Abstract
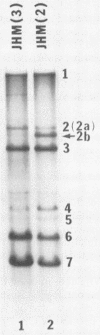
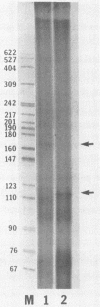
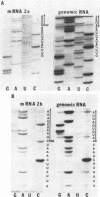
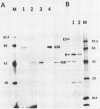
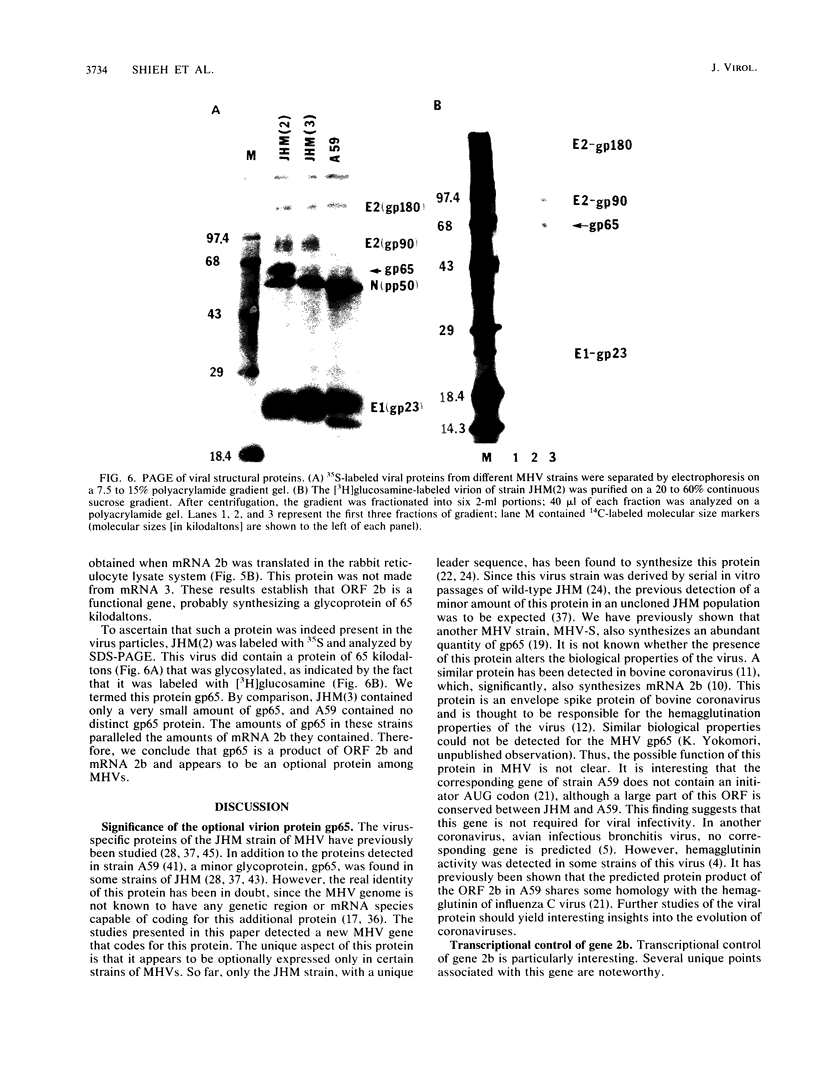
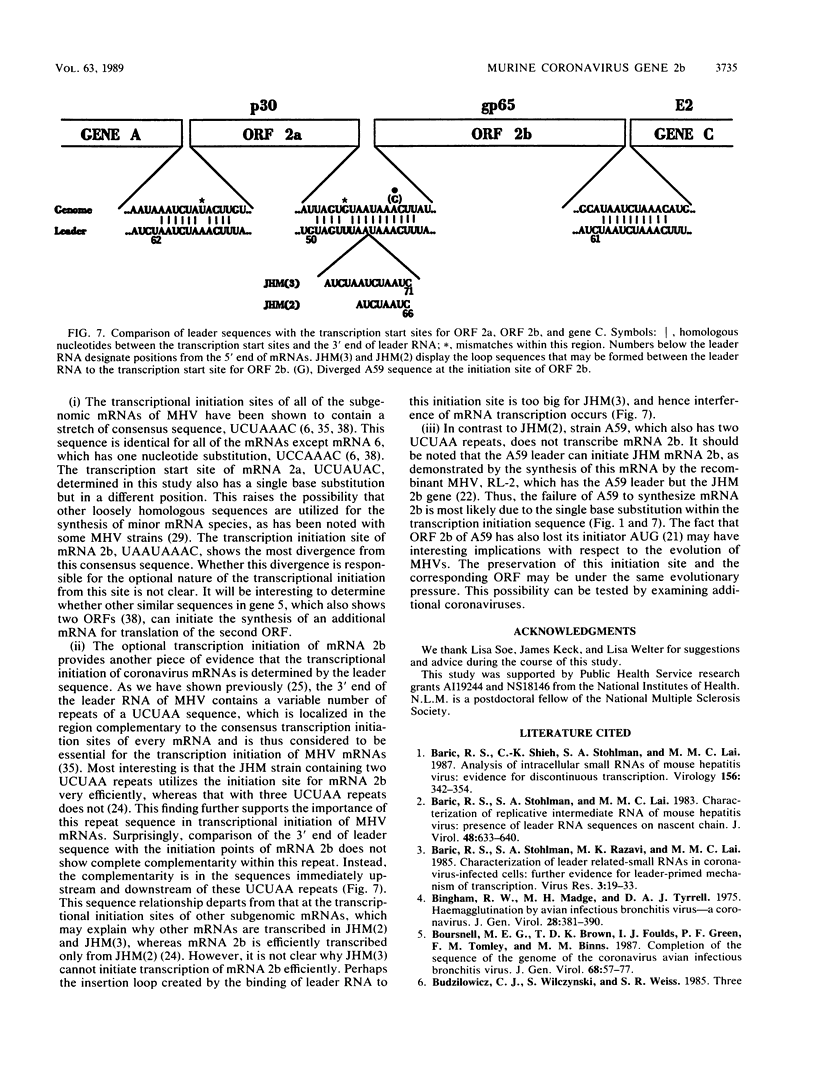
We have previously shown that some strains of the murine coronavirus mouse hepatitis virus (MHV) synthesize an additional mRNA species (mRNA 2b, previously called mRNA 2a) with a size intermediate between that of mRNAs 2 and 3, suggesting the presence of an optional transcriptional initiation site. This transcriptional start is dependent on the leader sequence of the virus strains. To study the mechanism of coronavirus transcriptional regulation, we have cloned and sequenced the region of the viral genome corresponding to the 5' unique coding region of mRNA 2 of the JHM strain of MHV. In addition to the open reading frame (ORF) predicted to encode the viral nonstructural protein p30, a second complete ORF, with the potential to encode a 439-amino-acid polypeptide, was discovered. The transcriptional initiation sites of both mRNA 2a (formerly called mRNA 2) and mRNA 2b were determined by primer extension studies and RNA sequencing. The data indicated that transcription of mRNA 2a initiated at a site, UCUAUAC, that resembled the consensus intergenic sequence. In contrast, the start signal of the optional mRNA 2b, UAAUAAAC, deviated from the consensus sequence. mRNA 2b is a functional mRNA, as shown by in vitro translation studies of mRNA and ORF 2b and by the detection of an additional viral structural protein, gp65, in the JHM strain that synthesized this mRNA. Although the A59 strain of MHV was found to retain ORF 2b, it lacked the correct transcriptional and translational start signals for this gene. This study has therefore identified an optional gene product for murine coronaviruses and provided insights into the mechanism of regulation of MHV RNA transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baric R. S., Shieh C. K., Stohlman S. A., Lai M. M. Analysis of intracellular small RNAs of mouse hepatitis virus: evidence for discontinuous transcription. Virology. 1987 Feb;156(2):342–354. doi: 10.1016/0042-6822(87)90414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Lai M. M. Characterization of replicative intermediate RNA of mouse hepatitis virus: presence of leader RNA sequences on nascent chains. J Virol. 1983 Dec;48(3):633–640. doi: 10.1128/jvi.48.3.633-640.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R. S., Stohlman S. A., Razavi M. K., Lai M. M. Characterization of leader-related small RNAs in coronavirus-infected cells: further evidence for leader-primed mechanism of transcription. Virus Res. 1985 Jul;3(1):19–33. doi: 10.1016/0168-1702(85)90038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham R. W., Madge M. H., Tyrrell D. A. Haemagglutination by avian infectious bronchitis virus-a coronavirus. J Gen Virol. 1975 Sep;28(3):381–390. doi: 10.1099/0022-1317-28-3-381. [DOI] [PubMed] [Google Scholar]
- Boursnell M. E., Brown T. D., Foulds I. J., Green P. F., Tomley F. M., Binns M. M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987 Jan;68(Pt 1):57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Keck J. G., Hogue B. G., Brian D. A., Lai M. M. Temporal regulation of bovine coronavirus RNA synthesis. Virus Res. 1988 Mar;9(4):343–356. doi: 10.1016/0168-1702(88)90093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B., Brian D. A. Bovine coronavirus structural proteins. J Virol. 1982 May;42(2):700–707. doi: 10.1128/jvi.42.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B., Potts B. J., Brian D. A. Bovine coronavirus hemagglutinin protein. Virus Res. 1985 Feb;2(1):53–59. doi: 10.1016/0168-1702(85)90059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Regulation of protein synthesis in virus-infected animal cells. Adv Virus Res. 1986;31:229–292. doi: 10.1016/S0065-3527(08)60265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Baric R. S., Brayton P. R., Stohlman S. A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Brayton P. R., Armen R. C., Patton C. D., Pugh C., Stohlman S. A. Mouse hepatitis virus A59: mRNA structure and genetic localization of the sequence divergence from hepatotropic strain MHV-3. J Virol. 1981 Sep;39(3):823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus leader-RNA-primed transcription: an alternative mechanism to RNA splicing. Bioessays. 1986 Dec;5(6):257–260. doi: 10.1002/bies.950050606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Stohlman S. A. Comparative analysis of RNA genomes of mouse hepatitis viruses. J Virol. 1981 May;38(2):661–670. doi: 10.1128/jvi.38.2.661-670.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M., Stohlman S. A. RNA of mouse hepatitis virus. J Virol. 1978 May;26(2):236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge J., Langridge P., Bergquist P. L. Extraction of nucleic acids from agarose gels. Anal Biochem. 1980 Apr;103(2):264–271. doi: 10.1016/0003-2697(80)90266-3. [DOI] [PubMed] [Google Scholar]
- Luytjes W., Bredenbeek P. J., Noten A. F., Horzinek M. C., Spaan W. J. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988 Oct;166(2):415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Fleming J. O., Keck J. G., Stohlman S. A., Lai M. M. RNA recombination of coronaviruses: localization of neutralizing epitopes and neuropathogenic determinants on the carboxyl terminus of peplomers. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6567–6571. doi: 10.1073/pnas.84.18.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Fujioka N., Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985 May;54(2):329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Lai M. M. Evolution of the 5'-end of genomic RNA of murine coronaviruses during passages in vitro. Virology. 1989 Mar;169(1):227–232. doi: 10.1016/0042-6822(89)90060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Soe L. H., Shieh C. K., Lai M. M. Discontinuous transcription generates heterogeneity at the leader fusion sites of coronavirus mRNAs. J Virol. 1988 Oct;62(10):3870–3873. doi: 10.1128/jvi.62.10.3870-3873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Stohlman S. A., Lai M. M. Leader sequences of murine coronavirus mRNAs can be freely reassorted: evidence for the role of free leader RNA in transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4204–4208. doi: 10.1073/pnas.83.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Fujiwara K. Defective interfering particles of mouse hepatitis virus. Virology. 1984 Feb;133(1):9–17. doi: 10.1016/0042-6822(84)90420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Hayami M., Fujiwara K. Characterization of small plaque mutants of mouse hepatitis virus, JHM strain. Microbiol Immunol. 1983;27(5):445–454. doi: 10.1111/j.1348-0421.1983.tb00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Hirano N., Fujiwara K. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology. 1984 Nov;139(1):138–151. doi: 10.1016/0042-6822(84)90335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pedersen F. S., Haseltine W. A. A micromethod for detailed characterization of high molecular weight RNA. Methods Enzymol. 1980;65(1):680–687. doi: 10.1016/s0076-6879(80)65066-6. [DOI] [PubMed] [Google Scholar]
- Rottier P. J., Spaan W. J., Horzinek M. C., van der Zeijst B. A. Translation of three mouse hepatitis virus strain A59 subgenomic RNAs in Xenopus laevis oocytes. J Virol. 1981 Apr;38(1):20–26. doi: 10.1128/jvi.38.1.20-26.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt I., Skinner M., Siddell S. Nucleotide sequence of the gene encoding the surface projection glycoprotein of coronavirus MHV-JHM. J Gen Virol. 1987 Jan;68(Pt 1):47–56. doi: 10.1099/0022-1317-68-1-47. [DOI] [PubMed] [Google Scholar]
- Shieh C. K., Soe L. H., Makino S., Chang M. F., Stohlman S. A., Lai M. M. The 5'-end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology. 1987 Feb;156(2):321–330. doi: 10.1016/0042-6822(87)90412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S. Coronavirus JHM: coding assignments of subgenomic mRNAs. J Gen Virol. 1983 Jan;64(Pt 1):113–125. doi: 10.1099/0022-1317-64-1-113. [DOI] [PubMed] [Google Scholar]
- Siddell S., Wege H., Barthel A., ter Meulen V. Coronavirus JHM: intracellular protein synthesis. J Gen Virol. 1981 Mar;53(Pt 1):145–155. doi: 10.1099/0022-1317-53-1-145. [DOI] [PubMed] [Google Scholar]
- Skinner M. A., Ebner D., Siddell S. G. Coronavirus MHV-JHM mRNA 5 has a sequence arrangement which potentially allows translation of a second, downstream open reading frame. J Gen Virol. 1985 Mar;66(Pt 3):581–592. doi: 10.1099/0022-1317-66-3-581. [DOI] [PubMed] [Google Scholar]
- Soe L. H., Shieh C. K., Baker S. C., Chang M. F., Lai M. M. Sequence and translation of the murine coronavirus 5'-end genomic RNA reveals the N-terminal structure of the putative RNA polymerase. J Virol. 1987 Dec;61(12):3968–3976. doi: 10.1128/jvi.61.12.3968-3976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Delius H., Skinner M., Armstrong J., Rottier P., Smeekens S., van der Zeijst B. A., Siddell S. G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2(10):1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturman L. S. I. Structural proteins: effects of preparative conditions on the migration of protein in polyacrylamide gels. Virology. 1977 Apr;77(2):637–649. doi: 10.1016/0042-6822(77)90488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Massa P. T., ter Meulen V. Characterization of a variant virus isolated from neural cell culture after infection of mouse coronavirus JHMV. Virology. 1986 Nov;155(1):267–270. doi: 10.1016/0042-6822(86)90187-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege H., Müller A., ter Meulen V. Genomic RNA of the murine coronavirus JHM. J Gen Virol. 1978 Nov;41(2):217–227. doi: 10.1099/0022-1317-41-2-217. [DOI] [PubMed] [Google Scholar]
- Wege H., Wege H., Nagashima K., ter Meulen V. Structural polypeptides of the murine coronavirus JHM. J Gen Virol. 1979 Jan;42(1):37–47. doi: 10.1099/0022-1317-42-1-37. [DOI] [PubMed] [Google Scholar]
- Zimmern D., Kaesberg P. 3'-terminal nucleotide sequence of encephalomyocarditis virus RNA determined by reverse transcriptase and chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4257–4261. doi: 10.1073/pnas.75.9.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]