Abstract
We have reported previously that the hepatitis B virus oncoprotein, HBx, can bind to the C terminus of p53 and inhibit several critical p53-mediated cellular processes, including DNA sequence-specific binding, transcriptional transactivation, and apoptosis. Recognizing the importance of p53-mediated apoptosis for maintaining homeostasis and preventing neoplastic transformation, here we further examine the physical interaction between HBx and p53 as well as the functional consequences of this association. In vitro binding studies indicate that the ayw and adr viral subtypes of HBx bind similar amounts of glutathione S-transferase-p53 with the distal C terminus of HBx (from residues 111 to 154) being critical for this interaction. Using a microinjection technique, we show that this same C-terminal region of HBx is necessary for sequestering p53 in the cytoplasm and abrogating p53-mediated apoptosis. The transcriptional transactivation domain of HBx also maps to its C terminus; however, a comparison of the ability of full-length and truncated HBx protein to abrogate p53-induced apoptosis versus transactivate simian virus 40- or human nitric oxide synthase-2 promoter-driven reporter constructs indicates that these two functional properties are distinct and thus may contribute to hepatocarcinogenesis differently. Collectively, our data indicate that the distal C-terminal domain of HBx, independent of its transactivation activity, complexes with p53 in the cytoplasm, partially preventing its nuclear entry and ability to induce apoptosis. These pathobiological effects of HBx may contribute to the early stages of hepatocellular carcinogenesis.
Keywords: tumor suppressor gene, hepatocarcinogenesis, programmed cell death, nitric oxide
Hepatitis B virus (HBV) is a major risk factor associated with the development of hepatocellular carcinoma (HCC) (reviewed in ref. 1), with the HBV X ORF being frequently integrated and expressed (2–4). Whereas HBx is capable of neoplastically transforming rodent cells (5, 6) and causing HCCs in transgenic mice (7, 8), its oncogenic mechanism is unclear.
The p53 tumor suppressor protein is involved in numerous cellular processes that are critical for maintaining the genomic integrity of cells (reviewed in refs. 9–12). p53 is functionally inactivated by structural mutations, viral proteins, and endogenous cellular mechanisms in the majority of human cancers. Although the molecular pathogenesis of human HCC can involve the somatic mutational inactivation of the p53 gene, especially in geographic areas where dietary aflatoxin B1 exposure is a liver cancer risk (13–15), the absence of p53 mutations in the majority of HCC cases (13, 14, 16, 17) suggests that its inactivation may be achieved by another mechanism(s). Evidence is now accumulating to indicate that HBx may contribute to hepatocarcinogenesis by blocking p53 function (18–21). In this study, we present data consistent with the hypothesis that HBx, via its distal C-terminal domain, binds to and partially sequesters p53 in the cytoplasm, resulting in the abrogation of p53-mediated apoptosis.
MATERIALS AND METHODS
Plasmids.
The plasmid constructs encoding GST-WTp53 and full-length HBx of the adr subtype (pSPX46) have been described previously (19). The following plasmids under control of the T3 or T7 promoter were used for in vitro translation of HBx of the ayw subtype: SK1–154x, encoding full-length HBx; SK1–110x, encoding the first 110 amino acids of the HBx ORF; and SK61–154x, encoding amino acids 61–154 of HBx. For microinjection and transfection studies, the following cytomegalovirus (CMV)-driven expression vectors were used: CMV-x1, encoding full-length HBx of the adr subtype (19, 21); CMV-1–154X, encoding full-length HBx (ayw subtype); CMV-30–154X, encoding amino acids 30–154 of HBx (ayw subtype); and CMV-61–154X, encoding amino acids 61–154 of HBx (ayw subtype). pactβgal, a gift of J. Yuan (Harvard University), encodes a β-galactosidase (β-gal) gene under the control of chicken β-actin promoter (22). pGreen-Lantern was obtained from Gibco/BRL. For transcriptional transactivation assays, an SV40 promoter-driven luciferase construct, pGL2 (Promega), and a human NOS2 promoter-driven luciferase reporter construct, pNOS2(3.8)luc (23), were used.
Binding Assay.
Preparation of fusion protein, in vitro translation of 35S-labeled proteins, and binding assays were carried out as described previously (19). To reference input for binding, aliquots representing 20% the volume of the different in vitro translated HBx used for the binding studies were immunoprecipitated by anti-HBx polyclonal antibody (19). Each construct was tested in at least three independent binding assays. Mean percent binding of the different HBx constructs is presented relative to full-length HBx of the ayw subtype (SK1–154X). Student’s t test was performed to assess statistical significance.
Cell Culture and Microinjection.
Low passage primary normal human fibroblasts (GMO7532) were obtained and cultured as previously described (21). Normal human hepatocytes were isolated from nontransplantable liver tissue from a 4-year-old male (donor 1) and a 36-year-old-male (donor 2) (Clonetics, San Diego). The cells were seeded directly onto grids and then microinjected within 48 h of plating. Plasmids, at 100 μg/ml in PBS, were injected into nuclei of cells using a glass microcapillary needle. For the mapping studies, each plasmid combination was microinjected into at least 50 cells per experiment, analyzed 24 h after microinjection, and tested in at least three independent experiments. Only those experiments with greater than 10 immunopositive cells present per condition were included in the analysis. The human liver cancer cell line, HepG2, was obtained from American Type Culture Collection and cultured in Eagle’s minimal essential medium supplemented with 10% fetal bovine serum.
Immunocytochemical Analysis of p53 and HBx.
Cells were fixed and immunostained as previously described (21) using anti-p53 polyclonal CM-1 antibody (1:200 dilution; Signet Laboratories, Dedham, MA) followed by Texas red-conjugated antirabbit IgG (1:200; Vector Laboratories), and/or 1:10 diluted 146x and 227x HBx monoclonal antibodies followed by FITC-conjugated antimouse Ig antibody (1:200 dilution; Vector Laboratories). Cellular localization of p53 and full-length HBx was evaluated in fibroblasts using an MRC 600 confocal microscope (Bio-Rad). β-gal was visualized using 1:50 diluted β-gal monoclonal antibody (Oncogene Science) followed by FITC-conjugated anti-mouse Ig antibody (1:200 dilution; Vector Laboratories). Nuclei were stained with 4′,6-diamidino-2-phenylindole (Sigma).
Transient Transcriptional Transactivation Assay.
HepG2 cells were seeded and transfected as previously described (23). Each 60-mm plate of cells, tested in triplicate, was cotransfected with 0.5–7.5 μg of expression vector encoding either full-length or truncated HBx and 500 ng of either pGL2 or pNOS2(3.8)luc reporter constructs. All dishes within an experiment were transfected with the same total amount of DNA by the addition of CMVneo (B. Vogelstein, Johns Hopkins Oncology Center). Preparation of cell extracts, measurement of resonance light units, and determination of total protein were carried out as described previously (23). Data are presented as fold activation by HBx relative to the neomycin control vector. The reported dose–response results reflect representative data from a single experiment, whereas the comparison of the transactivation activity of the different HBx deletion mutants represents data from three separate transfections.
RESULTS
The Distal C-Terminal Region of HBx Is Necessary for Efficient In Vitro Binding to GST-p53.
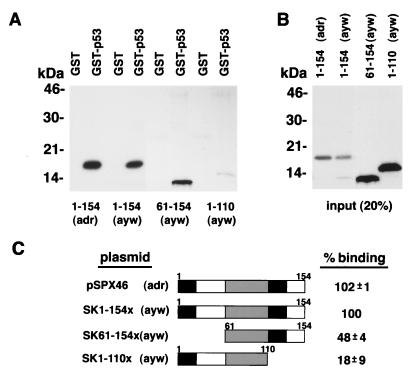
Consistent with our previous report (19), full-length HBx of the adr subtype (pSPX46) binds specifically to GST-p53 (Fig. 1 A and C). We further show in Fig. 1 that HBx of the ayw subtype (SK1–154x) binds a similar level of GST-p53 as the adr subtype. When two deletion mutants derived from HBx of the ayw subtype were analyzed, we found that an N-terminal deletion mutant, SK61–154x, retained on average 48% of the full-length HBx binding (P < 0.001), whereas a C-terminal deletion mutant, SK1–110x, consistently exhibited significantly lower levels of binding compared with both full-length HBx (18%; P < 0.001) and the N-terminal deletion mutant (P < 0.002). Similar levels of the different in vitro translated HBx proteins were used within each binding study (Fig. 1B) with their specificity being demonstrated by lack of binding to glutathione S-transferase (GST) (Fig. 1A, lanes 1, 3, 5, 7).
Figure 1.
The C-terminal domain of HBx is critical for in vitro association with GST-p53. (A) In vitro translated full-length HBx protein (lanes 1–4) and HBx deletion mutants (lanes 5–8) were incubated with glutathione-Sepharose beads loaded with either GST-p53 (lanes 2, 4, 6, 8) or GST (lanes 1, 3, 5, 7). Lanes 1–4 and 5–8, along with their respective binding input, are representative data from two independent assays. (B) To reference input for binding, 20% of the volume of the various in vitro translated HBx proteins used for binding were immunoprecipitated by anti-HBx antibody. (C) Schematic representation of full-length and truncated HBx, as described in Materials and Methods, along with a summary of their binding to p53. Percent binding represents the mean ± SD from at least three independent binding assays with values made relative to SK1–154x.
The Distal C-Terminal Domain of HBx Is Critical for Inhibition of p53-Mediated Apoptosis.
A microinjection strategy was employed using normal human fibroblasts to map the region of HBx necessary for abrogating p53-mediated apoptosis. Twenty-four hours after microinjection of a CMV-driven p53 expression vector into the nuclei of fibroblasts, 26% of the p53-immunopositive cells were apoptotic as measured by morphological criteria of 4′,6-diamidino-2-phenylindole-stained nuclei (Table 1). AnnexinV staining was used as a confirmatory assay for apoptosis (data not shown). Consistent with our recently published data (21), coinjection of p53 and full-length HBx genes of two different viral subtypes (CMV-x1, adr; CMV-1–154X, ayw) resulted in significant abrogation of p53-induced apoptosis (Table 1). The adr subtype was more efficient at blocking p53-mediated apoptosis with 7% of the cells being apoptotic, whereas 14% of the cells coexpressing p53 and the ayw subtype of HBx were apoptotic. This differential protective effect is not likely due to dissimilar levels of HBx protein expression, as a quantitative comparison of the HBx immunostaining intensity in fibroblasts microinjected with either CMV-x1 (adr subtype) or CMV-1–154X (ayw subtype) showed no significant difference (data not shown). When HBx deletion mutants, missing either the first 29 (CMV-30–154X) or 60 (CMV-61–154X) amino acids, were coinjected with p53, efficient abrogation of apoptosis relative to full-length HBx of the ayw subtype (CMV-1–154X) was observed (Table 1). In contrast, cells coexpressing p53 and HBx deletion mutants lacking either the last 44 (CMV-1–110X) or 57 (CMV-1–97X) amino acids exhibited high levels of apoptosis (19 and 20%, respectively), which were not significantly different than the percent of apoptotic cells following microinjection of p53 expression vector alone. Only very low levels of apoptosis were observed in uninjected fibroblasts or those microinjected with β-gal expression vector (Table 1). Twenty-four hours after the microinjection of an expression vector encoding full-length HBx of the ayw subtype (CMV-1–154X), we observed only a background level of apoptosis, which was assessed by the percent of apoptotic fibroblasts 24 h after microinjection of a β-gal expression vector (data not shown).
Table 1.
The C-terminal domain of the hepatitis B viral X gene is critical for inhibition of p53-mediated apoptosis
| Microinjected expression vector(s) | Percent apoptotic cells* | n† |
|---|---|---|
| — | 0.09 | 1,150 |
| β-gal | 1 ± 1 | 140 |
| p53 | 26 ± 7 | 912 |
| p53 + CMV-x1 (adr) | 7 ± 2 (P < 0.0001) | 289 |
| p53 + CMV-1-154X (ayw) | 14 ± 3 (P < 0.004) | 569 |
| p53 + CMV-30-154X (ayw) | 12 ± 2 (P < 0.001) | 277 |
| p53 + CMV-61-154X (ayw) | 13 ± 2 (P < 0.003) | 264 |
| p53 + CMV-1-110X (ayw) | 19 ± 7 (P > 0.100) | 674 |
| p53 + CMV-1-97X (ayw) | 20 ± 11 (P > 0.100) | 406 |
Fibroblasts with condensed and fragmented nuclei as well as cytoplasmic blebbing characteristic of cells undergoing apoptosis. Values represent mean ± SD. P values are for Student’s t test comparing p53-mediated apoptosis in the presence versus absence of the different HBx constructs.
n, number of p53 immunopositive cells scored following microinjection of the various expression vector combinations.
HBx Abrogates p53-Mediated Apoptosis in Normal Human Hepatocytes.
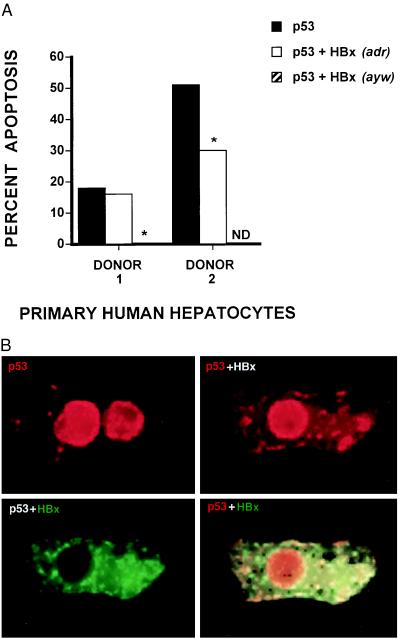
To evaluate the impact of HBx expression upon p53-induced apoptosis in liver, primary human hepatocytes isolated from nontransplantable liver from two donors were microinjected with wild-type p53 expression vector alone or together with an expression vector encoding full-length HBx. Overexpression of p53 induced apoptosis in hepatocytes from both donors; however, donor 2 hepatocytes were more sensitive than donor 1 hepatocytes, with percentage values of 51 and 18, respectively. Consistent with our findings in normal human fibroblasts, HBx inhibited p53-mediated apoptosis in primary human hepatocytes from the two donors (Fig. 2A). To assess the background level of apoptosis, pGreen-Lantern expression vector was injected into hepatocytes isolated from donor 1. When a total of 46 green lantern-expressing hepatocytes were analyzed, we saw no evidence of apoptosis.
Figure 2.
Effects of HBx expression on p53-mediated apoptosis and on p53 localization in normal human hepatocytes. Primary hepatocytes were microinjected with a p53 expression vector or coinjected with p53 and HBx (adr and ayw subtypes for donor 1; adr subtype for donor 2) expression vectors. (A) After 24 h, cells were immunostained for p53 and scored for apoptosis as described in Materials and Methods. Bar values represent the percentage of apoptotic hepatocytes from one to three individual experiments. The total number of p53 immunopositive cells scored for donors 1 and 2 were 108 and 119, respectively. ND, not determined. ∗, Fisher’s exact test comparing the levels of p53-mediated apoptosis in the absence versus presence of HBx expression, P ≤ 0.036. In the case of p53 ± HBx (adr) with donor 2, a χ2 test was performed because greater than 100 cells were analyzed, P ≤ 0.046. (B) Twenty-four hours after microinjection with p53 expression vector alone (Upper Left) or coinjection with p53 and full-length HBx expression vectors (Upper Right and Lower), hepatocytes were simultaneously immunostained for p53 (Texas red) and HBx (FITC) as described in Materials and Methods. Yellow regions (Lower Right) reflect overlapping areas of p53 and HBx immunostaining in a single hepatocyte also shown (Upper Right and Lower Left).
HBx via Its Distal C-Terminal Region May Sequester p53 in the Cytoplasm.
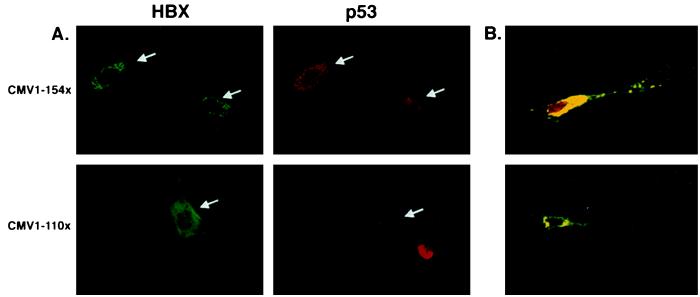
As previously described (21), microinjection of wild-type p53 expression vector into primary human fibroblasts results in elevated p53 levels accumulating predominantly in the nucleus or in both the nucleus and cytoplasm (data not shown). When observed, p53 cytoplasmic immunostaining was typically diffuse in appearance and always associated with intense nuclear p53 immunostaining (data not shown). Following the coinjection of p53 and full-length HBx expression vectors, p53 immunostaining was observed predominantly in both the nucleus and cytoplasm of fibroblasts, with the cytoplasmic staining being more intense and punctate in appearance (Fig. 3A). Fibroblasts with exclusively cytoplasmic p53 and HBx immunostaining also were occasionally observed (Fig. 3B Lower). As shown in Fig. 3, p53 colocalizes with HBx in the cytoplasm of fibroblasts coexpressing p53 and full-length HBx proteins. In contrast, p53 does not associate with HBx in cells coexpressing p53 and the C-terminal HBx deletion mutant encoded by CMV-1–110X (Fig. 3A). As with fibroblasts, p53 protein was predominantly nuclear in hepatocytes overexpressing only wild-type p53, whereas it tended to be more cytoplasmic when overexpressed with HBx (Fig. 2B). Moreover, many coinjected hepatocytes exhibited very similar cytoplasmic staining patterns for p53 and HBx proteins, suggesting an in vivo association (Fig. 2B).
Figure 3.
HBx via its distal C-terminal region sequesters p53 to the cytoplasm. (A) Normal human fibroblasts were microinjected with a p53 expression vector and either full-length HBx (CMV-1–154X) or a deletion mutant missing the last 44 amino acids (CMV-1–110X) followed by incubation for 24 h. Immunostaining was performed as described in Materials and Methods. (B) Confocal microscopic analysis of normal human fibroblasts coinjected with p53 and full-length HBx expression vectors. Yellow regions represent areas of colocalization. (Upper) Representative example of the degree of cytoplasmic sequestration typically observed in fibroblasts overexpressing p53 and HBx. (Lower) Fibroblast with all detectable p53 colocalizing with HBx in the cytoplasm.
Transcriptional Transactivation Activity of HBx Is Not Correlated with Abrogation of p53-Mediated Apoptosis.
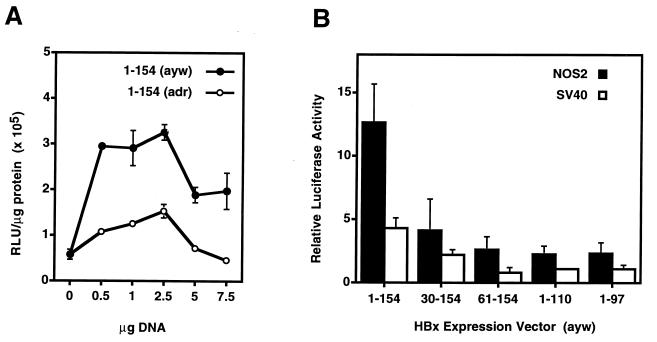
The adr and ayw subtypes of full-length HBx were compared regarding their ability to transcriptionally transactivate an SV40 promoter-driven luciferase reporter construct in human liver cells. Whereas the adr subtype more efficiently abrogated p53-mediated apoptosis (Table 1), the ayw subtype of HBx was a more potent transcriptional transactivator than the adr subtype over a wide range of DNA concentrations in HepG2 cells (P ≤ 0.003; Fig. 4A), as well as in two other human liver cell lines, Hep3B and AKN-1 (data not shown). Although full-length HBx transcriptionally transactivates both reporter constructs, all of the HBx deletion mutants exhibited significantly lower transactivational activity than full-length HBx (P ≤ 0.016, SV40; P ≤ 0.005, NOS2) (Fig. 4B). Particularly noteworthy are mutants 30–154 and 61–154, which despite their weak transcriptional transactivating potential, efficiently blocked p53-mediated apoptosis (Table 1).
Figure 4.
Ability of full-length HBx (A; adr versus ayw subtypes) and various HBx deletion mutants (B; ayw subtype) to transcriptionally transactivate SV40- and/or human NOS2 promoter-driven luciferase reporter constructs in HepG2 cells. Thirty-six to 48 hours after transfection, whole cell lysates were prepared, and resonance light units per μg protein were determined as described in Materials and Methods. (A) Representative data from a single experiment testing each sample in triplicate (Student’s t test; all data points, P ≤ 0.003). (B) Bar values represent the mean ± SD of resonance light units per μg protein relative to the CMV–neomycin control vector from three independent experiments (Student’s t test: all data points for SV40, P ≤ 0.016 and for NOS2, P ≤ 0.005).
DISCUSSION
Data are accumulating to indicate that HBx may contribute to hepatocarcinogenesis by binding to p53 and inhibiting several p53-mediated cellular processes critical for maintaining the genomic integrity of cells. In this study, in vitro binding data indicate that the distal 44 C-terminal amino acids of HBx are necessary for efficient binding to p53. Even though GST-p53 fusion protein behaves like wild-type p53 in terms of its sequence-specific DNA binding (data not shown), we cannot be certain whether the binding of full-length and truncated HBx protein to GST-p53 is the same as native p53. Complementing our in vitro binding data, however, are recent far-Western blotting results, indicating that p53 binds to HBx between amino acids 102 and 136 (51), as well as our finding in this study that p53 colocalizes in the cytoplasm of human fibroblasts with full-length HBx but not with an HBx deletion mutant missing the distal 44 amino acids.
Using a microinjection mapping strategy, we have demonstrated that in normal human fibroblasts, the same distal C-terminal region critical for binding GST-p53 in vitro also is essential for abrogating p53-mediated apoptosis. Recognizing that HBx functions by binding to other proteins, and that the complement of proteins is likely different in different cell types, we evaluated the effect of HBx expression on p53-induced apoptosis in primary human hepatocytes. Our data from two individual donors indicate that HBx also abrogates p53-mediated apoptosis in hepatocytes. We (unpublished data) and others (24) have demonstrated recently that prolonged overexpression of HBx induces apoptosis in normal human fibroblasts. Our data indicate that HBx mediates apoptosis at least in part via a p53-independent pathway and with different kinetics than p53-mediated apoptosis (unpublished data). Therefore, the ability of HBx to induce apoptosis does not necessarily exclude its proposed involvement in the abrogation of p53-mediated apoptosis.
The mechanism(s) for inhibition of p53-mediated apoptosis by HBx is likely a complex issue. Based on our findings in both normal human hepatocytes and fibroblasts, HBx may interfere with p53-mediated apoptosis by sequestering p53 in the cytoplasm. As lower amounts of nuclear p53 seem to favor G1 arrest, whereas higher levels induce apoptosis (25), partial cytoplasmic sequestration of p53 by HBx may sufficiently reduce the concentration of nuclear p53, resulting in the inhibition of apoptosis. Complementary to our model is a recent report describing high levels of cytoplasmic HBx and low levels of wild-type p53 in hepatocytes during chronic active hepatitis, a condition associated with an increased risk of HCC (26). Moreover, cytoplasmic sequestration of p53 by HBx has been reported in hepatocytes of HBx transgenic mice (27). The great majority of p53 staining in the livers of hepatocellular carcinoma patients is located in the nuclei of tumor cells (26, 28, 29). However, in many of these cases, accumulation of nuclear p53 correlates with a mutant p53 genotype, typically a late event in hepatocarcinogenesis (30, 31). Nuclear p53 could also be inactive, as recently demonstrated in teratocarcinoma cells (32). Thus, nuclear p53 in hepatocellular carcinomas does not necessarily exclude the possibility that HBx may sequester p53 in the cytoplasm early in the carcinogenic process.
In addition to cytoplasmic sequestration, HBx may abrogate p53-mediated apoptosis by influencing the transcriptional transactivation activity of p53. Consistent with this possibility are our previous data indicating that HBx reduces p53-mediated p21waf1 expression (21), inhibits p53 sequence-specific DNA binding (19), and blocks transcriptional transactivation by p53 of a reporter gene containing multiple p53-responsive elements (19). It is unlikely, however, that HBx abrogates p53-mediated apoptosis solely by inhibiting the transcriptional transactivation activity of p53, as p53-mediated apoptosis may not require the activation of downstream genes (33–36), and overexpression of a number of p53 downstream effectors, including p21waf1, BAX, FAS, GADD45, cyclin G, and IGF-BP3, does not induce apoptosis (refs. 21, 33, 37 and unpublished data). Alternatively, HBx may inhibit p53-dependent apoptosis by disrupting protein–protein interactions between p53 and other cellular factors in its apoptotic pathway (33, 38) or by directly interacting with proteins associated with DNA transcription and repair such as XPB and XPD (ref. 38 and unpublished data).
Numerous studies indicate that the transcriptional transactivation property of HBx contributes to hepatocarcinogenesis (reviewed in refs. 1 and 3). Although capable of binding single-stranded DNA (39), HBx seems to transcriptionally transactivate through protein–protein interactions with cellular transcriptional factors or effectors of signal transduction pathways (40–42). As abrogation of p53-mediated apoptosis by HBx is dependent on its C-terminal region, we tested whether this protective effect positively correlates with the transcriptional transactivation activity of HBx, which is also localized toward the C terminus (43, 44). Our observations that (i) N-terminal deletion mutants only weakly transactivated SV40- and NOS2 promoter-driven reporter constructs yet they efficiently blocked p53-mediated apoptosis and (ii) full-length HBx of the ayw viral subtype was consistently a stronger cotransactivator in liver cells, whereas the adr subtype more efficiently blocked p53-mediated apoptosis in fibroblasts, indicate that these functional properties of HBx are distinct. However, in the single case of hepatocytes in which both viral subtypes were tested, the ayw subtype more efficiently abrogated p53-mediated apoptosis. It is presently unclear why the adr subtype more efficiently blocks p53-mediated apoptosis in fibroblasts whereas the ayw subtype abrogates p53-mediated apoptosis better in hepatocytes. It is also unknown why hepatocytes from the two different donors exhibited different susceptibility to p53-mediated apoptosis. Two possible explanations are differences in the age of the donor or the differentiation state of the cells.
In this report, we demonstrate that the distal C-terminal region of HBx is critical for in vitro binding to GST-p53, sequestering p53 in the cytoplasm and abrogating p53-mediated apoptosis. Because the HBx gene is frequently integrated into the genome of HCC (2, 45), inhibition of p53-mediated apoptosis by HBx may provide a clonal selective advantage for hepatocytes expressing this integrated viral gene (21). In HBV-associated HCC, rearrangements or truncations of the N- and C-terminal domains of HBx have been reported (46–50). However, in the case of the C terminus, only small deletions (i.e., 10 amino acids) in the poorly conserved extreme distal region are affected presumably due to viral integration (48–52). Considering the multistage pathogenesis of HCC and the numerous biological properties of HBx, it is likely that this viral protein has additional oncogenic mechanisms. Data in the present study indicate that HBx may also contribute to hepatocarcinogenesis by transcriptionally transactivating NOS2, an enzyme that can produce consistent high levels of the putative endogenous mutagen, nitric oxide (53–55). This potentially novel oncogenic mechanism of HBx warrants further investigation, considering chronic HBV infection is a major risk factor associated with the development of HCC (reviewed in ref. 1), HBx is expressed and exhibits cotransactivation function in many HCCs (4, 45, 47), and NOS2 can be induced in human and rodent hepatocytes (56–58).
Acknowledgments
We thank B. Vogelstein for the CMV-driven wild-type p53 and neomycin expression vectors, J. Huibregtse for the GST-p53 vector, H. Thoenen for the CMV-driven construct, and J. Yuan for the β-gal expression vector. We are grateful to D. Dudek for the editorial assistance.
ABBREVIATIONS
- HBV
hepatitis B virus
- HBx
hepatitis B virus X protein
- HCC
hepatocellular carcinoma
- GST
glutathione S-transferase
- CMV
cytomegalovirus
- β-gal
β-galactosidase
- SV40
simian virus 40
- NOS2
inducible nitric oxide synthase
References
- 1.Robinson W S. Annu Rev Med. 1994;45:297–323. doi: 10.1146/annurev.med.45.1.297. [DOI] [PubMed] [Google Scholar]
- 2.Unsal H, Yakicier C, Marcais C, Kew M, Volkmann M, Zentgraf H, Isselbacher K J, Ozturk M. Proc Natl Acad Sci USA. 1994;91:822–826. doi: 10.1073/pnas.91.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caselmann W H. J Hepatol. 1995;22:34–37. [PubMed] [Google Scholar]
- 4.Paterlini P, Poussin K, Kew M, Franco D, Brechot C. Hepatology. 1995;21:313–321. [PubMed] [Google Scholar]
- 5.Shirakata Y, Kawada M, Fujiki Y, Sano H, Oda M, Yaginuma K, Kobayashi M, Koike K. Jpn J Cancer Res. 1989;80:617–621. doi: 10.1111/j.1349-7006.1989.tb01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Höhne M, Schaefer S, Seifer M, Feitelson M A, Paul D, Gerlich W H. EMBO J. 1990;9:1137–1145. doi: 10.1002/j.1460-2075.1990.tb08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim C M, Koike K, Saito I, Miyamura T, Jay G. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 8.Slagle B L, Lee T H, Medina D, Finegold M J, Butel J S. Mol Carcinogen. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 9.Levine A J, Momand J, Finlay C A. Nature (London) 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 10.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 11.Harris C C. J Natl Cancer Inst. 1996;88:1442–1455. doi: 10.1093/jnci/88.20.1442. [DOI] [PubMed] [Google Scholar]
- 12.Kinzler K W, Vogelstein B. N Engl J Med. 1994;331:49–50. doi: 10.1056/NEJM199407073310113. [DOI] [PubMed] [Google Scholar]
- 13.Hsu I C, Metcalf R A, Sun T, Welsh J A, Wang N J, Harris C C. Nature (London) 1991;350:427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- 14.Bressac B, Kew M, Wands J, Ozturk M. Nature (London) 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 15.Soini Y, Chia S C, Bennett W P, Groopman J D, Wang J S, DeBenedetti V M, Cawley H, Welsh J A, Hansen C, Bergasa N V, Jones E A, DiBisceglie A M, Trivers G E, Sandoval C A, Calderon I E, Munoz Espinosa L E, Harris C C. Carcinogenesis. 1996;17:1007–1012. doi: 10.1093/carcin/17.5.1007. [DOI] [PubMed] [Google Scholar]
- 16.Murakami Y, Hayashi K, Sekiya T. Cancer Res. 1991;51:3356–3361. [PubMed] [Google Scholar]
- 17.Hosono S, Chou M J, Lee C S, Shih C. Oncogene. 1993;8:491–496. [PubMed] [Google Scholar]
- 18.Feitelson M A, Zhu M, Duan L X, London W T. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 19.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nathan C, Xie Q W. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang X W, Gibson M K, Vermeulen W, Yeh H, Forrester K, Sturzbecher H W, Hoeijmakers J H J, Harris C C. Cancer Res. 1995;55:6012–6016. [PubMed] [Google Scholar]
- 22.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 23.Forrester K, Ambs S, Lupold S E, Kapust R B, Spillare E A, Weinberg W C, Felley-Bosco E, Wang X W, Geller D A, Billiar T R, Harris C C. Proc Natl Acad Sci USA. 1996;93:2442–2447. doi: 10.1073/pnas.93.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chirillo P, Pagano S, Natoli G, Puri P L, Burgio V L, Balsano C, Levrero M. Proc Natl Acad Sci USA. 1997;94:8162–8167. doi: 10.1073/pnas.94.15.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Ko L J, Jayaraman L, Prives C. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 26.Greenblatt M S, Feitelson M A, Zhu M, Bennett W P, Welsh J A, Jones R, Borkowski A, Harris C C. Cancer Res. 1997;57:426–432. [PubMed] [Google Scholar]
- 27.Ueda H, Ullrich S J, Gangemi J D, Kappel C A, Ngo L, Feitelson M A, Jay G. Nat Genet. 1995;9:41–47. doi: 10.1038/ng0195-41. [DOI] [PubMed] [Google Scholar]
- 28.Okuda T, Hirohashi K, Kinoshita H, Wakasa K, Sakurai M. World J Surg. 1996;20:215–220. doi: 10.1007/s002689900033. [DOI] [PubMed] [Google Scholar]
- 29.Schaff Z, Sarosi I, Hsia C C, Kiss A, Tabor E. Eur J Cancer. 1995;31A:1847–1850. doi: 10.1016/0959-8049(95)00380-2. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka S, Toh Y, Adachi E, Matsumata T, Mori R, Sugimachi K. Cancer Res. 1993;53:2884–2887. [PubMed] [Google Scholar]
- 31.Teramoto T, Satonaka K, Kitazawa S, Fujimori T, Hayashi K, Maeda S. Cancer Res. 1994;54:231–235. [PubMed] [Google Scholar]
- 32.Lutzker S G, Levine A J. Nat Med. 1996;2:804–810. doi: 10.1038/nm0796-804. [DOI] [PubMed] [Google Scholar]
- 33.Wang X W, Vermeulen W, Coursen J D, Gibson M, Lupold S E, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers J H J, Harris C C. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 34.Caelles C, Helmberg A, Karin M. Nature (London) 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 35.Haupt Y, Rowan S, Shaulian E, Vousden K H, Oren M. Genes Dev. 1995;9:2170–2183. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 36.Rowan S, Ludwig R L, Haupt Y, Bates S, Lu X, Oren M, Vousden K H. EMBO J. 1996;15:827–838. [PMC free article] [PubMed] [Google Scholar]
- 37.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 38.Qadri I, Conaway J W, Conaway R C, Schaack J, Siddiqui A. Proc Natl Acad Sci USA. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qadri I, Ferrari M E, Siddiqui A. J Biol Chem. 1996;271:15443–15450. doi: 10.1074/jbc.271.26.15443. [DOI] [PubMed] [Google Scholar]
- 40.Maguire H F, Hoeffler J P, Siddiqui A. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 41.Lucito R, Schneider R J. J Virol. 1992;66:983–991. doi: 10.1128/jvi.66.2.983-991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natoli G, Avantaggiati M L, Chirillo P, Costanzo A, Artini M, Balsano C, Levrero M. Mol Cell Biol. 1994;14:989–998. doi: 10.1128/mcb.14.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arii M, Takada S, Koike K. Oncogene. 1992;7:397–403. [PubMed] [Google Scholar]
- 44.Kumar V, Jayasuryan N, Kumar R. Proc Natl Acad Sci USA. 1996;93:5647–5652. doi: 10.1073/pnas.93.11.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsubara K, Tokino T. Mol Biol Med. 1990;7:243–260. [PubMed] [Google Scholar]
- 46.Feitelson M A. Lab Invest. 1994;71:324–349. [PubMed] [Google Scholar]
- 47.Wei Y, Etiemble J, Fourel G, Vitvitski-Trepo L, Buendia M A. J Med Virol. 1995;45:82–90. doi: 10.1002/jmv.1890450116. [DOI] [PubMed] [Google Scholar]
- 48.Takada S, Koike K. Proc Natl Acad Sci USA. 1990;87:5628–5632. doi: 10.1073/pnas.87.15.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wollersheim M, Debelka U, Hofschneider P H. Oncogene. 1988;3:545–552. [PubMed] [Google Scholar]
- 50.Yu M W, Chen C J. Crit Rev Oncol Hematol. 1994;17:71–91. doi: 10.1016/1040-8428(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 51.Lin Y, Nomura T, Yamashita T, Dorjsuren D, Tang H, Murakami S. Cancer Res. 1997;57:5137–5142. [PubMed] [Google Scholar]
- 52.Neuland C Y, Blattner W A, Mann D L, Fraser M C, Tsai S, Strong D M. J Natl Cancer Inst. 1983;71:1143–1150. [PubMed] [Google Scholar]
- 53.Wink D A, Kasprzak K S, Maragos C M, Elespuru R K, Misra M, Dunams T M, Cebula T A, Koch W H, Andrews A W, Allen J S, Keefer L K. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 54.Nguyen T, Brunson D, Crespi C L, Penman B W, Wishnok J S, Tannenbaum S R. Proc Natl Acad Sci USA. 1992;89:3030–3034. doi: 10.1073/pnas.89.7.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.deRojas-Walker T, Tamir S, Ji H, Wishnok J S, Tannenbaum S R. Chem Res Toxicol. 1995;8:473–477. doi: 10.1021/tx00045a020. [DOI] [PubMed] [Google Scholar]
- 56.Nussler A K, Di Silvio M, Billiar T R, Hoffman R A, Geller D A, Selby R, Madariaga J, Simmons R L. J Exp Med. 1992;176:261–264. doi: 10.1084/jem.176.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geller D A, Di Silvio M, Nussler A K, Wang S C, Shapiro R A, Simmons R L, Billiar T R. J Surg Res. 1993;55:427–432. doi: 10.1006/jsre.1993.1164. [DOI] [PubMed] [Google Scholar]
- 58.Liu R H, Jacob J R, Hotchkiss J H, Cote P J, Gerin J L, Tennant B C. Carcinogenesis. 1994;15:2875–2877. doi: 10.1093/carcin/15.12.2875. [DOI] [PubMed] [Google Scholar]