Abstract
Visual transduction in Drosophila is a G protein-coupled phospholipase C-mediated process that leads to depolarization via activation of the transient receptor potential (TRP) calcium channel. Inactivation-no-afterpotential D (INAD) is an adaptor protein containing PDZ domains known to interact with TRP. Immunoprecipitation studies indicate that INAD also binds to eye-specific protein kinase C and the phospholipase C, no-receptor-potential A (NORPA). By overlay assay and site-directed mutagenesis we have defined the essential elements of the NORPA–INAD association and identified three critical residues in the C-terminal tail of NORPA that are required for the interaction. These residues, Phe-Cys-Ala, constitute a novel binding motif distinct from the sequences recognized by the PDZ domain in INAD. To evaluate the functional significance of the INAD–NORPA association in vivo, we generated transgenic flies expressing a modified NORPA, NORPAC1094S, that lacks the INAD interaction. The transgenic animals display a unique electroretinogram phenotype characterized by slow activation and prolonged deactivation. Double mutant analysis suggests a possible inaccessibility of eye-specific protein kinase C to NORPAC1094S, undermining the observed defective deactivation, and that delayed activation may similarly result from NORPAC1094S being unable to localize in close proximity to the TRP channel. We conclude that INAD acts as a scaffold protein that facilitates NORPA–TRP interactions required for gating of the TRP channel in photoreceptor cells.
Transmembrane signaling utilizing heterotrimeric GTP-binding proteins is ubiquitous in the nervous system. Drosophila visual transduction provides an ideal system for the molecular dissection of the G protein-mediated phospholipase C (PLC) mechanism (1–3). In this signal transduction pathway, light stimulates rhodopsin, which activates an eye-specific Gαq (4, 5). Activated Gαq triggers NORPA (no-receptor-potential A), a phospholipase C-β (6, 7) to catalyze the breakdown of phospholipids to generate inositol trisphosphate and diacylglycerol. Both Gαq and NORPA are essential for visual signaling, as mutants missing either of these two proteins display no response to light (7, 8). The end result of this visual signaling is the opening of cation channels leading to depolarization of photoreceptors. Both TRP (transient receptor potential) and TRPL (TRP-like) are cation channels that are activated in the visual transduction process. These two proteins share homology with α-subunits of voltage-gated calcium and sodium channels in vertebrates (9). The mechanism by which these two channels are gated remains to be explored. Recently TRP has been suggested to be the homolog of the store-operated calcium channel in vertebrates (10, 11). The store-operated calcium channel in the plasma membrane is responsible for replenishing the internal calcium stores depleted during phosphoinositide-mediated process.
Inactivation-no-afterpotential D (INAD) is a photoreceptor-specific protein containing multiple PDZ repeats (12–14). We previously reported that INAD interacts with the TRP calcium channel that is involved in light-induced depolarization (15). Furthermore, we localized the interacting domains to PDZ3 of INAD and the C-terminal tail of TRP containing an internal Ser/Thr-Xaa-Val motif (16). The functional consequence of interaction is revealed in the InaDp215 mutant. We showed that InaDp215 displays aberrant recovery because it expresses a mutant INAD that exhibits no association with TRP. Based on these findings, we proposed that INAD plays a role in regulating the TRP calcium channel. Consistent with this hypothesis, we observed that the abnormal phenotype of InaDp215 is dependent on extracellular calcium; removal of calcium masks the defective electrophysiology (12). Moreover, the double mutants containing both trp and InaD mutations display an electroretinogram (ERG) similar to that of trp, suggesting INAD acts either at or downstream of TRP. The calcium influx via TRP is involved in both positive and negative regulation of visual transduction (17, 18).
In the Drosophila visual system, INAD has been shown to be a component of a signal transduction complex consisting of at least four proteins, INAD, TRP, NORPA, and INAC [eye-specific PKC (eye-PKC)], based on copurification and immunoprecipitation studies (19, 20). The presence of signal transduction complexes has also been observed in other systems, such as A-kinase anchoring protein 79 (AKAP79) complexed with protein kinases A and C, and phosphatase 2B (21). However, the in vivo functional significance of the complex formation remains to be addressed. We have taken advantage of this system in combination with the available transgenic approaches to gain insight into the physiological consequence of protein–protein interactions within the signaling complex. Our initial experiments concentrated on determining the location of respective interacting domains in INAD and NORPA. We have identified a novel tripeptide motif in the C terminus of NORPA that associates with INAD. Furthermore, we have investigated the in vivo functional consequence of the INAD–NORPA interaction by generating transgenic flies expressing a modified NORPA that displays no association with INAD. Electrophysiological characterization of the norpA transgenic flies reveals abnormal ERG with slow kinetics, suggesting that the association between INAD and NORPA is important in the regulation of activation and deactivation of visual transduction. The transgenic flies were further examined in the background of INAD complex-specific mutants to gain insight into the molecular basis of the defects. These results demonstrate the important role of a PDZ-containing adaptor protein to regulate signal transduction.
EXPERIMENTAL PROCEDURES
Generation of 35S-Labeled Protein and Ligand Overlay Assay.
35S-labeled proteins were generated by coupled in vitro transcription and translation by using the TNT system (Promega) in the presence of [35S]methionine. The [35S]INAD overlay assays were carried out in a buffer containing 1× PBS/1.2 mM CaCl2/1 mM EGTA/1% BSA/0.2% Triton X-100 and wash with 1× PBS/5% milk powder/0.2% Triton as described (16).
Overexpression of NORPA Fusion Proteins in Bacteria.
Fusion protein constructs were generated in pET8C or pGEMEX1 vectors. Expression of fusion proteins was induced by infection with the CE6 bacteriophage expressing T7 RNA polymerase (22) as described (12).
Generation of Anti-NORPA and Anti-PKC Polyclonal Antibodies.
Polyclonal anti-NORPA and anti-PKC antibodies were generated by immunizing rabbits according to a standard protocol (23). The antigens used were NORPA(553–1074) and eye-PKC(562–700), obtained from bacteria by overexpression.
Extraction of Fly Heads.
Fly heads were isolated and homogenized with buffer A (50 mM Na phosphate/5 mM EDTA/1% Triton X-100, pH 7.0/1 mM benzamidine/1 mM benzamide) by Polytron (Brinkmann; 6 times for 5 sec), and incubated for 30 min at 4°C with constant agitation. The supernatant (buffer A extract) was collected following 100,000 × g centrifugation. The pellet was resuspended in buffer B (0.2% SDS/2% Nonidet P-40/100 mM Na phosphate, pH 7.0/10 mM EDTA) and incubated for 30 min at 4°C. Following 100,000 × g centrifugation, the supernatant was saved as the buffer B extract. The buffer B extracts were used in immunoprecipitation assays.
Immunoprecipitation.
Assays (23) were carried out by using buffer B extracts and protein A-Sepharose (Sigma). The immunoprecipitates were washed with buffer B, denatured, and analyzed on a 10% SDS/PAGE gel. Western blot analysis was carried out as indicated (12).
Immunofluorescence and Confocal Microscopy.
Flies reared at 21°C for <24 h posteclosion were used for the immunolocalization experiments according to Porter and Montell (24) with some modifications. After fixation, tissues were rinsed in 0.1 M phosphate (pH 7.4), immersed in OCT (Tissue-Tek, Miles) and rapidly frozen in liquid N2. Sections (8–10 μM) were prepared on a Jung CM3000 cryostat (Leica, Deerfield, IL) and lightly fixed in 0.5% formaldehyde/1× PBS for 10 min. Sections were incubated with primary antibodies followed by fluorescent labeled secondary antisera (Jackson ImmunoResearch). Confocal images were collected on a Zeiss LSM 140, processed in Adobe photoshop 3.0, and montaged in canvas 3.5.
P-Element Mediated Germ-Line Transformation.
The mutant or wild-type (wt) norpA cDNA was subcloned into a modified pCaSpeR 4 vector (25) that contains the Drosophila hsp70 promoter. The P-element construct and a transposase plasmid were injected into the W1118 embryos to generate transgenic flies (26). Flies with the transgene integrated into the second or third chromosome were selected and crossed with norpAp24 for further analysis.
ERG Recordings.
ERG recordings were carried out as described (12). Light stimulation was delivered by a fiber optic light source (Oriel, Stamford, CT). Signals were amplified by means of a WPI (Sarasota, FL) Dam 60 preamplifier and digitized by using Superscope II (GW Instruments, Somerville, MA).
RESULTS
INAD Interacts with NORPA and TRP.
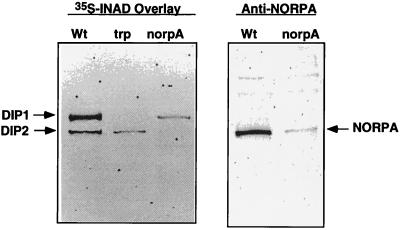
We showed previously that INAD interacts with two retinal proteins in vitro by [35S]INAD overlay assay (Fig. 1). The 150-kDa protein or DIP1 (INAD-interacting protein 1) corresponds to the trp gene product. The second INAD-interacting protein, DIP2, is about 130 kDa in size, which could be either NORPA or TRPL. Further analysis using [35S]INAD overlay reveals that DIP2 is greatly reduced in a norpA mutant (Fig. 1 Left). NORPA is a polypeptide of 1,095 residues (6) with an estimated molecular mass of 125 kDa and it comigrates with DIP2 by Western blot analysis (Fig. 1 Right). Based on these findings and immunoprecipitation (Fig. 2), we conclude that DIP2 is NORPA. These results confirmed the previously published work of Huber et al. (19) and Chevesich et al. (20).
Figure 1.
INAD interacts with NORPA and TRP by ligand overlay assay. Shown on the left is an autoradiogram of [35S]INAD overlay assay using wt, trpp301, and norpAp16 retinal extracts. Shown on the right is a Western blot of wt and norpAp16 retinal extracts probed with anti-NORPA antibodies.
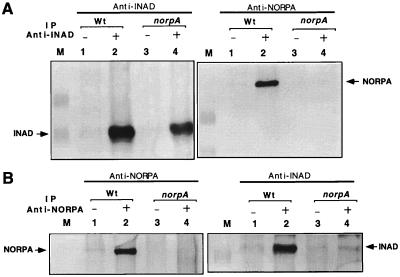
Figure 2.
Stable association between INAD and NORPA in wt photoreceptors. (A) Anti-INAD antibodies immunoprecipitate both INAD and NORPA. Shown are Western blots of anti-INAD immunoprecipitation assays in wt and norpAp16 extracts. (B) Anti-NORPA antisera immunoprecipitate both NORPA and INAD. Shown are Western blots of anti-NORPA immunoprecipitation assays. Preimmune antisera were used as a negative control (lanes 1 and 3). M, protein size standards.
Stable Association Between INAD and NORPA in Photoreceptors.
To explore whether the in vitro interaction between INAD and NORPA is also present in vivo, we further investigated the association in fly extracts. In wt head extracts, anti-INAD antibodies immunoprecipitate both INAD and NORPA (Fig. 2A Right, lane 2) whereas pre-immune serum does not (lane 1). The coisolation of NORPA with INAD indicates that these two proteins are associated in wt photoreceptors. The identity of the INAD-interacting protein as NORPA was further supported by its absence in norpA extracts, where anti-INAD precipitates INAD (Fig. 2A Left, lane 4) but not NORPA (Fig. 2A Right, lane 4). Furthermore, we show that anti-NORPA antibodies precipitate both NORPA (Fig. 2B Left, lane 2) and INAD (Fig. 2B Right, lane 2) from wt extracts but not from norpA extracts (Fig. 2B, lanes 4), providing evidence of the interaction in vivo (Fig. 2B, lanes 4). To further investigate if the association is activity-dependent, we analyzed extracts prepared from dark-adapted wt and ninaD flies. The ninaD mutation affects retinal metabolism, leading to a drastic reduction of rhodopsins (27). However, the INAD–NORPA interaction appears to be preexisting or constitutive, and is not dependent on functional visual transduction, as this association is also detected in dark-adapted wt flies and in the ninaD mutants (data not shown).
NORPA Interacts Through a Domain that Spans the C-Terminal Half of INAD.
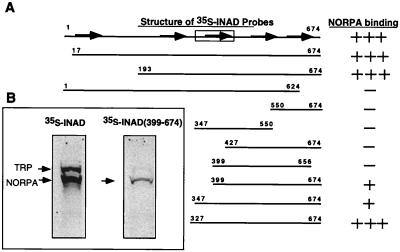
To gain insight into the molecular basis of the INAD–NORPA interaction, we used overlay assays to identify the sites of association. We generated several 35S-methionine-labeled probes containing overlapping regions of INAD to evaluate the association with NORPA from wt retinas. INAD is a polypeptide of 674 residues containing five PDZ domains that span residues 19–94 (PDZ1), 254–323 (PDZ2), 371–443 (PDZ3), 493–565 (PDZ4), and 589–655 (PDZ5) (F.M.A. and B.H.S., unpublished data). As summarized in Fig. 3A, we mapped the high affinity NORPA–interacting site to INAD(327–674). However, we also detected low affinity association using INAD(399–674) (Fig. 3B Right) and INAD(347–674). This INAD sequence, responsible for the high affinity interaction, encompasses the last three PDZ repeats.
Figure 3.
Localization of the NORPA-interacting domain in INAD. (A) The C-terminal half of INAD interacts with NORPA. Shown is a summary of various INAD sequences tested for the interaction with NORPA by overlay assay. (B) The full-length INAD binds to retinal NORPA with high affinity and INAD(399–674) associates with a reduced affinity. The five PDZ repeats of INAD are depicted as arrows and the TRP-interacting region spanning residues 347–450 is indicated as a box.
INAD Interacts with the C-Terminal Region of NORPA.
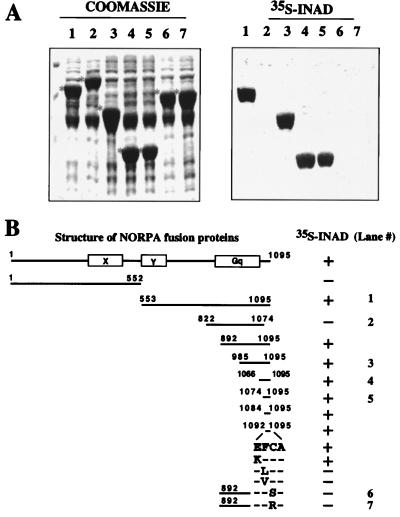
NORPA is a phospholipase C-β. It contains the characteristic X and Y domains that share homology with other members of the phospholipase C family (28). To define the INAD-interacting site, we examined the association between [35S]INAD and several NORPA fusion proteins overexpressed in bacteria. Shown on the right in Fig. 4A are the overlay results. Coomassie staining of the corresponding gel containing bacterial extracts is shown on the left, with each fusion protein marked with a star on the left. We show that NORPA(aa 553–1095; Fig. 4A, lane 1), NORPA(aa 985–1095; Fig. 4A, lane 3), NORPA(aa 1066–1095; Fig. 4A, lane 4), and NORPA(aa 1074–1095 Fig. 4A, lane 5), interact with INAD. In contrast, NORPA(aa 822–1074; Fig. 4A, lane 2) displays no association. Moreover, T7 gene 10, the backbone of the fusion proteins, shows no binding to [35S]INAD (data not shown). Further mapping analyses summarized in Fig. 4B localize the INAD-interacting domain to the last three amino acids, FCA, in the C terminus of NORPA (Fig. 4B). We used site-directed mutagenesis to further identify critical residues required for the NORPA–INAD association in the tripeptide interacting motif. We found that the phenylalanine residue (F), when mutated to either a leucine (L) or a valine (V), leads to a loss of interaction (Fig. 4B). Similarly, mutations that convert the penultimate cysteine residue (C) to either a serine (S) or an arginine (R) also eliminate the association with INAD (Fig. 4A Right, lanes 6 and 7).
Figure 4.
INAD interacts with the C-terminal region of NORPA. (A) Shown on the left is Coomassie staining of a protein gel containing bacterial extracts expressing several overlapping NORPA sequences as fusion proteins of T7 gene 10 or alone. Each fusion protein is indicated with a star on the left. The corresponding autoradiogram of [35S]INAD overlay assay is shown on the right. (B) Shown is a summary of [35S]INAD overlay results.
Transgenic Flies Expressing NORPAC1094SDisplay a Unique ERG Phenotype Characteristic of Slow Kinetics.
To gain insight into the functional consequence of the INAD–NORPA association in vivo, we examined the visual electrophysiology of the transgenic flies expressing a modified norpA in which the codon for the cysteine residue at 1,094 was converted to that of a serine. This point mutation results in a loss of the INAD interacting activity as shown by the in vitro overlay assay (Fig. 4). Transgenic flies were generated by introducing norpAC1094S under the control of the Drosophila hsp70 promoter, into the germ line of the embryo. As a control, we also generated transgenic flies expressing a wt copy of the norpA cDNA. The expression of norpA in flies can be induced by heat shock treatment starting at late pupal stages, when endogenous proteins involved in visual transduction begin to express (12).
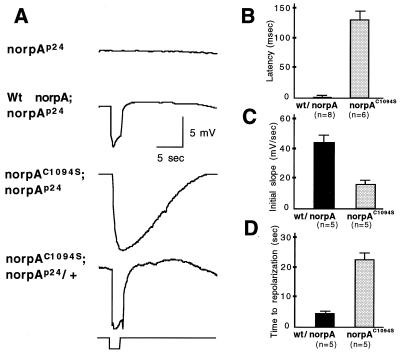
Fig. 5A shows ERG recordings of norpAp24, wt norpA, and norpAC1094S following heat shock treatment. norpAp24, a null allele, exhibits no response to a 2-sec orange light pulse due to the absence of NORPA (7). Expression of wt norpA in the norpAp24 background restores the light response with fast kinetics of depolarization and repolarization, similar to that of wt flies. In contrast, transgenic flies expressing norpAC1094S in the norpAp24 background have an ERG with slower kinetics (Fig. 5A). However, in wt background the expression of norpAC1094Sleads to a wt physiology, suggesting that the modified NORPA does not interfere with the activity of wt protein. We compared latencies (Fig. 5B), initial slopes (rate to reach 50% response following the initiation, Fig. 5C), and declining phase (time to resting potential, Fig. 5D) of ERGs between wt norpA and norpAC1094S transgenic flies. We observed long latencies (wt, 10 ± 5; norpAC1094S, 125 ± 30 msec) and reduced initial slopes (wt, 44 ± 5; norpAC1094S, 16 ± 3 mV/sec) in norpAC1094S flies. These findings suggest that activation of the visual response is delayed in norpAC1094S. Furthermore, following the termination of light, repolarization is much prolonged as the time to resting potential is also lengthened (wt, 4.5 ± 0.4; norpAC1094S, 22.6 ± 2 sec; Fig. 5D). This suggests a possible deactivation defect in the norpAC1094S transgenic flies. This phenotype of defective activation and seemingly defective deactivation is not due to low level of expression of protein (see below, Fig. 7A), because expression of a similar level wt norpA leads to a wt ERG in the transgenic flies (data not shown).
Figure 5.
Transgenic flies expressing norpAC1094S display abnormal ERG with slower kinetics. (A) Shown are ERG recordings of norpAp24, transgenic flies expressing wt norpA, and norpAC1094S in norpAp24 or wt genetic background in response to a 2-sec pulse of the orange light. Three independent lines were analyzed, and consistent results were obtained. The kinetics of ERG were examined using the following parameters: latency, defined as time elapsed between stimulation and initiation of ERG (B); initial slope of the response, defined as the rate to reach 50% of the maximal response following the initiation of the response (C); and time to repolarization, time to reach resting potential following the termination of stimulation (D) in wt norpA and norpAC1094S;norpAp24. Both latency (B) and time to repolarization (D) are lengthened, and initial slope (C) is reduced in norpAC1094S;norpAp24 lines.
Figure 7.
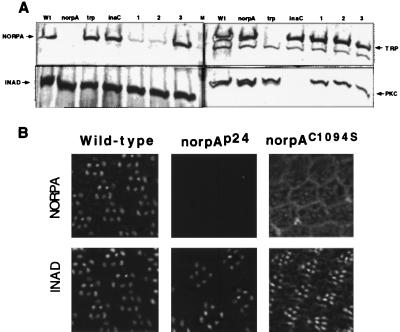
Expression and subcellular distribution of NORPAC1094S. (A) Expression of NORPAC1094S in transgenic flies. Shown are Western blots of retinal extracts from norpAC1094S transgenic lines in norpAp24 (lanes 1 and 2) or wt background (lane 3), and extracts from wt (wt) and various mutants probed with antibodies as indicated. Each lane represents extracts from one fly head. M, prestained protein molecular weight standards, 150, 112, 84 kDa. (B) Random distribution of NORPAC1094S in the compound eye. Shown is immunolocalization of NORPA and INAD in wt, norpAp24 and norpAC1094S;norpAp24 eyes. In wt retinas, both NORPA and INAD are co-localized in the rhabdomere. However, NORPAC1094S appears to be distributed in cytoplasm as well as in rhabdomeres of norpAC1094S following heat shock treatment.
Double Mutant Analysis Reveals Molecular Basis of Activation and Deactivation Defects in norpAC1094S.
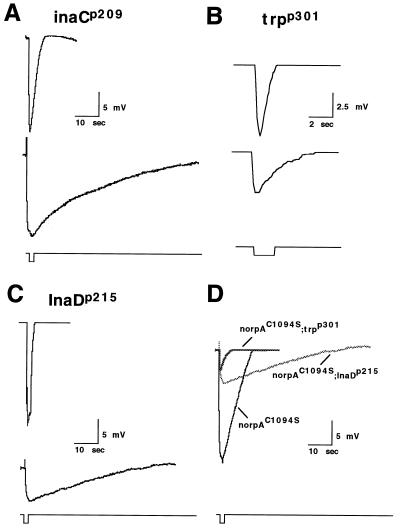
The activation and deactivation defects resulting from a loss of the INAD–NORPA interaction were investigated via electrophysiological analysis of norpAC1094S alone, and in the genetic backgrounds of INAD complex-specific mutants (i.e. inaCp209, InaDp215, and trpp301). Interestingly, the norpAC1094S phenotype is very similar to that of inaCp209 (Fig. 6 A and D). The inaC gene encodes eye-PKC and mutants such as inaCp209 that lack the gene product display an abnormal visual phenotype involving defective deactivation and light adaptation (29, 30). Shown in Fig. 6A are ERGs of inaCp209 (Upper) and norpAC1094S;inaCp209 (Lower) flies. We show that the deactivation waveform of the double mutant is similarly delayed as in the norpAC1094S fly but requires more time to completely repolarize. This is most likely attributed to the additional failure of quantum bump termination normally seen in inaC flies, as shown (30). It is important to note, however, that the latency to activation in the norpAC1094S;inaCp209 fly is almost identical to that of the norpAC1094S fly (data not shown).
Figure 6.
Double mutants analysis of norpAC1094S by ERG. norpAC1094S was crossed into inaCp209, InaDp215, and trpp301 mutant backgrounds and analyzed. (A) inaCp209 (Upper), norpAC1094S;inaCp209 (Lower) (B) trpp301 (Upper), norpAC1094S; trpp301 (Lower). (C) InaDp215 (Upper), norpAC1094S;InaDp215 (Lower) (D) Superimposition of ERG from norpAC1094S, norpAC1094S;InaDp215, and norpAC1094S;trpp301. Flies of 1-day posteclosion were subject to heat shock and analyzed. Flies were dark adapted for 3 min prior to the recording using a 2-sec orange light. Multiple animals were recorded and consistent results obtained. Note the difference in the time course and amplitude of the response in trpp301 and norpAC1094S;trpp301.
The contribution from TRP and TRPL channel activity in the visual response of norpAC1094S flies was uncovered using norpAC1094S;trpp301 double mutants. The trpp301 mutant lacks functional TRP channels and cannot maintain a sustained response to light (ref. 15, see also Fig. 6B Upper). As expected, the norpAC1094S;trpp301 flies display a much reduced response to light (Fig. 6B Lower) relative to our other mutants, suggesting that most of the current seen in the norpAC1094S and norpAC1094S;inaCp209 flies was primarily composed of TRP conductance. In addition, the response seen in trpp301, which is primarily composed of TRPL current, is also drastically affected when norpAC1094S is introduced into the background. The latency of activation of the TRPL channels was approximately twice that of the latency seen in the other norpAC1094S mutants.
To determine how the mislocalization of TRP affects the visual response in a system where NORPA is also mislocalized, we introduced norpAC1094S into the InaDp215 background, where TRP is not able to bind to the INAD complex (16). ERG analysis of norpAC1094S;InaDp215 double mutant flies revealed a profound loss of the visual response (amplitude of response) as compared with norpAC1094S single mutants (Fig. 6 C Lower and D). To insure that this was not due to the absence of TRP channels, we performed Western blot analysis and found that the level of TRP in norpAC1094S;InaDp215 flies was of wt levels (data not shown). Furthermore, this weakened response was still larger than the response of mutants with no functional TRP (Fig. 6D), suggesting that some TRP channels are indeed activated. Together, our findings indicate that activation of the depolarizing current in norpAC1094S flies is partially abolished when TRP is no longer closely associated with INAD. Similarly, the reduced response of TRPL in the norpAC1094S background suggests that NORPA may act to directly regulate TRPL conductances, in addition to TRP.
Random Subcellular Distribution of NORPAC1094S in Photoreceptor Cells.
The expression and subcellular localization of the modified NORPA were determined. Fig. 7A shows Western blot analysis from flies of the following genotypes: wt (Wt), norpAp24 (norpA), trpp301 (trp), inaCp209 (inaC), norpAC1094S; norpAp24 (two lines, lane 1 and 2) and norpAC1094S; + (lane 3). The level of NORPAC1094S in transgenic flies is about 5–10% of wt (Fig. 7A Left, lanes 1 and 2). This low level of expression may be due to reduced stability of the modified NORPA. However, the level of INAD (Fig. 7A Left), PKC (Fig. 7A Right), and TRP (Fig. 7A Right) is similar to that of wt. Moreover, flies expressing norpAC1094S in wt genetic background contain wt level of NORPA (Fig. 7A Left, lane 3). By an overlay assay we showed that NORPAC1094S from transgenic flies fails to associate with [35S]INAD (data not shown).
We also examined the subcellular distribution of the modified NORPA by immunofluorescence using confocal microscopy. Unlike that of wt, we found no preferential localization of NORPAC1094S in rhabdomeres (Fig. 7B). However, the rhabdomeric distribution of INAD is not altered in norpAC1094S flies.
DISCUSSION
Scaffold proteins such as AKAPs are involved in the targeting or assembling of macromolecular complexes to facilitate protein–protein interactions that may be critical for regulation of signal transduction. In Drosophila photoreceptors, INAD, a novel adaptor protein with five PDZ domains, has been shown to interact with multiple polypeptides involved in the visual cascade (19, 20). We showed previously that INAD interacts with the TRP calcium channel, and a loss of the INAD–TRP association leads to slower recovery of the visual response, possibly due to defective TRP channel activity (16). The fact that INAD also associates with NORPA, the effector of the visual cascade, is intriguing. NORPA belongs to the phospholipase C-β family with high similarity to PLC-β4 isoform (31, 32). PLC-β is activated by the α-subunit of the heterotrimeric G proteins such as Gq and G11, or the βγ subunits (for reviews see ref. 33). To date, little is known about the regulation of PLC-β in vivo.
Numerous studies on Drosophila visual transduction have shown that disruption of any components in the INAD complex results in significant changes in the fly’s response to light (6, 15, 18, 30). Therefore, the stable interaction between NORPA and INAD within the complex assuredly has a major functional importance. Indeed, colocalization of INAD and NORPA in the rhabdomere is required in the modulation of visual response kinetics, as revealed in transgenic flies expressing a non-INAD associating NORPA. Our findings indicate that a lack of the interaction results in prolonged time courses for activation and possibly deactivation. Specifically, the electrophysiological defects detected in the visual response of norpAC1094S transgenic flies include longer latencies to activation, slower initial slopes, and more prolonged repolarizing phases than that of wt flies and other INAD complex-specific mutants. The increased latencies and the smaller initial slopes imply a general slower activation of TRP channels in response to light stimulus. Similarly, the delayed repolarization defects are likely to be because of either an accumulation of late activated channels, a defect in the deactivation of TRP channels, or a mixture of both.
The norpAC1094S recordings indicate that the localization of NORPA is the most important factor in normal activation kinetics. The specific channel activating phenomenon of NORPA also seems to be independent of INAD and eye-PKC, as seen in our single and double mutants, suggesting possible direct interactions of NORPA with TRP for activation. In terms of activation, it is important to note that a loss of the INAD–NORPA interaction only affects latency and initial slope, and does not lead to a complete loss of the activation phenomenon. Once again, this strongly supports the notion that NORPA acts directly to regulate TRP’s activation, and that it or its effector expectedly requires a longer time to “find” TRP and produce its effects on activation when mislocalized.
There are several possible mechanisms that may account for the deactivation defects. Our finding that the double mutant norpAC1094S;inaCp209 displays a more severe deactivation than either single mutants, suggests that the delayed deactivation in norpAC1094S and inaCp209 may result from two independent processes. It may be that norpAC1094S, unlike inaCp209, has no deactivation defects on the single-cell/single-channel level and that the large residual conductance seen after a stimulus, on a macroscopic level, is carried by late-activated TRP channels. Late-activated TRP channels could thereby mask the normal deactivation of single channels. It is also possible that eye-PKC is partially active in norpAC1094S and that the deactivation is synergistically lengthened in the absence of eye-PKC (norpAC1094S;inaCp209). The exact mechanism by which eye-PKC regulates deactivation is not known. It is known, however, that eye-PKC is an integral part of the INAD complex and may phosphorylate NORPA, INAD, TRP, and/or other unidentified substrates. The mislocalization of NORPA may lead to improper activation of PKC that may phosphorylate TRP, which in turn acts to help repolarize the cell. It is also possible that PKC may facilitate down-regulation by phosphorylating NORPA. Future analysis using patch clamp techniques will allow distinction among these possible mechanisms.
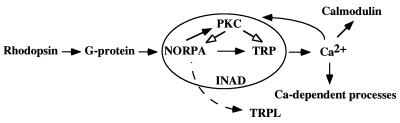
One of the most interesting, yet unresolved dilemmas in Drosophila visual transduction is the mechanism for cation channel gating. There are two cation channels, TRP and TRPL, that contribute to the light-induced depolarization. The calcium influx via TRP constitutes the major component of light-induced currents, whereas TRPL, a nonselective cation channel (34), contributes only to a minor fraction (35). Our analysis of norpAC1094S;trpp301 is consistent with this notion. The mechanism by which these cation channels are gated has been subjected to intense investigation. Several diffusible molecules generated upon the activation of NORPA, such as inositol triphosphate and calcium, have been proposed as potential second messengers, although no satisfactory candidates have been elucidated. Besides the diffusible compounds as second messengers, it is also likely that gating of channels can be regulated via protein–protein interactions. One such example is the activation of GirK, the potassium channel in heart, by direct interaction with the βγ-subunit of the GTP-binding proteins (36). The elusive nature of the second messenger in the visual cascade leads us to speculate that novel protein–protein interactions may account for the activation of channels. Indeed, our double mutant analysis strongly supports this hypothesis. We show that the ERG response in norpAC1094S is drastically reduced in the presence of the InaDp215 mutation. InaDp215 expresses a modified INAD that lacks the interaction with TRP. Consequently, both TRP and NORPA are not associated with INAD in norpAC1094S;InaDp215 double mutants. Because the activation of TRP is greatly diminished in the double mutants, this suggests that NORPA may be involved in activation of the TRP channel. Based on these results, we propose that INAD acts as a scaffold protein to facilitate the NORPA–TRP interactions required for gating of the TRP calcium channel (Fig. 8). We conclude that the association of INAD with these two interacting proteins is essential for the controlled activation and deactivation of light-induced currents.
Figure 8.
A model for visual transduction in Drosophila. Shown is a simplified visual cascade highlighting the protein–protein interactions in the INAD complex. In this model, G protein activates NORPA which leads to generation of diacylglycerol and opening of TRP. An increase in cytoplasmic calcium may activate calmodulin and Ca-dependent processes to modulate the visual response. Eye-PKC, activated by Ca2+ and diacylglycerol, may phosphorylate NORPA and TRP to terminate the activity. How TRPL is gated is not known.
Acknowledgments
We thank Dr. Alan Brash and members of the laboratory for critical reading of the manuscript. We thank Dr. W. Pak (Purdue University) for the norpA stocks and Dr. R. Shortridge (State University of New York, Buffalo) for the norpA cDNA. We thank Ryan Collins for excellent technical assistance. This work was supported by a grant from the National Institutes of Health (EY09743). B.H.S. is an Established Investigator of the American Heart Association.
ABBREVIATIONS
- AKAP
A-kinase anchoring protein
- INAD
InaD, inactivation-no-afterpotential D
- INAC
inaC, inactivation-no-afterpotential C
- NORPA
norpA, no-receptor-potential A
- PKC
protein kinase C
- eye-PKC
eye-specific PKC
- TRP
trp, transient receptor potential
- ninaD
neither-inactivation-nor-afterpotential D
- ERG
electroretinogram
- wt
wild type
Note
While this manuscript was in review, Tsunoda et al. (37) reported the identification of an InaD mutant, InaD2, that shows an ERG phenotype similar to that of norpAC1094S.
References
- 1.Hurley J B. J Bioenerg Biomembr. 1992;24:219–226. doi: 10.1007/BF00762680. [DOI] [PubMed] [Google Scholar]
- 2.Selinger Z, Doza Y N, Minke B. Biochim Biophys Acta. 1993;1179:283–99. doi: 10.1016/0167-4889(93)90084-3. [DOI] [PubMed] [Google Scholar]
- 3.Ranganathan R, Malicki D M, Zuker C S. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y-J, Dobbs M B, Verardi M L, Hyde D R. Neuron. 1990;5:889–898. doi: 10.1016/0896-6273(90)90349-k. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y-J, Shah S, Suzuki E, Zars T, O’Day P M, Hyde D R. Neuron. 1994;13:1143–1157. doi: 10.1016/0896-6273(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 6.Bloomquist B, Shortridge R, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak W. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 7.Pearn M T, Randall L L, Shortridge R D, Burg M, Pak W L. J Biol Chem. 1996;271:4937–4945. doi: 10.1074/jbc.271.9.4937. [DOI] [PubMed] [Google Scholar]
- 8.Scott K, A, Becker A, Sun Y, Hardy R, Zuker C. Neuron. 1995;115:919–927. doi: 10.1016/0896-6273(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 9.Phillips A M, Bull A, Kelly L E. Neuron. 1992;8:631–642. doi: 10.1016/0896-6273(92)90085-r. [DOI] [PubMed] [Google Scholar]
- 10.Hardie R C, Minke B. Trends Neurosci. 1993;16:371–376. doi: 10.1016/0166-2236(93)90095-4. [DOI] [PubMed] [Google Scholar]
- 11.Clapham D E. Neuron. 1996;16:1069–1072. doi: 10.1016/s0896-6273(00)80132-4. [DOI] [PubMed] [Google Scholar]
- 12.Shieh B-H, Niemeyer B. Neuron. 1995;14:201–210. doi: 10.1016/0896-6273(95)90255-4. [DOI] [PubMed] [Google Scholar]
- 13.Huber A, Sander P, Paulsen R. J Biol Chem. 1996;271:11710–11717. doi: 10.1074/jbc.271.20.11710. [DOI] [PubMed] [Google Scholar]
- 14.Ponting C P, Phillips C. Trends Biochem Sci. 1995;20:102–103. doi: 10.1016/s0968-0004(00)88973-2. [DOI] [PubMed] [Google Scholar]
- 15.Hardie R C, Minke B. Neuron. 1992;8:643–651. doi: 10.1016/0896-6273(92)90086-s. [DOI] [PubMed] [Google Scholar]
- 16.Shieh B-H, Zhu M-Y. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 17.Hardie R C. Proc R Soc London B. 1991;245:203–210. [Google Scholar]
- 18.Ranganathan R, Harris G L, Stevens C F, Zuker C S. Nature (London) 1991;354:230–235. doi: 10.1038/354230a0. [DOI] [PubMed] [Google Scholar]
- 19.Huber A, Sander P, Gobert A, Bahner M, Hermann R, Paulsen R. EMBO J. 1996;15:7036–7045. [PMC free article] [PubMed] [Google Scholar]
- 20.Chevesich J, Kreuz A J, Montell C. Neuron. 1997;18:95–105. doi: 10.1016/s0896-6273(01)80049-0. [DOI] [PubMed] [Google Scholar]
- 21.Klauck T M, Faux M C, Labudda K, Langeberg L K, Jaken S, Scott J D. Science. 1996;271:1589–1592. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 22.Studier F W, Moffatt B A. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 24.Porter J A, Montell C. J Cell Biol. 1993;122:601–612. doi: 10.1083/jcb.122.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thummel C S, Pirrotta V. Drosophila Information Service. 1992;71:150. [Google Scholar]
- 26.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson R S, O’Tousa J, Scavarda N J, Randall L L, Pak W L. Symp Soc Exp Biol. 1983;36:477–501. [PubMed] [Google Scholar]
- 28.Rhee S G, Choi K D. J Biol Chem. 1992;267:12393–12396. [PubMed] [Google Scholar]
- 29.Smith D P, Ranganathan R, Hardy R W, Marx J, Tsuchida T, Zuker C S. Science. 1991;254:1478–1484. doi: 10.1126/science.1962207. [DOI] [PubMed] [Google Scholar]
- 30.Hardie R C, Peretz A, Suss-Toby E, Rom-Glas A, Bishop S A, Selinger Z, Minke B. Nature (London) 1993;363:634–637. doi: 10.1038/363634a0. [DOI] [PubMed] [Google Scholar]
- 31.Kim M J, Bahk Y Y, Min D S, Lee S J, Ryu S H, Suh P G. Biochem Biophys Res Commun. 1993;194:706–712. doi: 10.1006/bbrc.1993.1879. [DOI] [PubMed] [Google Scholar]
- 32.Lee C W, Park D J, Lee K H, Kim C G, Rhee S G. J Biol Chem. 1993;268:21318–21327. [PubMed] [Google Scholar]
- 33.Exton J H. Annu Rev Physiol. 1994;56:349–369. doi: 10.1146/annurev.ph.56.030194.002025. [DOI] [PubMed] [Google Scholar]
- 34.Sinkins W G, Vaca L, Hu Y, Kunze D L, Schilling W P. J Biol Chem. 1996;271:2955–2960. doi: 10.1074/jbc.271.6.2955. [DOI] [PubMed] [Google Scholar]
- 35.Niemeyer B A, Suzuki E, Scott K, Jalink K, Zuker C S. Cell. 1996;85:651–659. doi: 10.1016/s0092-8674(00)81232-5. [DOI] [PubMed] [Google Scholar]
- 36.Logothetis D E, Kurachi Y, Galper J, Neer E J, Clapham D E. Nature (London) 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 37.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, et al. Nature (London) 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]