Abstract
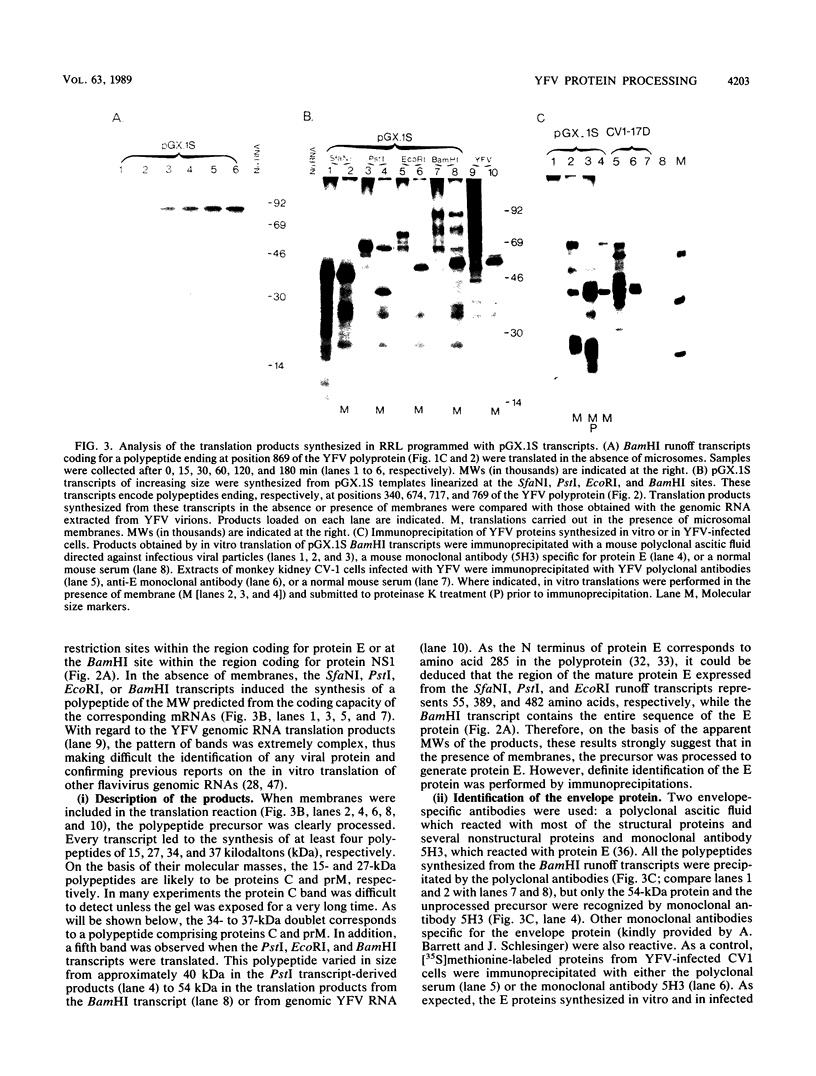
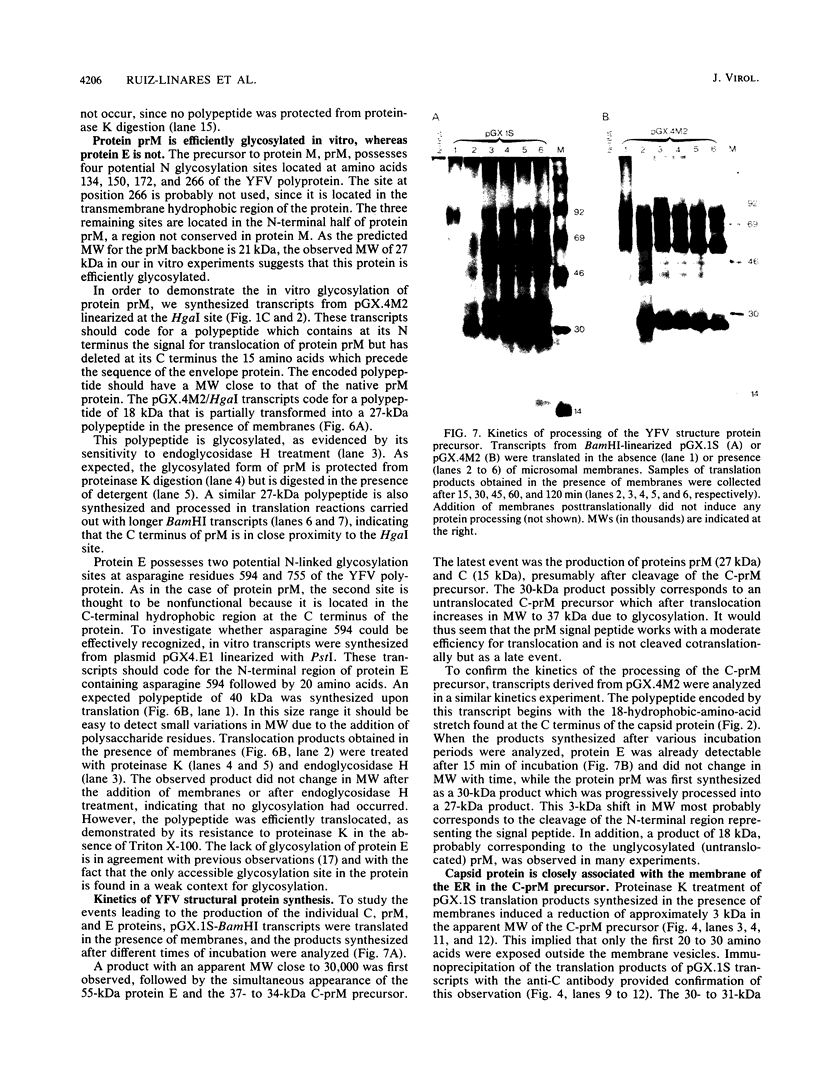
The yellow fever virus (YFV) cDNA segment coding for the part of the precursor polyprotein generating the structural proteins C (capsid), prM (precursor to the membrane protein M), and E (envelope) was expressed in vitro by using the T7 promoter-polymerase transcription system coupled to translation in rabbit reticulocyte lysates. A polypeptide of the expected molecular weight was observed to accumulate in the assay and was processed into proteins C, prM, and E only when dog pancreas microsomal membranes were added to the translation system. Proteins prM and E were translocated inside the endoplasmic reticulum, where prM underwent glycosylation. Regions essential for translocation of these proteins were localized to the 18- and 15-amino-acid C-terminal hydrophobic regions of proteins C and prM, respectively. Translocation of protein prM appeared to be less efficient than that of protein E. Maturation of these proteins followed different kinetics, indicating that the prM signal is probably cleaved off more slowly. A polypeptide composed of proteins C and prM, similar to the NVx polypeptide described in yellow fever virus-infected cells, was also produced in the in vitro system in the presence of membranes. No mature protein M was detected, suggesting that the cleavage of prM to M is a late processing event mediated by a protease different from endoplasmic reticulum signalases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell J. R., Kinney R. M., Trent D. W., Lenches E. M., Dalgarno L., Strauss J. H. Amino-terminal amino acid sequences of structural proteins of three flaviviruses. Virology. 1985 May;143(1):224–229. doi: 10.1016/0042-6822(85)90110-2. [DOI] [PubMed] [Google Scholar]
- Biedrzycka A., Cauchi M. R., Bartholomeusz A., Gorman J. J., Wright P. J. Characterization of protease cleavage sites involved in the formation of the envelope glycoprotein and three non-structural proteins of dengue virus type 2, New Guinea C strain. J Gen Virol. 1987 May;68(Pt 5):1317–1326. doi: 10.1099/0022-1317-68-5-1317. [DOI] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boege U., Heinz F. X., Wengler G., Kunz C. Amino acid compositions and amino-terminal sequences of the structural proteins of a flavivirus, European Tick-Borne Encephalitis virus. Virology. 1983 Apr 30;126(2):651–657. doi: 10.1016/s0042-6822(83)80020-8. [DOI] [PubMed] [Google Scholar]
- Cardiff R. D., Lund J. K. Distribution of dengue-2 antigens by electron immunocytochemistry. Infect Immun. 1976 Jun;13(6):1699–1709. doi: 10.1128/iai.13.6.1699-1709.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle E., Leidner U., Nowak T., Wengler G., Wengler G. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology. 1986 Feb;149(1):10–26. doi: 10.1016/0042-6822(86)90082-6. [DOI] [PubMed] [Google Scholar]
- Castle E., Nowak T., Leidner U., Wengler G., Wengler G. Sequence analysis of the viral core protein and the membrane-associated proteins V1 and NV2 of the flavivirus West Nile virus and of the genome sequence for these proteins. Virology. 1985 Sep;145(2):227–236. doi: 10.1016/0042-6822(85)90156-4. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Cleaves G. R. Identification of dengue type 2 virus-specific high molecular weight proteins in virus-infected BHK cells. J Gen Virol. 1985 Dec;66(Pt 12):2767–2771. doi: 10.1099/0022-1317-66-12-2767. [DOI] [PubMed] [Google Scholar]
- Coia G., Parker M. D., Speight G., Byrne M. E., Westaway E. G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988 Jan;69(Pt 1):1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- Crawford G. R., Wright P. J. Characterization of novel viral polyproteins detected in cells infected by the flavivirus Kunjin and radiolabelled in the presence of the leucine analogue hydroxyleucine. J Gen Virol. 1987 Feb;68(Pt 2):365–376. doi: 10.1099/0022-1317-68-2-365. [DOI] [PubMed] [Google Scholar]
- Desprès P., Cahour A., Dupuy A., Deubel V., Bouloy M., Digoutte J. P., Girard M. High genetic stability of the region coding for the structural proteins of yellow fever virus strain 17D. J Gen Virol. 1987 Aug;68(Pt 8):2245–2247. doi: 10.1099/0022-1317-68-8-2245. [DOI] [PubMed] [Google Scholar]
- Desprès P., Cahour A., Wychowski C., Girard M., Bouloy M. Expression of the yellow fever virus envelope protein using hybrid SV40/yellow fever viruses. Ann Inst Pasteur Virol. 1988 Jan-Mar;139(1):59–67. doi: 10.1016/s0769-2617(88)80006-6. [DOI] [PubMed] [Google Scholar]
- Deubel V., Kinney R. M., Trent D. W. Nucleotide sequence and deduced amino acid sequence of the nonstructural proteins of dengue type 2 virus, Jamaica genotype: comparative analysis of the full-length genome. Virology. 1988 Jul;165(1):234–244. doi: 10.1016/0042-6822(88)90677-0. [DOI] [PubMed] [Google Scholar]
- Deubel V., Kinney R. M., Trent D. W. Nucleotide sequence and deduced amino acid sequence of the structural proteins of dengue type 2 virus, Jamaica genotype. Virology. 1986 Dec;155(2):365–377. doi: 10.1016/0042-6822(86)90200-x. [DOI] [PubMed] [Google Scholar]
- Deubel V., Schlesinger J. J., Digoutte J. P., Girard M. Comparative immunochemical and biological analysis of African and South American yellow fever viruses. Arch Virol. 1987;94(3-4):331–338. doi: 10.1007/BF01310727. [DOI] [PubMed] [Google Scholar]
- Friedlander M., Blobel G. Bovine opsin has more than one signal sequence. 1985 Nov 28-Dec 4Nature. 318(6044):338–343. doi: 10.1038/318338a0. [DOI] [PubMed] [Google Scholar]
- Garoff H. Using recombinant DNA techniques to study protein targeting in the eucaryotic cell. Annu Rev Cell Biol. 1985;1:403–445. doi: 10.1146/annurev.cb.01.110185.002155. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lyapustin V. N., Svitkin YuV, Ugarova TYu, Lashkevich V. A., Agol V. I. A tentative model of formation of structural proteins of tick-borne encephalitis virus (flavivirus). FEBS Lett. 1986 May 12;200(2):314–316. doi: 10.1016/0014-5793(86)81159-0. [DOI] [PubMed] [Google Scholar]
- Mandl C. W., Guirakhoo F., Holzmann H., Heinz F. X., Kunz C. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J Virol. 1989 Feb;63(2):564–571. doi: 10.1128/jvi.63.2.564-571.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl C. W., Heinz F. X., Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology. 1988 Sep;166(1):197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Melancon P., Garoff H. Reinitiation of translocation in the Semliki Forest virus structural polyprotein: identification of the signal for the E1 glycoprotein. EMBO J. 1986 Jul;5(7):1551–1560. doi: 10.1002/j.1460-2075.1986.tb04396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monckton R. P., Westaway E. G. Restricted translation of the genome of the flavivirus Kunjin in vitro. J Gen Virol. 1982 Nov;63(Pt 1):227–232. doi: 10.1099/0022-1317-63-1-227. [DOI] [PubMed] [Google Scholar]
- Mueckler M., Lodish H. F. The human glucose transporter can insert posttranslationally into microsomes. Cell. 1986 Feb 28;44(4):629–637. doi: 10.1016/0092-8674(86)90272-2. [DOI] [PubMed] [Google Scholar]
- Ozden S., Poirier B. Dengue virus induced polypeptide synthesis. Brief report. Arch Virol. 1985;85(1-2):129–137. doi: 10.1007/BF01317012. [DOI] [PubMed] [Google Scholar]
- Perara E., Lingappa V. R. A former amino terminal signal sequence engineered to an internal location directs translocation of both flanking protein domains. J Cell Biol. 1985 Dec;101(6):2292–2301. doi: 10.1083/jcb.101.6.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C. M., Aebersold R., Teplow D. B., Pata J., Bell J. R., Vorndam A. V., Trent D. W., Brandriss M. W., Schlesinger J. J., Strauss J. H. Partial N-terminal amino acid sequences of three nonstructural proteins of two flaviviruses. Virology. 1986 May;151(1):1–9. doi: 10.1016/0042-6822(86)90098-x. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Lenches E. M., Eddy S. R., Shin S. J., Sheets R. L., Strauss J. H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985 Aug 23;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Ruiz-Linares A., Bouloy M., Girard M., Cahour A. Modulations of the in vitro translational efficiencies of Yellow Fever virus mRNAs: interactions between coding and noncoding regions. Nucleic Acids Res. 1989 Apr 11;17(7):2463–2476. doi: 10.1093/nar/17.7.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger J. J., Brandriss M. W., Monath T. P. Monoclonal antibodies distinguish between wild and vaccine strains of yellow fever virus by neutralization, hemagglutination inhibition, and immune precipitation of the virus envelope protein. Virology. 1983 Feb;125(1):8–17. doi: 10.1016/0042-6822(83)90059-4. [DOI] [PubMed] [Google Scholar]
- Speight G., Coia G., Parker M. D., Westaway E. G. Gene mapping and positive identification of the non-structural proteins NS2A, NS2B, NS3, NS4B and NS5 of the flavivirus Kunjin and their cleavage sites. J Gen Virol. 1988 Jan;69(Pt 1):23–34. doi: 10.1099/0022-1317-69-1-23. [DOI] [PubMed] [Google Scholar]
- Spiess M., Lodish H. F. An internal signal sequence: the asialoglycoprotein receptor membrane anchor. Cell. 1986 Jan 17;44(1):177–185. doi: 10.1016/0092-8674(86)90496-4. [DOI] [PubMed] [Google Scholar]
- Stohlman S. A., Wisseman C. L., Jr, Eylar O. R., Silverman D. J. Dengue virus-induced modifications of host cell membranes. J Virol. 1975 Oct;16(4):1017–1026. doi: 10.1128/jvi.16.4.1017-1026.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y. V., Lyapustin V. N., Lashkevich V. A., Agol V. I. Differences between translation products of tick-borne encephalitis virus RNA in cell-free systems from Krebs-2 cells and rabbit reticulocytes: involvement of membranes in the processing of nascent precursors of flavivirus structural proteins. Virology. 1984 Jun;135(2):536–541. doi: 10.1016/0042-6822(84)90207-1. [DOI] [PubMed] [Google Scholar]
- Svitkin Y. V., Ugarova T. Y., Chernovskaya T. V., Lyapustin V. N., Lashkevich V. A., Agol V. I. Translation of tick-borne encephalitis virus (flavivirus) genome in vitro: synthesis of two structural polypeptides. Virology. 1981 Apr 15;110(1):26–34. doi: 10.1016/0042-6822(81)90004-0. [DOI] [PubMed] [Google Scholar]
- Walter P., Lingappa V. R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Wengler G., Beato M., Wengler G. In vitro translation of 42 S virus-specific RNA from cells infected with the flavivirus West Nile virus. Virology. 1979 Jul 30;96(2):516–529. doi: 10.1016/0042-6822(79)90108-9. [DOI] [PubMed] [Google Scholar]
- Westaway E. G., Brinton M. A., Gaidamovich SYa, Horzinek M. C., Igarashi A., Käriäinen L., Lvov D. K., Porterfield J. S., Russell P. K., Trent D. W. Flaviviridae. Intervirology. 1985;24(4):183–192. doi: 10.1159/000149642. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]