by PCR.
Binding of Polypeptide Oligomers to Soluble HLA-DR1 Molecules.
Soluble HLA-DR1 molecules were prepared from baculovirus-infected cells as described (14) and kindly were provided by the laboratory of D. Wiley. The binding studies were carried out by incubating 3.75 μl of HLA-DR1 (1 mg/ml) with 0.5 μl of oligomer (1 mg/ml) in PBS for 16 h at 37°C. SDS/PAGE separation was done at 4°C.
Western Blotting and Staining with HLA-DR-Specific Antibodies.
The proteins were transferred by tank blotting from the gel onto polyvinylidene difluoride (PVFD) membranes (Bio-Rad) by using 25 mM Tris/192 mM glycine/0.1% SDS. The blots were incubated for 1 h with rabbit sera specific for HLA-DR molecules (anti-HLA-DRαβ: 1:1000; anti-HLA-DRα: 1:500; anti-HLA-DRβ: 1:500, kindly provided by H. Ploegh) and with goat anti-rabbit immunoglobulin G coupled to horseradish peroxidase (DAKO; 1:8000). The blot was developed by autoradiography after incubation with enhanced chemiluminescence according to the manufacturer’s manual (Amersham).
Cell Lines and T Cell Clones.
THP1 cells were obtained from American Type Culture Collection (TIB 202) and maintained in RPMI 1640 medium/10% fetal calf serum (the density always was kept below 500,000 cells/ml). The T cell clone HA1.7 (kindly provided by J. Lamb; ref. 12) was maintained in 96-well round bottom plates in RPMI 1640 medium/10% human serum (BioWhitaker) supplemented with 5 units/ml interleukin 2 (Boehringer Mannheim). Restimulation of this clone was carried out every 2 weeks with 10,000 radiated (6000 rad) heterologous PBMC in the presence of 5 μg/ml phytohemagglutinin (Wellcome) and 5 units/ml interleukin 2.
Fluorescence-Activated Cell Sorter (FACS) Analysis and Induction of Cell Surface Expression.
THP1 cells (25,000) were incubated for 40 h in 96-well flat bottom plates with antigen and interferon-γ (Collaborative Biomedical Products, Bedford, MA) in RPMI 1640 medium/10% fetal calf serum. The following antibodies were used for the staining: L243 (anti-HLA-DR), B7/21 (anti-HLA-DP), anti-HLA-DQ-FITC (Becton Dickinson), anti-ICAM-1 (Becton Dickinson), and goat anti-mouse immunoglobulin G F(ab′)2–fluorescein isothiocyanate (Cappel).
Proliferation Assays.
Proliferation assays were performed in 96-well round bottom plates by using 150,000 radiated (6000 rad) HLA-DR1-expressing PBMC and 50,000 HA1.7 T cells per well in RPMI 1640 medium/10% human serum. In experiments in which antigen-pulsed targets were used, the PBMC were preincubated for 15 min with the indicated amounts of antigen in serum-free Hybridoma medium (GIBCO) and washed thoroughly before the addition of T cells. [3H]Thymidine (1 μCi/well) was added after 48 h, and the assay was harvested after 72 h and counted in a microbetaplate reader (Wallac, Gaithersburg, MD).
RESULTS AND DISCUSSION
Production of T Cell Epitope Oligomers.
The T cell epitope oligomers used in these experiments are linear polypeptides, consisting of alternating repeats of a T cell epitope and a spacer (Fig. 1A). They represent “artificial” proteins with molecular mass of up to 76 kDa and are produced recombinantly in Escherichia coli. The epitope spacer repeats used contain up to 32 copies of the HA306–318 T cell epitope (HA) connected by spacer sequences, which consist of 3, 6, or 9 tandem repeats of the tetrapeptide -G-P-G-G- (termed S3, S6, and S9, respectively). In addition to these epitope spacer repeats, the full length construct contains only 13 additional amino acids. They are encoded by the expression vector and include a His tag used for the purification of the protein. The DNA encoding the epitope spacer repeats was constructed out of small synthetic oligonucleotide units. Sequence and structure of the two basic elements, the HA306–318 T cell epitope and the S3 spacer, are shown in Fig. 1A. The repeating unit of a HAS3 oligomer encodes 26 amino acids (HA13, S12, and one additional Gly at the joint to the next unit).
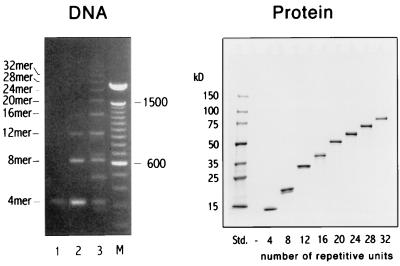
The full length DNA constructs were generated in the following way: First, tetrameric epitope spacer units were produced by several rounds of cloning, extending, and recloning of inserts by using a modified cloning vector (Fig. 1B). A complete size series of oligonucleotide oligomers then was generated in a single step by fragment condensation, i.e., by directly connecting the tetrameric oligonucleotide units to each other (Fig. 2, Left). Separated by size, the reaction products yield a ladder of bands spaced by the size of one tetrameric unit, with the individual bands corresponding to oligonucleotides encoding 4–32 repetitive HAS3 units. The individual bands were isolated, cloned into an expression vector, and expressed in a bacterial host. The polypeptide oligomers were isolated with Ni2+-nitrilotriacetic acid (NTA) agarose and finally purified by RP-HPLC. An SDS/PAGE separation of the size series of HAS3 polypeptide oligomers is shown in the right panel of Fig. 2.
Figure 2.
Production and characterization of T cell epitope oligomers. (Left) A size separation of the reaction products of a fragment condensation of the HAS3 4 mer is shown. To identify the products and to control the reaction, titrated amounts of ligase were used [lanes: 1, no ligase; 2, 250 units/ml; 3, 1200 units/ml; M, 100-bp marker (GIBCO)]. The ladder of bands corresponds to oligonucleotides with increasing number of repetitive units (n; indicated on the left side of the gel). The theoretical molecular mass can be calculated by MM = n × 78 bp (one HAS3 unit). Some bands do not match the theoretical MM and probably correspond to cyclic ligation products (e.g., lane 3: ≈480 bp). (Right) An SDS/PAGE separation of the respective polypeptide oligomers is shown. The number of repetitive units is indicated at the bottom of each lane. Their theoretical molecular mass can be calculated by MM = n × 2332 (one HAS3 unit) + 1574 (13 amino acids encoded by the vector).
Binding of HA306–318 Oligomers to Soluble HLA-DR1 Molecules.
The binding capability of the T cell epitope oligomers was tested by incubation with “empty” soluble HLA-DR1 molecules (14) and analyzed by SDS/PAGE. Because the oligomers were supposed to bind multiple HLA-DR1 molecules, a series of discrete bands corresponding to complexes of various stoichiometric ratios was expected to appear (Fig. 3).
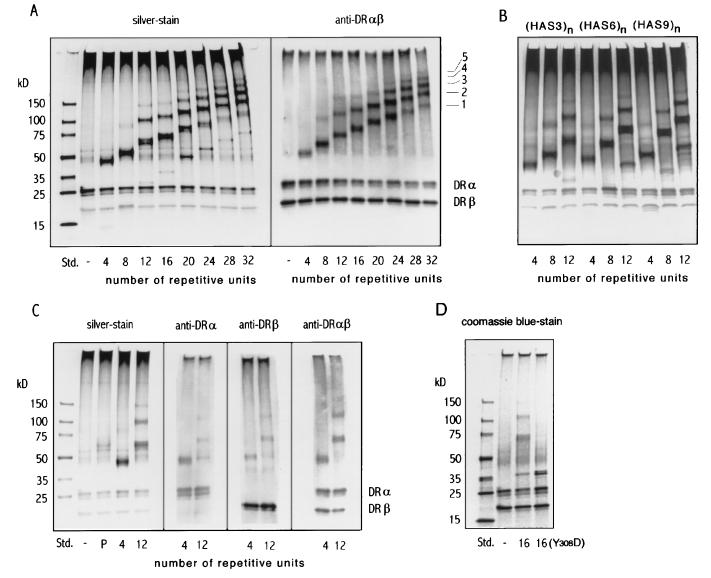
Figure 3.
Interaction of HAS3 polypeptide oligomers with soluble HLA-DR1 molecules. SDS/PAGE separations of oligomers after incubation with soluble “empty” HLA-DR1 molecules are shown. (A) Separation of the HLA-DR1 complexes formed with the HAS3 size series (Fig. 2). The bands were visualized by silver staining (Left) or, after Western blotting, by staining with rabbit anti-HLA-DRαβ serum (Right). The bands corresponding to the HLA-DRα and the HLA-DRβ chain are indicated. The putative stoichiometric ratio of bands referring to HLA-DR1/oligomer complexes are indicated and are labeled 1–5. [In the left panel, faint bands that correspond to uncomplexed oligomer are also evident at the appropriate positions]. (B) Effect of spacer length variation on HLA-DR1 binding. The epitope spacing of each series is indicated above the gel, and the oligomer length is labeled below each lane. The gel was visualized by silver staining. For this experiment, 0.5 μg of HAS3, 0.7 μg of HAS6, and 0.9 μg of HAS9 oligomers were used, corresponding to equimolar quantities of the HA306–318 epitope contained in oligomers (≈0.2 nmol). (C) Characterization of HLA-DR1/oligomer complexes with HLA-DR-specific antibodies. The HLA-DR1 molecules were incubated previously with the HA306–318 peptide (P), the HAS3 4-mer (4), and the HAS3 12-mer (12) or without antigen (−). The bands were visualized either directly by silver staining or by staining Western blots with rabbit sera specific for the HLA-DRα chain, the β chain, or the HLA-DRαβ complex. (D) SDS/PAGE separation of HLA-DR1 molecules incubated with the HAS3 16-mer or with the Y308D 16-mer (stained with Coomassie blue).
Visualized by silver staining, each oligomer, in fact, yielded a ladder of bands, with the number of these bands increasing with the number of epitopes contained by the oligomer (Fig. 3A Left). The specific staining of these bands by an anti-HLA-DRαβ antiserum confirmed that they correspond to complexes of class II MHC molecules (Fig. 3A Right). The stoichiometric ratios of HLA-DR to oligomer in these complexes presumably range from 1:1 up to 5:1 HLA-DR1-to-oligomer (represented by the fastest and slowest migrating bands, respectively). Additional MHC molecules appear to be added at every length increase of ≈5–8 HAS3 units. Only a single major band is detectable for the HAS3 4-mer. However, it usually is accompanied by a faint secondary band indicating that a very small fraction of the 4-mer also is associated with two HLA-DR1 molecules.
Within a series, the number and density of bands, corresponding to complexes with higher stoichiometric ratios, increased gradually with the length of the oligomer. Oligomer length, however, did not seem to have a substantial impact on the binding efficiency. The total amount of HLA-DR bound to the oligomers was affected only slightly, i.e., the summation of density scans of the bands revealed similar values for all oligomers (data not shown). Also, the distance of the epitope did not seem to effect the binding of soluble HLA-DR1 (Fig. 3B). Irrespective of whether the HA306–318 epitopes were spaced by 13 (HAS3), by 25 (HAS6) or by 37 amino acids (HAS9), essentially the same number (and amount) of class II MHC molecules were found to be associated with the respective oligomer. Notably, when analyzing the apparent molecular mass of the HLA-DR1/oligomer complexes, an anomaly of the gel migration was evident (Fig. 3C, left). Although the molecular mass of the 4-mer is higher than that of the peptide, the respective band of the HLA-DR1 complex was observed at an apparent molecular mass of 46 kDa (calculated size 57 kDa: 11-kDa 4-mer + 46-kDa HLA-DR) whereas the peptide complex migrated at 62 kDa (calculated 47 kDa). The reduction of the apparent molecular mass is reminiscent of the conversion from the “floppy” to the “compact” form (15, 16) and was observed for all MHC molecules bound to the oligomers, i.e., the spacing between individual bands of the ladders was equivalent to an addition of only 32 kDa.
To establish that, in fact, HLA-DRαβ heterodimers, and not separate α- or β-chains, were bound to the oligomers, Western blots were carried out and stained with polyclonal rabbit sera specific for the HLA-DRα or the β chain. The same bands, which were stained with the antibody specific for the αβ complex, also stained positive with the antibodies specific for the individual chains (Fig. 3C). To further ensure that the interaction with the oligomer occurs within the peptide binding site of the class II molecules, a modified HAS3 oligomer was constructed in which the tyrosine 308 residues of the HA306–318 epitopes were substituted by aspartate residues. The Asp substitution of Tyr308 affects an important “anchor” residue, and it has been shown that this substitution virtually abolishes the binding of the peptide to the MHC molecule (13). In contrast to the unsubstituted 16-mer, the substituted Y308D 16-mer failed to form any bands that corresponded to monomeric or multimeric MHC/oligomer complexes (Fig. 3D).
Induction of the Expression of Cell Surface Molecules in THP1 Cells.
The monocyte cell line THP1 has been used in previous studies to document class II MHC-induced signal transduction (17). In this cell line, the cross-linking of class II MHC molecules, for example with staphylococcal enterotoxin A, triggers or enhances the expression of a number of molecules relevant for the immune response. To test whether THP1 cells also respond to HA306–318 epitope oligomers, the expression levels of several cell surface molecules were monitored by FACS analysis (Fig. 4). The cell surface molecules analyzed included the class II MHC molecules HLA-DR, -DP, and -DQ as well as the intercellular adhesion molecule 1 (ICAM-1).
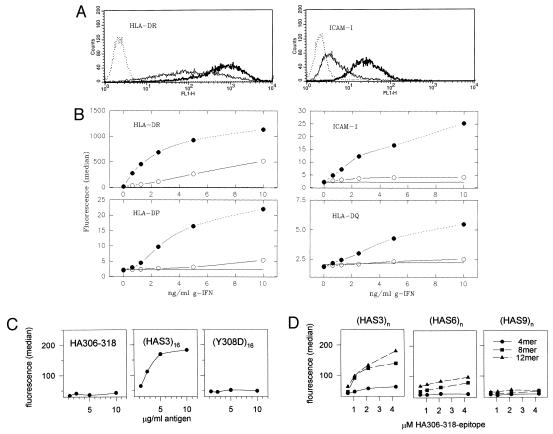
Figure 4.
Induction of cell surface expression by HA306–318 oligomers. The cell surface expression of MHC class II molecules and ICAM-1 were determined by FACS analysis after incubation of THP1 cells with oligomers. (A) FACS profiles displaying the cell surface expression of HLA-DR and ICAM-1 are shown. The THP1 cells were incubated without (solid thin line) or with 10 μg/ml HAS3 16 mer (solid bold line) in the presence of 2.5 ng/ml (HLA-DR) or 10 ng/ml interferon-γ (ICAM-1). The dashed line corresponds to an isotype-matched control. (B) The dose-response curves of interferon-γ for the induction of four cell surface molecules: HLA-DR, -DP, -DQ, and ICAM-1 in the absence (open circles) or presence of 10 μg/ml HAS3 16 mer (closed circles) are shown. Data are plotted as the median fluorescence under each condition. The line without symbols represents the median fluorescence measured with an isotype-matched control antibody. (C) The median of the cell surface expression of HLA-DR molecules after incubation with HA306–318 peptide, the HAS3 16-mer, or a 16-mer containing the Y308D substitutions is shown. THP1 cells were incubated with the indicated amounts of peptide or oligomer in the presence of 0.5 ng/ml interferon-γ. (D) Influence of oligomer length and spacing. The cell surface expression of HLA-DR after incubation with 4-mers (circles), 8-mers (square), and 12-mers (triangle) of the S3, S6, and S9 series is shown. The same conditions as in C were used; the x axis refers to the molar concentration of the HA306–318 epitope.
After incubation of the cells with the HAS3 16 mer, an increase of surface expression of all three isotypes of class II MHC molecules was observed (Fig. 4 A and B). The surface expression was 5- to 10-fold greater than that found in the absence of the oligomer. Class II MHC molecules are known to be inducible by interferon-γ, and the oligomer-dependent enhancement was, in fact, strictly dependent on the presence of this cytokine. For the demonstration of signal transduction, however, the up-regulation of ICAM-1 is of particular importance. ICAM-1 is an adhesion molecule that is not expected to interact directly with the oligomers, but also here an interferon-γ-dependent elevation in cell surface expression was observed similar to that of the class II MHC molecules (Fig. 4 A and B).
ICAM-1 also was found to be inducible by treatment with another MHC class II crosslinker, staphylococcal enterotoxin A (data not shown). In contrast to the oligomers, staphylococcal enterotoxin A does not enhance the expression levels of the three MHC class II isotypes, and, under the conditions of this experiment, neither staphylococcal enterotoxin A nor the oligomers were able to effect the expression of B7-1 or B7-2 (data not shown). The latter have been reported to be inducible by signaling through class II MHC molecules (18). To ensure that also the oligomer-induced effect is caused by MHC cross-linking, control experiments were carried out in which the effect of the 16 mer was compared with that of the peptide and the substituted Y308D 16 mer (Fig. 4C). In contrast to the 16-mer, little or no induction was evident for both the peptide and the Y308D 16-mer.
Induction experiments using size series of HA306–318-oligomers (Fig. 4D) further revealed that the level of cell surface expression increases gradually with the number of epitope repeats per oligomer and that only a little effect is observed after incubation with 4-mers [a more significant increase is usually seen for the HAS3 4-mer at higher concentrations of interferon-γ (not shown)]. In contrast to the binding of soluble HLA-DR1 molecules, here also the spacer length had a substantial impact. Comparing oligomers of the S3, the S6, and the S9 series, the increase in spacer length resulted in a decreasing ability to induce cell surface expression. The activity of oligomers connected by the 25-aa long S6 spacer was significantly reduced and almost negligible if the epitopes were separated by the 37-aa long S9 spacer. Nevertheless, in all three series, the influence of repetition was evident. Oligomers longer than the 12-mer of the S6 or S9 series have not been analyzed yet but within the HAS3 series a maximal induction was observed usually with the 12-mer up to the 20 mer and seemed to decline with larger sizes (data not shown).
Effect of HA306–318 Oligomers on T Cell Recognition.
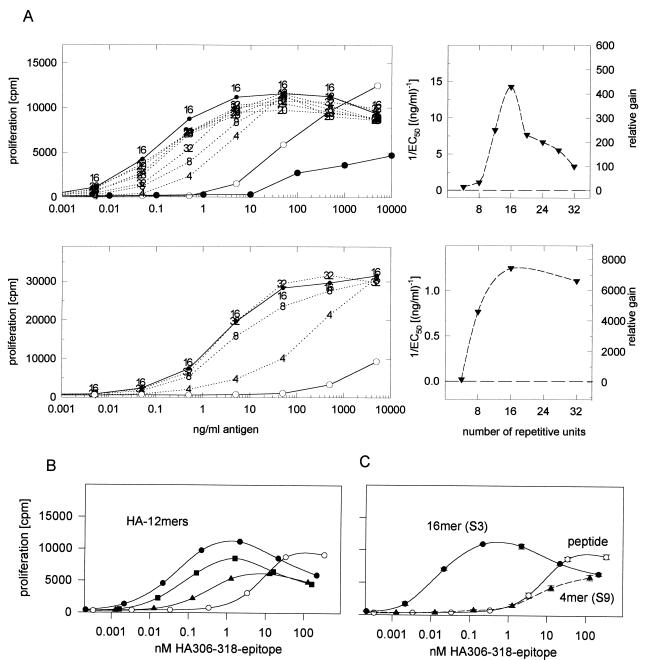
The influence of epitope oligomerization on the T cell response was tested by using the T cell clone HA1.7 as responder and HLA-DR1-expressing PBMC as APC (Fig. 5). The dose–response curves of the size series of HAS3 oligomers, the HA306–318 peptide, and the hemagglutinin protein are shown in Fig. 5A. The proliferation assays were carried out in two ways: The antigens were either present continuously during the stimulation of the HA1.7 cells (Fig. 5A, upper) or pulsed only briefly (15 min) onto the PBMC (Fig. 5A, lower). In both experiments, the oligomers were found to be substantially more efficient as T cell stimulators than the peptide or the protein. When the antigen was present continuously, the amounts required to induce half-maximal proliferation (EC50) was up to 500 times lower for the most effective oligomer (16 mer: 0.07 ng/ml) than for the peptide (35 ng/ml). Compared with the protein, the enhancement was even more significant. It amounted to 3–5 logs (because of the weak T cell response triggered by the protein, the EC50 value was more difficult to determine and was estimated to be 100–1000 ng/ml). An even larger difference in the sensitivity between peptide and oligomer was observed in the pulse experiment. Here, the EC50 of the 16-mer (3 ng/ml) was found to be ≈4 logs lower than that of the peptide (>10,000 ng/ml; under these conditions, no response to the protein usually was detected).
Figure 5.
Proliferation of the T cell clone HA1.7 challenged with various HA306–318 oligomers, the peptide, or the hemagglutinin protein. (A, left) Oligomers of the HAS3 series (see Fig. 2) were tested, and the dose–response curve was compared with that of the peptide (open circle) and the protein (closed circle). The antigens were present continuously during the 72-h assay (Upper) or pulsed onto the PBMC for only 15 min (Lower). Right panels: The reciprocal of the EC50 (left axis) of each oligomer is plotted against the length (i.e., the number of repetitive units) of the oligomer (the dashed line represents the 1/EC50 value of the peptide). The relative gain in sensitivity compared with the peptide [EC50(P)/EC50(n)] is indicated on the right. (B) Effect of spacer length. The influence of spacer length was tested with a set of 12-mers, connected either with the S3 (closed circle), the S6 (closed square), or the S9 spacer (closed triangle) or with the peptide (open circle). The antigens were continuously present during the experiment, and the proliferative response is plotted against the molar concentration of HA306–318 epitopes to compensate for variations in the molecular mass due to the different spacer lengths. (C) Comparison of the T cell response triggered by the HAS3 16-mer, the HAS9 4-mer, and the peptide. The figure summarizes the effect of distance and repetition on the antigenicity of oligomers. The dose-response curves of the weakest (HAS9 4-mer) and the most potent oligomers (HAS3 16-mer) are shown and compared with that of the peptide.
The influence of repetition is illustrated in a plot of the reciprocal EC50 values against the number of repetitive units per oligomer (Fig. 5A Upper Right and Lower Right). The relative gain in sensitivity (indicated on the right axis) describes a curve, which, in the case of the continuous exposure experiment, is characterized by a defined maximum (16-mer). The steepest slope of the curve is observed within the size range from the 4-mer [1/EC50: 10 (ng/ml)−1] to the 12-mer [1/EC50: 240 (ng/ml)−1]. Here, the reciprocal EC50 values appear to be almost exponentially related to the repetition number of the oligomer. The curve derived from the pulse experiment does not show this maximum but rather resembles a saturation curve. This change in shape, caused by the very short exposure time, suggests that longer oligomers bind faster to the target cells than the shorter ones. The greatest shift in the EC50 was observed for the peptide. Consequently, in the pulse experiment, the relative gains of the oligomers were even larger, indicating that, under these conditions, oligomers are up to 7000 times more effective than the peptide.
To address the influence of epitope spacing, HA1.7 T cells were challenged with 12-mers of the S3, S6, and S9 spacer series (Fig. 5B). Analogous to the induction experiments, also for T cell stimulation, oligomers with the S3 spacer seemed to exhibit the strongest biological activity. Both the EC50 and the maximal proliferation appeared to be affected by the increase in spacing. Whereas the EC50 of the HAS6 12-mer was only approximately three to four times higher than that of the HAS3 12-mer, the effectiveness of the HAS9 12-mer dropped by a factor of 50–100. The maximal proliferation decreased in the same manner and was particularly evident for the oligomer with the S9 spacing.
Although oligomer/MHC clusters on the cell membrane were not visualized in the present study, circumstantial evidence supports their formation. The proliferation experiments with the T cell clone HA1.7 demonstrated that the oligomerization of the HA306–318 peptide dramatically amplified the antigenicity of the epitope. It is already a well established fact that TCRs interact more efficiently with multimerized than with single MHC/peptide complexes. For example, tetrameric MHC/antigen complexes, generated by the cross-linking of biotinylated MHC molecules with avidin, bind to the TCR so tightly that they can be used to identify T cells specific for the antigen (19), and also some functional studies suggest a preferred interaction with dimerized MHC molecules (20, 21). A kinetic proofreading model even postulates the formation of large TCR/MHC/antigen clusters during antigen recognition (22) so that the systematic influence of the oligomer structure—evident in the T cell response as well as in APC-signaling—might well reflect its impact on geometry and size of these arrays. The strongest biological effect was observed for the T cell response. The variation of distance repetition resulted in a modulation of the antigenicity over a range of more than 3 logs (Fig. 5C); the HA306–318 epitopes of the HAS9 4-mer were even slightly less potent than the free peptide but, oligomerized to a 16-mer and connected by the short S3 spacer, their EC50 value dropped by a factor of almost 1000. In general, an increase in activity was seen on increasing the number of epitope spacer units (e.g., from 4-mer to 16-mer). The maximum activity observed for the 16-mer (Fig. 5A Upper), however, suggested that, after the oligomer reached a certain length, other factors, such as diffusion, might have limited the effect. Additional effects, which, in principal, could also contribute to an enhancement of activity, are (i) the improvement of APC “fitness,” evident in the amplification of the interferon-γ induction of expression of surface molecules, (ii) the “proteinization” of the epitope, potentially allowing the epitope to use the antigen presentation pathways of “professional” APCs, and (iii) the increase in avidity. However, at least in the case of the HA306–318 oligomers, in particular the first two aspects are probably of minor relevance because the addition of the HA306–318-16-mer did not improve the antigen-specific response of T cell lines specific for other antigens and chloroquine had little effect on the recognition of HA306–318 oligomers (data not shown).
The HA306–318 peptide was selected as a model system to study the impact of epitope oligomerization. The effectiveness of other oligomers might depend on structural features such as the solubility, affinity, and protease resistance of the particular epitope, but if these data can be extended, a number of practical applications are apparent, including the enhancement of immune responses to peptide antigens derived from infectious agents or tumors. Many class II MHC-restricted epitopes already have been identified (23, 24) but most are only weakly immunogenic in vivo. Self-peptide epitopes involved in autoimmune diseases also are known, and “high zone tolerance” to these epitopes, by using either the peptide (25, 26) or altered peptide ligands (27, 28), is a possible therapeutic approach. In all of these applications, however, the concentrations of peptides were usually very high and may be substantially lowered if used in an oligomerized form.
Acknowledgments
We are very grateful to our colleagues for providing empty HLA-DR1 produced in baculovirus and hemagglutinin protein (Jun Park, Michael Eisen, and Don Wiley), rabbit anti HLA-DR sera (Hidde Ploegh), and T cell clone HA1.7 (Jonathan Lamb) and for blood donation (Hugh Reyburn, who also provided stimulating discussion). This research was supported by a grant from the National Institutes of Health (CA-47554) and by fellowships from the Deutsche Forschungsgemeinschaft to K.F. and O.R.
ABBREVIATIONS
- TCR
T cell receptor
- MHC
major histocompatibility complex
- PBMC
peripheral blood mononuclear cells
- FACS
fluorescence-activated cell sorter
- ICAM-1
intercellular adhesion molecule 1
- APC
antigen-presenting cells
References
- 1.Heldin C H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 2.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 3.Stern L J, Brown J H, Jardetzky T S, Gorga J C, Urban R G, Strominger J L, Wiley D C. Nature (London) 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 4.Fields B A, Malchiodi E L, Li H, Ysern X, Stauffacher C V, Schlievert P M, Karjalainen K, Mariuzza R A. Nature (London) 1996;384:188–192. doi: 10.1038/384188a0. [DOI] [PubMed] [Google Scholar]
- 5.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 6.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 7.Kaye J, Janeway C A., Jr J Exp Med. 1984;159:1397–1412. doi: 10.1084/jem.159.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reich Z, Boniface J J, Lyons D S, Borochov N, Wachtel E J, Davis M M. Nature (London) 1997;387:617–620. doi: 10.1038/42500. [DOI] [PubMed] [Google Scholar]
- 9.Symer D E, Dintzis R Z, Diamond D J, Dintzis H M. J Exp Med. 1992;176:1421–1430. doi: 10.1084/jem.176.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop G A. J Immunol. 1991;147:1107–1114. [PubMed] [Google Scholar]
- 11.Urban R G, Chicz R M, Vignali D A, Strominger J L. Chem Immunol. 1993;57:197–234. [PubMed] [Google Scholar]
- 12.Lamb J R, Eckels D D, Lake P, Woody J N, Green N. Nature (London) 1982;300:66–69. doi: 10.1038/300066a0. [DOI] [PubMed] [Google Scholar]
- 13.Jardetzky T S, Gorga J C, Busch R, Rothbard J, Strominger J L, Wiley D C. EMBO J. 1990;9:1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stern L J, Wiley D C. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 15.Sadegh-Nasseri S, Germain R N. Nature (London) 1991;353:167–170. doi: 10.1038/353167a0. [DOI] [PubMed] [Google Scholar]
- 16.Sadegh-Nasseri S, Stern L J, Wiley D C. Nature (London) 1994;370:647–650. doi: 10.1038/370647a0. [DOI] [PubMed] [Google Scholar]
- 17.Mehindate K, Thibodeau J, Dohlsten M, Kalland T, Sekaly R P, Mourad W. J Exp Med. 1995;182:1573–1577. doi: 10.1084/jem.182.5.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nabavi N, Freeman G J, Gault A, Godfrey D, Nadler L M, Glimcher L H. Nature (London) 1992;360:266–268. doi: 10.1038/360266a0. [DOI] [PubMed] [Google Scholar]
- 19.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 20.Abastado J P, Lone Y C, Casrouge A, Boulot G, Kourilsky P. J Exp Med. 1995;182:439–447. doi: 10.1084/jem.182.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dal Porto J, Johansen T E, Catipovic B, Parfiit D J, Tuveson D, Gether U, Kozlowski S, Fearon D T, Schneck J P. Proc Natl Acad Sci USA. 1993;90:6671–6675. doi: 10.1073/pnas.90.14.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKeithan T W. Proc Natl Acad Sci USA. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rammensee H G, Friede T, Stevanoviic S. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 24.Topalian S L, Gonzales M I, Parkhurst M, Li Y F, Southwood S, Sette A, Rosenberg S A, Robbins P F. J Exp Med. 1996;183:1965–1971. doi: 10.1084/jem.183.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diener E, Feldmann M. Cell Immunol. 1972;5:130–136. doi: 10.1016/0008-8749(72)90090-1. [DOI] [PubMed] [Google Scholar]
- 26.Liblau R S, Tisch R, Shokat K, Yang X, Dumont N, Goodnow C C, McDevitt H O. Proc Natl Acad Sci USA. 1996;93:3031–3036. doi: 10.1073/pnas.93.7.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brocke S, Gijbels K, Allegretta M, Ferber I, Piercy C, et al. Nature (London) 1996;379:343–346. doi: 10.1038/379343a0. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson L B, Greer J M, Sobel R A, Lees M B, Kuchroo V K. Immunity. 1995;3:397–405. doi: 10.1016/1074-7613(95)90169-8. [DOI] [PubMed] [Google Scholar]