Abstract
HLA-G is the putative natural killer (NK) cell inhibitory ligand expressed on the extravillous cytotrophoblast of the human placenta. Killing of the class I negative human B cell line 721.221 by NK cells is inhibited by the expression of HLA-G. This inhibition is dependent on a high level of HLA-G expression. In the present study, the nature of the receptors that mediate the inhibition has been studied with 140 NK cell lines from two donors and 246 NK clones from 5 donors by blocking the inhibition using monoclonal antibodies against the known NK inhibitory receptors: CD158a, CD158b, and CD94. Both CD94 and the two CD158 proteins can function as receptors, although the former clearly predominates. In many cases, a combination of antibodies to these receptors is required to achieve maximal reversal of inhibition. Moreover, in at least one-third of the NK cells that are inhibited by HLA-G, these antibodies alone or in combination do not reverse inhibition, strongly suggesting the existence of a third major unidentified receptor for HLA-G.
Soon after the cloning of class I major histocompatibility complex (MHC) cDNA, several large restriction enzyme fragments encoding unidentified class I MHC sequences were detected (1). The sequencing of one of these fragments (initially called HLA 6.0) later showed that it encoded an apparently intact class I MHC gene, although with a shortened cytoplasmic tail (2) and it was renamed HLA-G. However, no evidence of expression of this gene could be found in any tissue examined. Subsequently, however, its unique expression pattern was elucidated. The product of this gene was found expressed by the extravillous cytotrophoblast of the human placenta and by two tumor cell lines of trophoblastic origin, BeWo and JEG 3 (3, 4). The extravillous cytotrophoblast is formed from cells derived from the tip of those villi that contact the uterine decidua and then becomes an outer shell around the other layers of the placenta. These cells are directly opposed to maternal structures. They also migrate to and invade the maternal uterine spiral arteries, replacing their endothelium (5). These cells express HLA-G (6).
A major physiological need for the hemiallogeneic trophoblast and the fetus is the avoidance of rejection by the maternal immune system. The failure of the trophoblast to express the usual class I and class II MHC proteins that are ligands for T cells [with the exception of a small amount of HLA-C in the first trimester (7)] was recognized 20 years ago (8) and has been thought to be the major reason for the failure of fetal allograft rejection. However, more recently the role of a second type of peripheral blood effector cell, the natural killer (NK) cell, has been clarified (9). These cells, in contrast to peripheral T cells, are normally activated for lysis of targets that fail to express class I MHC proteins, but on recognition of appropriate allotypes of class I MHC ligands an inhibitory signal is delivered. Thus, the role of NK cells is to carry out immunosurveillance for cells that have down-regulated class I MHC proteins (10). Consequently, escape from attack by NK cells as the result of down regulation of class I MHC proteins is a second requirement for the trophoblast to avoid cytolysis. A similar predicament occurs when virus-infected cells down-regulate class I MHC proteins (11, 12).
HLA-G can function as an inhibitor of NK cells in vitro, and its expression pattern suggests that it may play an important role in this escape. Initially, relatively weak inhibition of NK lytic activity by HLA-G was observed using bulk peripheral blood NK cells as well as uncharacterized NK clones (13, 14). Subsequent work established that the recognition of cellular targets (phytohemagglutinin blasts) by NK cells is allospecific and linked to a dominant resistance gene in the MHC (15) and that HLA-C is a dominant inhibitory ligand (16). Two groups of HLA-C allotypes are distinguished by polymorphisms at residues 77 and 80 in the α1 domain of HLA-C of which group 1 is defined by Lys80 and group 2 by Asn80 (17). These observations permitted the establishment of lines and clones of NK cells called NK1 and NK2 cells that were inhibited by members of the HLA-C group 1 and group 2 molecules, respectively. The lytic activity of such lines and clones was completely inhibited by the presence of appropriate HLA-C ligands (16). Moreover, lysis of class I negative target cells by both NK1 and NK2 cell lines was inhibited comparably strongly by the expression of HLA-G after transfection (18).
The nature of the receptor(s) for HLA-G is of considerable interest. In the initial study (18), we showed that the inhibition could be completely reversed by class I MHC-specific monoclonal antibodies (mAbs) or alternatively partially reversed by HP3E4 or GL183, mAbs that bind to the NK cell inhibitory receptors (CD158a and CD158b, NKIR1 and NKIR2, respectively, p58 proteins) that contain two Ig domains. Additionally, NK cells that recognized the HLA-B molecule HLA-B51 using the three Ig domain receptors also recognized HLA-G as an inhibitory ligand, and this inhibition was reversed by mAb 5.133, which bound to the NKIR3 on these cells (19).
Other studies, however, have concluded that the dominant inhibitory receptor for HLA-G is a complex of CD94 and NKG2A, members of the C-type lectin family of receptors (20–22). In view of this discrepancy the present experiments were initiated to extend and clarify our published results.
MATERIALS AND METHODS
Cells and mAbs.
The MHC class I negative human B cell line 721.221 (23) was obtained from the American Type Culture Collection. mAb GL183 and EB6 were purchased from Immunotech (Luminy, France) and HP3D9 from PharMingen. mAb HP3E4 was a gift from M. Lopez-Botet (Madrid, Spain). NK lines were prepared from peripheral human blood by sorting CD3–CD56+ cells at 50 cells per well. NK clones were generated as described (17).
Transfection.
Stable 721.221 transfectants expressing HLA-C and HLA-G molecules were generated as described (17). Expression of class I molelcules was analyzed both by flow cytometry and by immunoprecipitation followed by SDS/PAGE or IEF analysis using the pan-anti-class I mAb W6/32. To positively identify the HLA-G transfectant, mAb G233 (24) (a gift of Y. W. Loke, Cambridge, U.K.) also was used.
Cytolytic Assays.
The cytolytic activity of NK lines and clones against the various transfectants was assessed in 5 hr 35S release assays (17). In all presented experiments the spontaneous release was less than 25% of maximal release. The range of the triplicates or duplicates was under 5% of their mean. In experiments where CD158 or CD94 specific mAbs were used to block MHC/NKIR interaction, mAb was included in the medium to a final concentration of 5 μg/ml unless indicated otherwise.
Class I MHC Turnover.
Turnover of class I MHC proteins at the cell surface was determined by FACS analysis as described (25).
RESULTS
Inhibition of NK Lines by HLA-G and Its Reversal by mAbs.
Initially 59 NK lines were established from one donor. Of these cell lines, 24 (40%) were strongly inhibited by the presence of HLA-G (i.e., the killing of the transfected cell line, 721.221/HLA-G, was below 25% of that of the untransfected 721.221 cells) and 16 (27%) showed some inhibition (i.e., killing of the transfectant was 25–75% of the 721.221 control), i.e., two-thirds of the NK lines were inhibited, at least to some extent. However, 13 cell lines (22%) were not inhibited significantly (inhibition less than 25%), and 6 lines (10%) were activated, lysing the HLA-G transfectant better than the 721.221 target by as much as 100%.
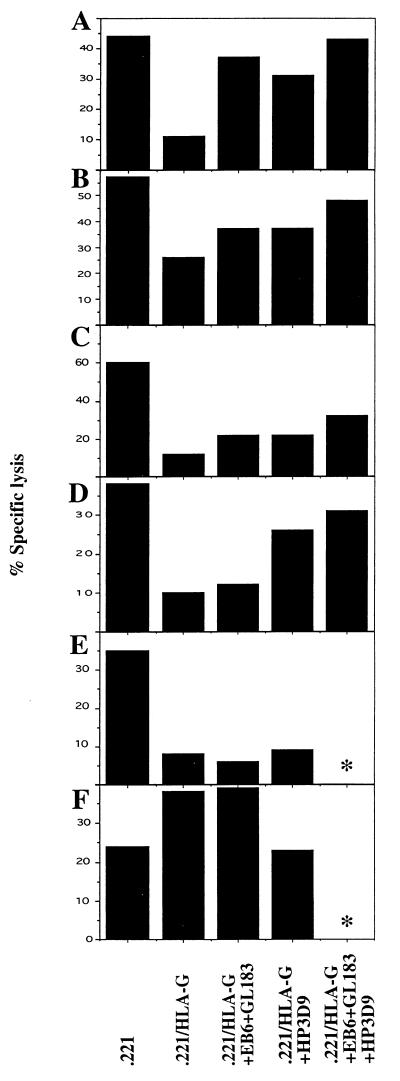
To investigate the involvement of the CD158 Ig superfamily receptors, and the CD94 C-type lectin receptor, cytotoxicity assays were performed in the presence of mAbs that block the function of these inhibitory receptors. Only one NK cell line (2%) showed near complete reversal of HLA-G induced inhibition after the addition of a mixture of anti-CD158 mAb (Fig. 1A), while four other NK lines (7%) showed partial reversal (Fig. 1 B and C). All five of these lines were also reversed comparably by anti-CD94 mAb. Combinations of mAb frequently appeared to be additive in their effects. Additionally, with 17 lines (29%) inhibition by HLA-G was partially reversed in the presence of an anti-CD94 mAb alone (Fig. 1D), i.e., a total of 38% were reversed by anti-CD94. Finally the inhibition could not be reversed by any of the mAb in the case of 18 lines (30%) (Fig. 1E) (the 32% remainder being lines that were either not inhibited or were activated by HLA-G). The activation of NK cell killing in the presence of HLA-G could be reversed in two lines by the addition of anti-CD94 mAb (Fig. 1F), while the other four lines activated by HLA-G killed the targets to the same extent in the presence of both anti-CD94 and anti-CD158 mAbs.
Figure 1.
Phenotypes among 59 NK cell lines established from one individual. See text for description of phenotypes and frequency of each. The E:T ratio used in the assay was 3:1 and the concentration of each mAb added was 5 μg/ml (targets and mAb listed at the bottom). ∗, Not done, amounts of NK cells were limiting.
In a second set of experiments, 81 NK cell lines were generated from another donor. Of these cell lines, 44 (54%) showed strong (more than 75%) and 24 (30%) significant (25–75%) inhibition of lysis on HLA-G transfected cells. Only 9 (13%) of the cell lines from this individual showed reversal of inhibition after the addition of the anti-CD94 mAb alone. Because these lines from the two donors were all complex, analysis of clones seemed likely to yield the most valuable data.
Distinct Inhibitory Phenotypes of NK Clones.
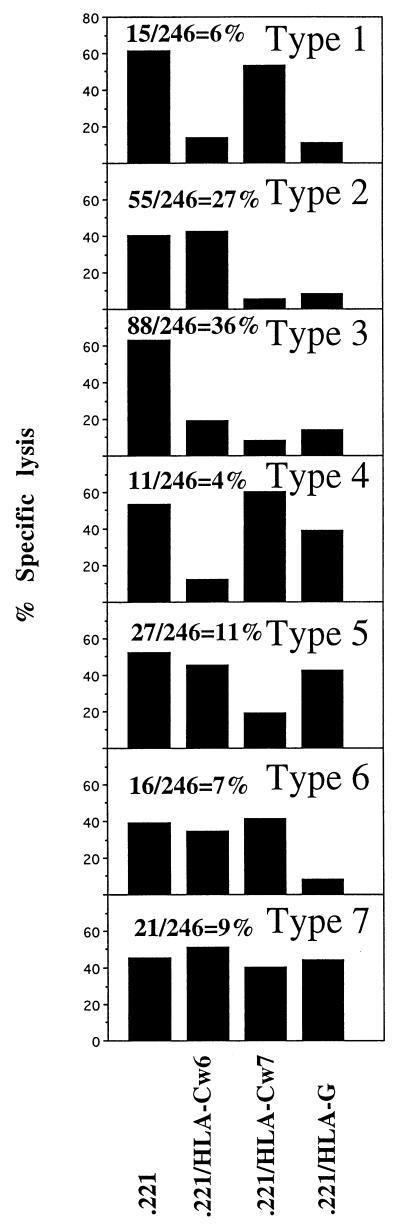
Clones (n = 246) were generated from five NK cell lines established from five individuals. The inhibition of these clones by HLA-C allotypes (Cw6, a group 1 allotype and Cw7, a group 2 allotype) and by HLA-G was examined. Based on the inhibition phenotypes, seven groups of NK clones could be defined, inhibited by: (1) HLA-Cw6 and HLA-G (6%); (2) HLA-Cw7 and HLA-G (27%); (3) HLA-Cw6, HLA-Cw7 and HLA G (36%); (4) only HLA-Cw6 (4%); (5) only HLA-Cw7 (11%); (6) only HLA-G (7%); and (7) none of these HLA molecules (9%) (Fig. 2). Thus, 46% of the clones were inhibited by HLA-Cw6, 74% by HLA-Cw7, 76% by HLA-G, and 9% were not inhibited by any of these molecules.
Figure 2.
Frequency of NK clones inhibited with distinct class I MHC allotype dependency. The 246 NK clones were prepared from five different donors and reacted with various [35S]methionine-labeled target cells for 5 hr. at E:T ratio of 1:1. One out of three representative experiments is shown. Percentages of the different type of clones are indicated.
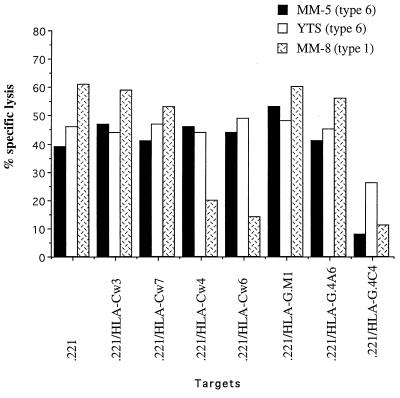
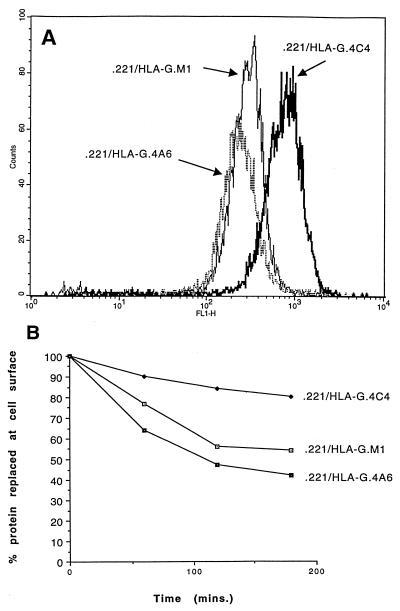
Examples of the analysis of two clones and an NK tumor cell line are shown in Fig. 3. Clone MM-8 is representative of clones inhibited by HLA Cw6 and other group 1 allotypes of HLA-C such as HLA-Cw4 and by HLA-G. Clones MM-5 and YTS [a subclone of the YT NK tumor cell line (26) provided by Z. Eshhar, Rehovoth, Israel] were inhibited only by HLA-G. The inhibition by HLA-G is notable in that three transfectants of 721.221 cells were employed that differed in the expression level of HLA-G over a 10-fold range (Fig. 4A). Only the highest expressing transfectant (721.221/HLA-G.4C4) was inhibitory (Fig. 3). The transfectants 721.221/HLA-G.M1 and/HLA-G.4A6 that had a 5-to 10-fold lower expression did not inhibit lysis. HLA-G is turned over at the cell surface at a low but measurable rate (25) with almost 50% turnover in the low expressors over a 3-hour period and only 10% turnover in the high expressor (Fig. 4B), suggesting that the system responsible for turnover may have been saturated at relatively low HLA-G expression levels. Thus, the ability of HLA-G to inhibit NK cells is correlated with a high level of expression and a low rate of turnover at the cell surface. It is unclear whether or how this turnover could be related to the difference in inhibition observed.
Figure 3.
Targets expressing high levels of HLA-G are protected from lysis by NK clones and a NK tumor line. NK clones that were prepared from donor MM and the NK tumor line YTS were incubated with various [35S]methionine-labeled target cells for 5 hr at E:T ratio of 1:1 for the clones and 5:1 for the tumor line. One out of three representative experiments is shown. The level of HLA-G expression correlates with the turnover of MHC class I proteins at the cell surface.
Figure 4.
HLA-G expression in transfectants. Various HLA-G transfectants were stained with mAb W6/32 (A) and assayed for class I MHC protein turnover after ligation with this mAb (22) (B).
Reversal of Inhibition of NK Clones by mAbs.
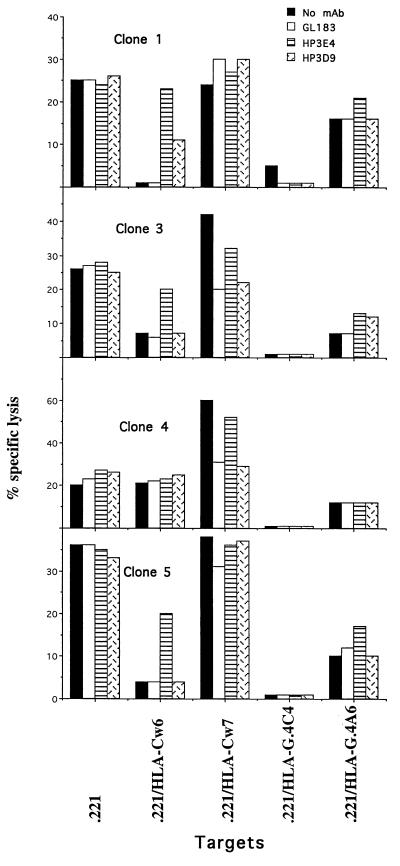
Several examples of clones examined by blocking with mAb are shown in Fig. 5. Clone 1 had an inhibitory receptor(s) for HLA-Cw6 and for HLA-G that resulted in virtually complete inhibition by these ligands. The interaction with HLA-Cw6 was reversed by the CD158a mAb HP3E4. However, partial reversal of inhibition by HLA-Cw6 was also obtained with the CD94 mAb HP3D9. Notably, none of the antibodies tested could reverse the inhibition by HLA-G. Clone 3 similarly had an inhibitory receptor for HLA-Cw6 that was partially reversed by HP3E4. However, it also expressed an activating receptor for HLA-Cw7 whose effect was blocked by the CD158b mAb GL183 (but not by HP3E4) and by HP3D9, the anti-CD94 mAb. Clone 4 expressed only an activating receptor for HLA-Cw7 with a similar reversal phenotype. Notably again, the inhibition of clones 3 and 4 by HLA-G was not reversed by any of the mAb tested. Finally, clone 5 was similar to clone 1 except that the reversal of the inhibition by HLA-Cw6 by HP3E4 was only partial and HP3D9 did not reverse at all.
Figure 5.
Reversal of inhibition by NK clones using monoclonal antibodies. NK clones were incubated with various [35S]methionine-labeled target cells for 5 hr at E:T ratio of 1:1, with or without mAb (GL183, HP3E4, HP3D9 5 μg/ml). One out of two representative experiments is shown.
Reversal of Inhibition of NK Clones by Combinations of mAbs.
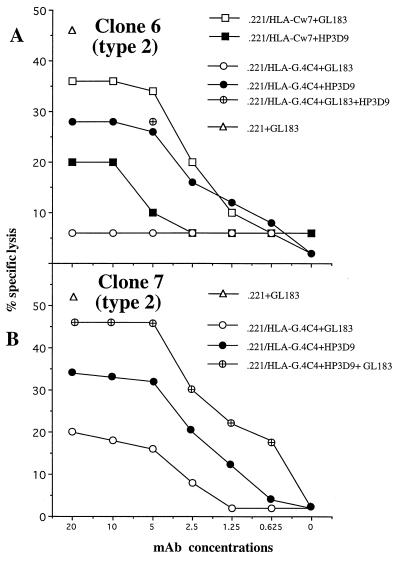
These data and data obtained earlier (see figure 1 and figure 3 legend in ref. 18) suggested that combinations of mAb might be required in some cases to obtain complete reversal of inhibition. Two examples of clones of this type are shown in Fig. 6. Clone 6 (Fig. 6A) was inhibited by both HLA-Cw7 and HLA-G and belongs to the second type of clone illustrated in Fig. 2. The inhibition by HLA-Cw7 was incompletely reversed by GL183 (anti-CD158b) and similarly partially reversed by HP3D9 (anti-CD94). However, the reversal of HLA-G inhibition by HP3D9 (anti-CD94) was incomplete and no reversal was obtained with GL183. Thus, this clone is similar to clone 1 in Fig. 4 but differs in the ability of anti-CD94 to reverse the HLA-G inhibition to a considerable extent.
Figure 6.
Reversal of inhibition of NK clones by combinations of mAbs. NK clones 6 (A) and 7 (B) were incubated with various [35S]methionine-labeled target cells for 5 hr at E:T ratio of 1:1, with or without increasing amounts of the mAb GL183, HP3E4, and HP3D9.
Clone 7 (Fig. 6B) is of considerable interest. This clone was inhibited by HLA-G (as well as by HLA-Cw7, data not shown) and the inhibition was partially reversed by either HP3D9 or GL183 but essentially completely reversed by the combination of the two mAb.
Finally, inhibition by HLA-G of the NK tumor clone YTS could not be reversed by any of the mAb employed or by the combination of all three mAb (data not shown). Similar results were reported with another subclone of the NK tumor line, YT2C2 (27). The failure of any mAb available to a known NK receptor alone or in combination to reverse inhibition by HLA-G has been observed in multiple other NK clones.
DISCUSSION
Several phenotypes were evident among the 140 NK cell lines and 246 NK clones examined. These included cells that were inhibited by HLA-G and whose inhibition could not be reversed by any of the available mAb to NK receptors or combinations of them, similar to subclones of the YT NK tumor cell line. Clearly, an unidentified receptor(s) or inhibition mechanism(s) for HLA-G must exist. Also notable was the existence of clones that were not inhibited by any of the three HLA molecules examined, HLA-Cw6, HLA-Cw7, and HLA-G. These clones could be examples of clones that can be inhibited only by HLA-B or HLA-A molecules but the possibility of the occurrence of clones not inhibited by any class I MHC molecule must be entertained.
As shown in previous work (20–22) and also in these experiments CD94 (or more precisely its complex with NKG2A) can clearly function as an inhibitory receptor for HLA-G. However, notably in the present experiments using unrelated donors only 56% and 15% of the NK cell lines that were inhibited by the HLA-G transfectant could be reversed by anti-CD94 mAb, and no clones were identified in which inhibition by the high expressing HLA-G could be completely reversed by anti-CD94 mAb alone. For example, clones 6 and 7 in Fig. 6 were only reversed by about 50%. This discrepancy with other reports could be explained by differences between individual donors, or differences in HLA-G expression levels between the transfectant cell lines used in various studies. Similarly, the reversal of inhibition by HLA-Cw6 or HLA-Cw7 by the CD158a and CD158b mAb HP3E4 and GL183 was often incomplete and in some of these cases a combination of an anti-CD158 mAb and an anti-CD94 mAb resulted in complete reversal (and compare clone JE50 in ref. 20).
Finally, clone 7 in Fig. 6 is particularly interesting. This clone was inhibited by HLA-G and the inhibition was partially reversed by both anti-CD158b (GL183) and anti-CD94 (HP3D9). Notably, the combination of both mAbs resulted in complete reversal. In the present study, this type of clone was rare but it illustrates that in some cases CD158 molecules can function as inhibitory receptors for HLA-G. The best interpretation of the inability to readily find bulk NK cell lines whose inhibition by HLA-G could be reversed by anti-CD158 mAb to the extent reported in the earlier experiments (18) is that those experiments were carried out with NK lines in which an atypical expansion of this type of clone had occurred, either in vivo or in vitro. In the present experiments this same phenotype was observed in only one NK cell line (Fig. 1A) and lines in which either CD94 or an unidentified molecule functioned as a receptor clearly predominated (Fig. 1E).
Those same properties are also evident in a few of the clones described in other recent papers documenting the importance of CD94 (20–22). For example, partial reversal of the inhibition by HLA-G of clone KK2 (22) or of the NK 4 clone (21) by anti-CD158a mAb was reported, and the inhibition of clone JE50 by HLA-Cw3 (20) was minimally reversed by either anti-CD158b or anti-CD94 mAb, but nearly completely reversed by the combination of the two mAb. In addition, low but measurable binding of CD158a fusion protein to HLA-G was observed (20), consistent with the observation that only a transfectant expressing a high level of HLA-G was able to inhibit (Figs. 3 and 4). Clones lacking CD94 that were nevertheless inhibited by HLA-G have also been reported by others (20–22, 27). In addition, the CD94 complex occurs both in an inhibitory configuration (CD94/NKG2A) and in an activating configuration (CD94/p39) (28), and clones expressing CD94 that were neither inhibited nor activated by HLA-G also have been observed (ref. 20 and type 7 in Fig. 2). Enhanced NK killing of HLA-G expressing targets has been observed by several laboratories (Fig. 1F and refs 13, 19, and 22). That HLA-G can also activate cell killing suggests that its expression is not the only mechanism by which the placenta avoids NK attack, an interpretation supported by the observation that the trophoblast cells lining the placental intervillous space and in intimate contact with maternal blood do not express HLA-G.
In conclusion, the present data suggest the existence of at least three types of inhibitory receptor for HLA-G: (1) An inhibitory receptor that recognizes only HLA-G and is present, for example, on the type 6 clones of Fig. 2. The function of this receptor was not abrogated by any of the known mAbs to NK receptors, (2) CD94, which can function as an inhibitory receptor on many NK clones either by itself or in combination with another receptor, (3) CD158a and CD158b, which appear to function as an inhibitory receptor on a small number of clones in association with HLA-G as illustrated in Fig. 6. An additional and intriguing possibility is that several of these inhibitory receptors may physically associate to form an inhibitory complex, possibly analogous to the activating complexes found associated with the T cell receptor. This possibility is evident for CD158a and CD158b the receptors for HLA-Cw6 and HLA-Cw7, as well as for the inhibitory complex(es) for which HLA-G is the ligand, and may also apply to the receptor(s) for UL18, the class I MHC homologue encoded by cytomegalovirus that inhibits NK cells (12, 29). The relationship of these putative complexes to two other NK cell markers CD16, the low affinity Fcγ III receptor, and CD56, the neural cell adhesion molecule (NCAM), both of which are also expressed on NK cells, is also worthy of further study.
Acknowledgments
We are very grateful to the individuals cited for gifts of mAb and cell lines, to Dr. L. Lanier for discussion, and to Drs. H.G. Rammensee and M. Lopez-Botet for sending preprints prior to publication. This work was supported by an National Institutes of Health Research Grant No. CA-47554. O.M. was supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship Grant No. DRG 1454.
ABBREVIATIONS
- NK
natural killer
- MHC
major histocompatibility complex
References
- 1.Orr H T, Bach F H, Ploegh H L, Strominger J L, Kavathas P, DeMars R. Nature (London) 1982;296:454–456. doi: 10.1038/296454a0. [DOI] [PubMed] [Google Scholar]
- 2.Geraghty D E, Koller B H, Orr H T. Proc Natl Acad Sci USA. 1987;84:9145–9149. doi: 10.1073/pnas.84.24.9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovats S, Main E K, Librach C, Stubblebine M, Fisher S J, DeMars R. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 4.Ellis S A, Palmer M S, McMichael A J. J Immunol. 1990;144:731–735. [PubMed] [Google Scholar]
- 5.Hamilton W J, Boyd J D. Nature (London) 1966;212:906–908. doi: 10.1038/212906a0. [DOI] [PubMed] [Google Scholar]
- 6.McMaster M T, Librach C L, Zhou Y, Lim K H, Janaptour M J, DeMars R, Kovats S, Damsky C, Fisher S J. J Immunol. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 7.King A, Boocock C, Sharkey A M, Gardner L, Beretta A, Siccardi A G, Loke Y W. J Immunol. 1996;156:2068–2076. [PubMed] [Google Scholar]
- 8.Faulk W P, Temple A. Nature(London) 1976;262:799–802. doi: 10.1038/262799a0. [DOI] [PubMed] [Google Scholar]
- 9.Parham, P., ed. (1997) Immunol. Rev. 157, 5–221.
- 10.Ljunggren H-G, Karre K. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 11.Farrell H E, Vally H, Lynch D M, Fleming P, Shellam G R, Scalzo A A, Davis-Poynter N J. Nature (London) 1997;386:510–514. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 12.Reyburn H T, Mandelboim O, Vales-Gomez M, Davis D M, Pazmany L, Strominger J L. Nature (London) 1997;386:514–517. doi: 10.1038/386514a0. [DOI] [PubMed] [Google Scholar]
- 13.Chumbley G, King A, Robertson K, Holmes N, Loke Y W. Cell Immunol. 1994;155:312–322. doi: 10.1006/cimm.1994.1125. [DOI] [PubMed] [Google Scholar]
- 14.Deniz G, Christmas S E, Brew R, Johnson P M. J Immunol. 1994;152:4255–4261. [PubMed] [Google Scholar]
- 15.Ciccone E, Pende D, Viale O, Tambussi G, Ferrini S, Biassoni R, Longo A, Guardiola J, Moretta A, Moretta L. J Exp Med. 1990;172:47–52. doi: 10.1084/jem.172.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colonna M, Borsellino G, Falco M, Ferrara G B, Strominger J L. Proc Natl Acad Sci USA. 1993;90:1200–1204. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandelboim O, Reyburn H T, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger J L. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pazmany L, Mandelboim O, Vales-Gomez M, Davis D M, Reyburn H T, Strominger J L. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 19.Munz C, Holmes N, King A, Loke Y W, Colonna M, Schild H, Rammensee H-G. J Exp Med. 1997;185:385–391. doi: 10.1084/jem.185.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Villar J J, Melero I, Navarro F, Carretero M, Bellon T, Llano M, Colonna M, Geraghty D E, Lopez-Botet M. J Immunol. 1997;158:5736–5743. [PubMed] [Google Scholar]
- 21.Soderstrom K, Corliss B, Lanier L L, Phillips J H. J Immunol. 1997;159:1072–1076. [PubMed] [Google Scholar]
- 22.Pende D, Sivori S, Accame L, Pareti L, Falco M, Geraghty D E, Le Buteiller P, Moretta L, Moretta A. Eur J Immunol. 1997;27:1875–1880. doi: 10.1002/eji.1830270809. [DOI] [PubMed] [Google Scholar]
- 23.Shimizu Y, DeMars R. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- 24.Loke Y W, King A, Burrows T, Gardner L, Bowen M, Hilby S, Howlett S, Holmes N, Jakobs D. Tissue Antigens. 1997;50:135–146. doi: 10.1111/j.1399-0039.1997.tb02852.x. [DOI] [PubMed] [Google Scholar]
- 25.Davis D M, Reyburn H T, Pazmany L, Chiu I, Mandelboim O, Strominger J L. Eur J Immunol. 1997;27:2714–2719. doi: 10.1002/eji.1830271035. [DOI] [PubMed] [Google Scholar]
- 26.Yoneda N, Tatsum,i E, Kawano S, Teshiwagara K, Oka T, Fukuda M, Yamaguchi N. Leukemia. 1992;6:136–142. [PubMed] [Google Scholar]
- 27.Rouas-Freiss N, Marchal R E, Kirszenbaum M, Dausset J, Carosella E D. Proc Natl Acad Sci USA. 1997;94:5249–5254. doi: 10.1073/pnas.94.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Villar J J, Carretero M, Navarro F, Melero I, Rodriguez A, Bottino C, Moretta A, Moretta L. J Immunol. 1996;157:5367–5374. [PubMed] [Google Scholar]
- 29.Cosman D, Fanger N, Borgrs L, Kubin M, Chin W, Peterson L, Hsu M-L. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]