Abstract
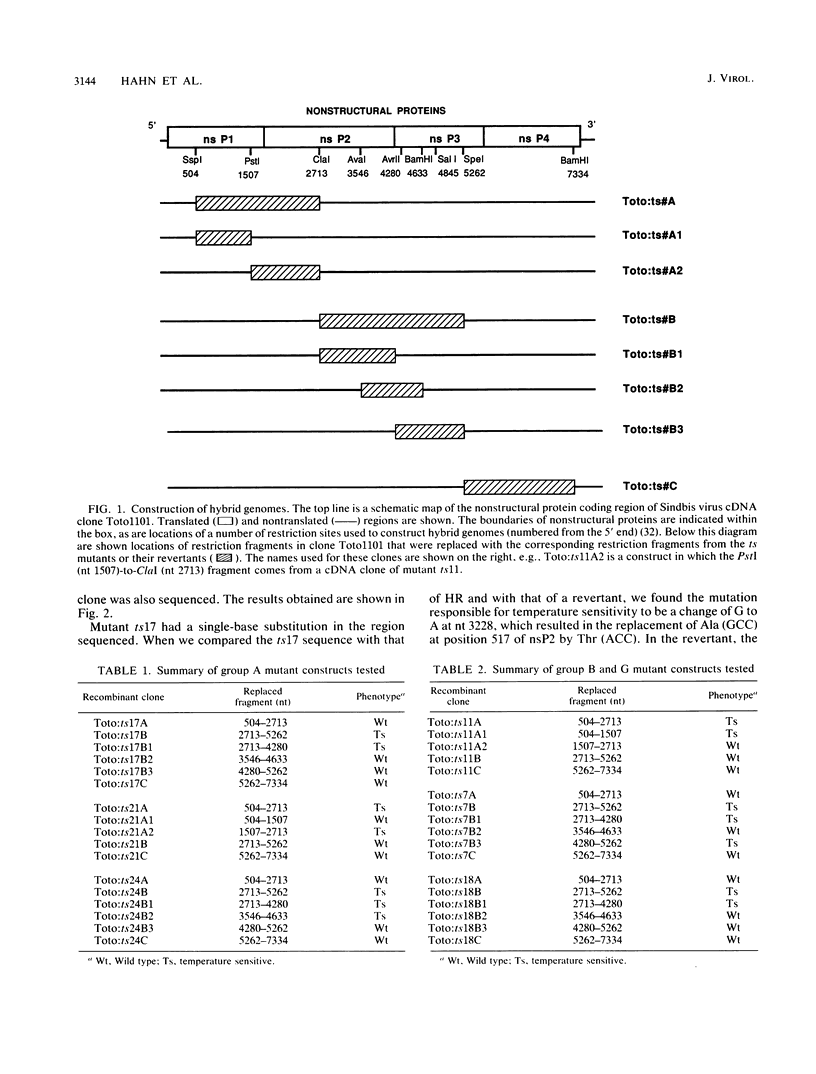
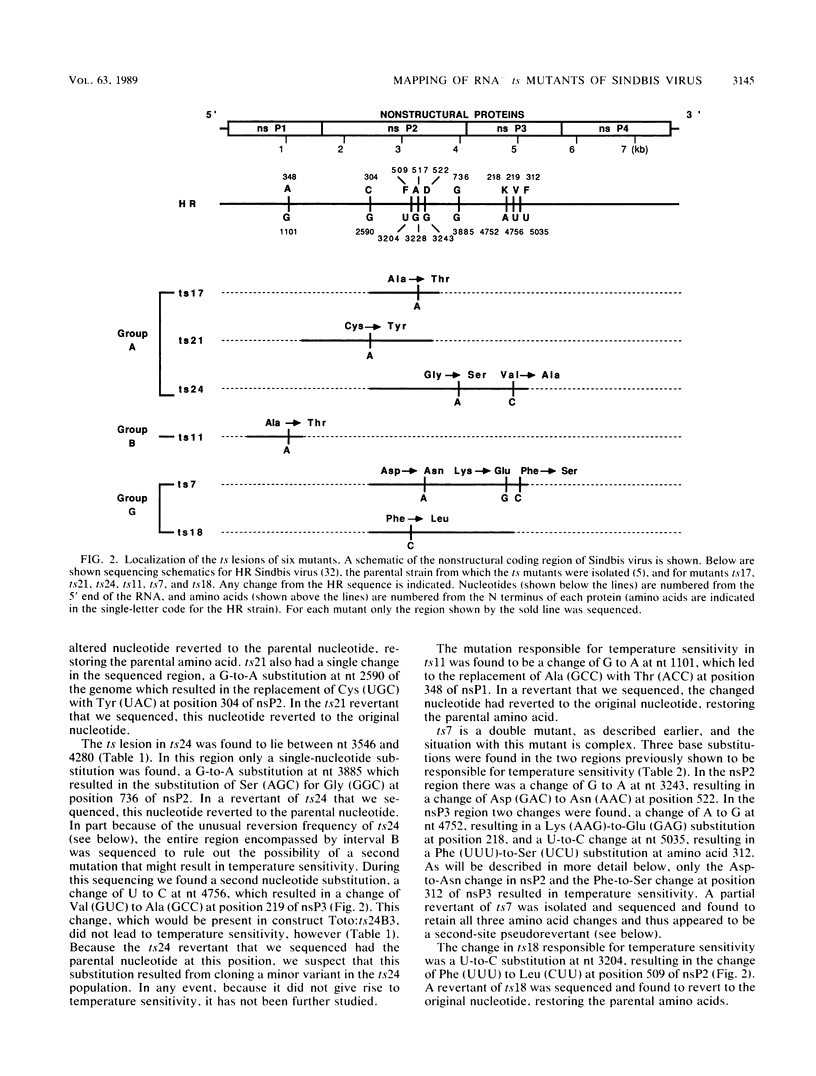
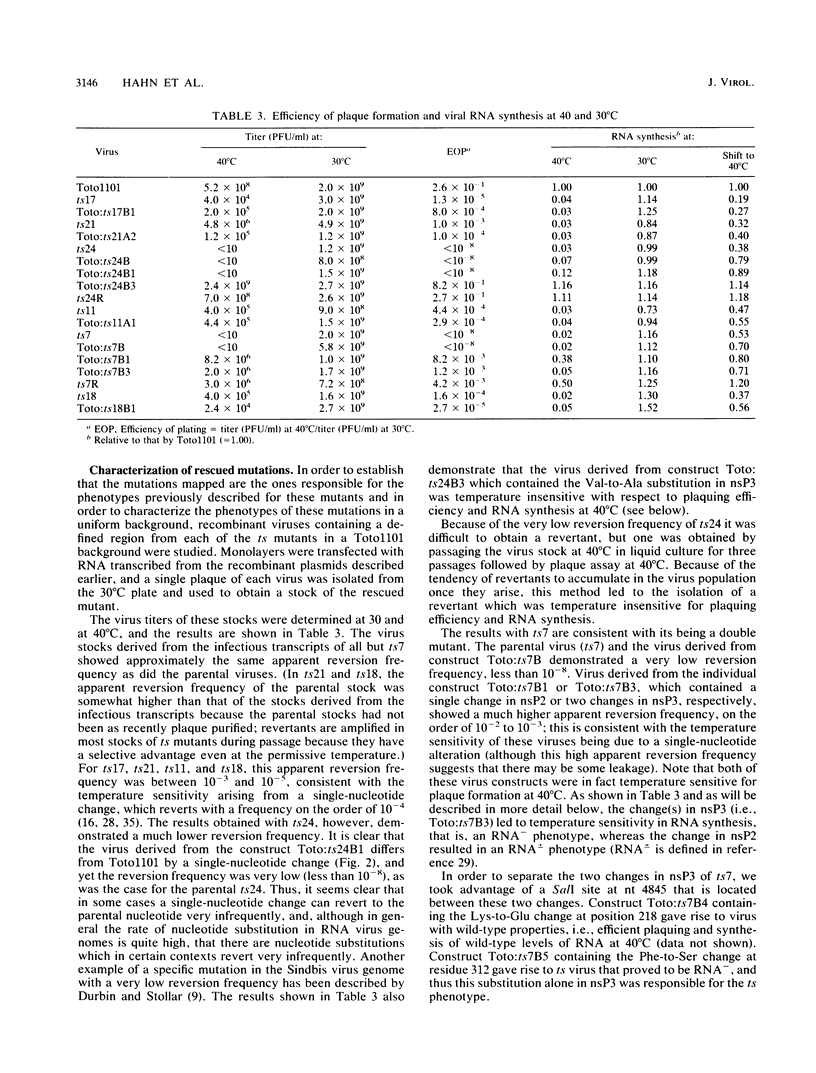
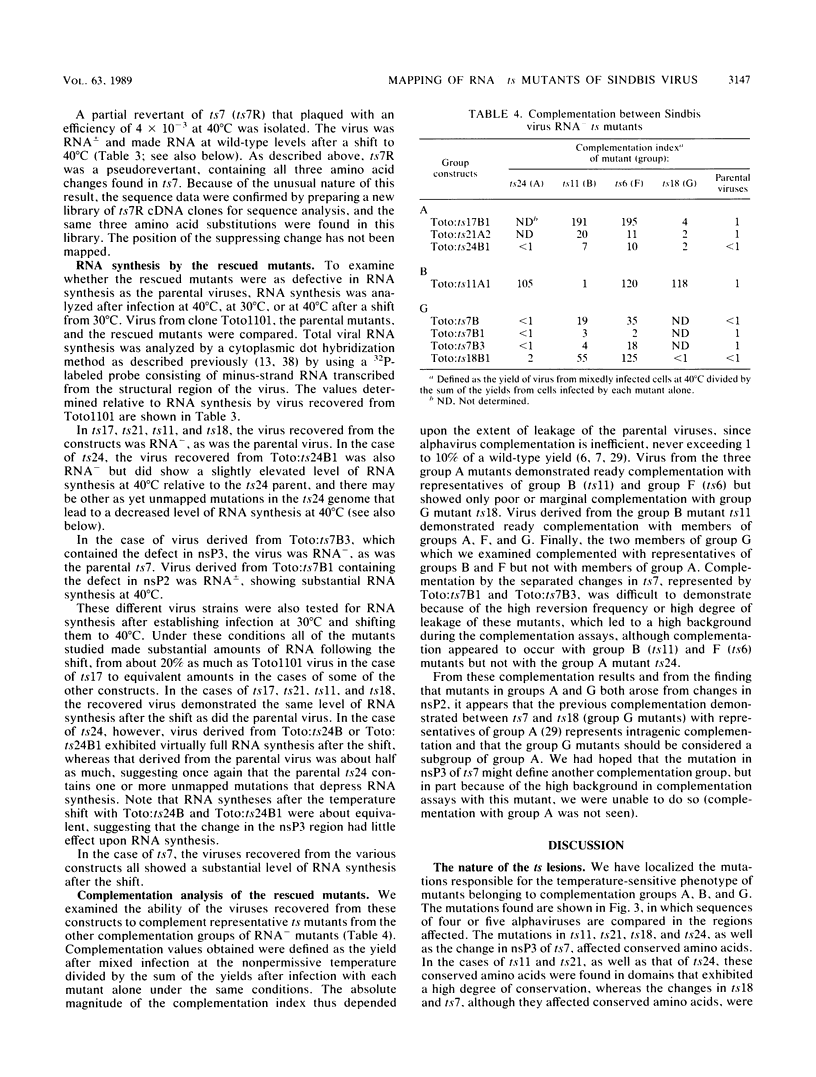
Four complementation groups of temperature-sensitive (ts) mutants of Sindbis virus that fail to make RNA at the nonpermissive temperature are known, and we have previously shown that group F mutants have defects in nsP4. Here we map representatives of groups A, B, and G. Restriction fragments from a full-length clone of Sindbis virus, Toto1101, were replaced with the corresponding fragments from the various mutants. These hybrid plasmids were transcribed in vitro by SP6 RNA polymerase to produce infectious RNA transcripts, and the virus recovered was tested for temperature sensitivity. After each lesion was mapped to a specific region, cDNA clones of both mutants and revertants were sequenced in order to determine the precise nucleotide change responsible for each mutation. Synthesis of viral RNA and complementation by rescued mutants were also examined in order to study the phenotype of each mutation in a uniform genetic background. The single mutant of group B, ts11, had a defect in nsP1 (Ala-348 to Thr). All of the group A and group G mutants examined had lesions in nsP2 (Ala-517 to Thr in ts17, Cys-304 to Tyr in ts21, and Gly-736 to Ser in ts24 for three group A mutants, and Phe-509 to Leu in ts18 and Asp-522 to Asn in ts7 for two group G mutants). In addition, ts7 had a change in nsP3 (Phe-312 to Ser) which also rendered the virus temperature sensitive and RNA-. Thus, changes in any of the four nonstructural proteins can lead to failure to synthesize RNA at a nonpermissive temperature, indicating that all four are involved in RNA synthesis. From the results presented here and from previous results, several of the activities of the nonstructural proteins can be deduced. It appears that nsP1 may be involved in the initiation of minus-strand RNA synthesis. nsP2 appears to be involved in the initiation of 26S RNA synthesis, and in addition it appears to be a protease that cleaves the nonstructural polyprotein precursors. It may also be involved in shutoff of minus-strand RNA synthesis. nsP4 appears to function as the viral polymerase or elongation factor. The functions of nsP3 are as yet unresolved.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Strauss E. G., Rice C. M., Strauss J. H., Haseloff J., Zimmern D. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J Virol. 1985 Feb;53(2):536–542. doi: 10.1128/jvi.53.2.536-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C., Bell J. R., Lenches E. M., Strauss E. G., Strauss J. H. Sequence analysis of two mutants of Sindbis virus defective in the intracellular transport of their glycoproteins. J Mol Biol. 1983 Jul 25;168(1):87–102. doi: 10.1016/s0022-2836(83)80324-6. [DOI] [PubMed] [Google Scholar]
- Barton D. J., Sawicki S. G., Sawicki D. L. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J Virol. 1988 Oct;62(10):3597–3602. doi: 10.1128/jvi.62.10.3597-3602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge B. W., Pfefferkorn E. R. Complementation between temperature-sensitive mutants of Sindbis virus. Virology. 1966 Oct;30(2):214–223. doi: 10.1016/0042-6822(66)90097-3. [DOI] [PubMed] [Google Scholar]
- Burge B. W., Pfefferkorn E. R. Isolation and characterization of conditional-lethal mutants of Sindbis virus. Virology. 1966 Oct;30(2):204–213. doi: 10.1016/0042-6822(66)90096-1. [DOI] [PubMed] [Google Scholar]
- Burge B. W., Pfefferkorn E. R. Temperature-sensitive mutants of Sindbis virus: biochemical correlates of complementation. J Virol. 1967 Oct;1(5):956–962. doi: 10.1128/jvi.1.5.956-962.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. K., Stollar V. Sequence analysis of the E2 gene of a hyperglycosylated, host restricted mutant of Sindbis virus and estimation of mutation rate from frequency of revertants. Virology. 1986 Oct 15;154(1):135–143. doi: 10.1016/0042-6822(86)90436-8. [DOI] [PubMed] [Google Scholar]
- Faragher S. G., Meek A. D., Rice C. M., Dalgarno L. Genome sequences of a mouse-avirulent and a mouse-virulent strain of Ross River virus. Virology. 1988 Apr;163(2):509–526. doi: 10.1016/0042-6822(88)90292-9. [DOI] [PubMed] [Google Scholar]
- Fuller F. J., Marcus P. I. Sindbis virus. I. Gene order of translation in vivo. Virology. 1980 Dec;107(2):441–451. doi: 10.1016/0042-6822(80)90311-6. [DOI] [PubMed] [Google Scholar]
- Hahn C. S., Strauss E. G., Strauss J. H. Sequence analysis of three Sindbis virus mutants temperature-sensitive in the capsid protein autoprotease. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4648–4652. doi: 10.1073/pnas.82.14.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y. S., Grakoui A., Rice C. M., Strauss E. G., Strauss J. H. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol. 1989 Mar;63(3):1194–1202. doi: 10.1128/jvi.63.3.1194-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy W. R., Strauss J. H. Processing the nonstructural polyproteins of Sindbis virus: study of the kinetics in vivo by using monospecific antibodies. J Virol. 1988 Mar;62(3):998–1007. doi: 10.1128/jvi.62.3.998-1007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J., Goelet P., Zimmern D., Ahlquist P., Dasgupta R., Kaesberg P. Striking similarities in amino acid sequence among nonstructural proteins encoded by RNA viruses that have dissimilar genomic organization. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4358–4362. doi: 10.1073/pnas.81.14.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Keränen S., Käriäinen L. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J Virol. 1979 Oct;32(1):19–29. doi: 10.1128/jvi.32.1.19-29.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist B. H., DiSalvo J., Rice C. M., Strauss J. H., Strauss E. G. Sindbis virus mutant ts20 of complementation group E contains a lesion in glycoprotein E2. Virology. 1986 May;151(1):10–20. doi: 10.1016/0042-6822(86)90099-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Levis R., Strauss J. H., Huang H. V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987 Dec;61(12):3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Strauss J. H. Synthesis, cleavage and sequence analysis of DNA complementary to the 26 S messenger RNA of Sindbis virus. J Mol Biol. 1981 Aug 15;150(3):315–340. doi: 10.1016/0022-2836(81)90550-7. [DOI] [PubMed] [Google Scholar]
- Sawicki D. L., Sawicki S. G. Functional analysis of the A complementation group mutants of Sindbis HR virus. Virology. 1985 Jul 15;144(1):20–34. doi: 10.1016/0042-6822(85)90301-0. [DOI] [PubMed] [Google Scholar]
- Sawicki D. L., Sawicki S. G., Keränen S., Käriäinen L. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J Virol. 1981 Aug;39(2):348–358. doi: 10.1128/jvi.39.2.348-358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D. L., Sawicki S. G. Short-lived minus-strand polymerase for Semliki Forest virus. J Virol. 1980 Apr;34(1):108–118. doi: 10.1128/jvi.34.1.108-118.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L., Käriäinen L., Keränen S. A Sindbis virus mutant temperature-sensitive in the regulation of minus-strand RNA synthesis. Virology. 1981 Nov;115(1):161–172. doi: 10.1016/0042-6822(81)90098-2. [DOI] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. The effect of loss of regulation of minus-strand RNA synthesis on Sindbis virus replication. Virology. 1986 Jun;151(2):339–349. doi: 10.1016/0042-6822(86)90054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D. A., Holland J. J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Lenches E. M., Strauss J. H. Mutants of sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virology. 1976 Oct 1;74(1):154–168. doi: 10.1016/0042-6822(76)90137-9. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Levinson R., Rice C. M., Dalrymple J., Strauss J. H. Nonstructural proteins nsP3 and nsP4 of Ross River and O'Nyong-nyong viruses: sequence and comparison with those of other alphaviruses. Virology. 1988 May;164(1):265–274. doi: 10.1016/0042-6822(88)90644-7. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984 Feb;133(1):92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J. H., Strauss E. G. Evolution of RNA viruses. Annu Rev Microbiol. 1988;42:657–683. doi: 10.1146/annurev.mi.42.100188.003301. [DOI] [PubMed] [Google Scholar]
- Takkinen K. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986 Jul 25;14(14):5667–5682. doi: 10.1093/nar/14.14.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. A., Bancroft F. C. Cytoplasmic dot hybridization. Simple analysis of relative mRNA levels in multiple small cell or tissue samples. J Biol Chem. 1982 Aug 10;257(15):8569–8572. [PubMed] [Google Scholar]


