Abstract
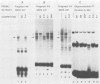
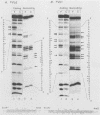
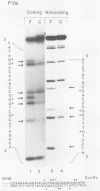
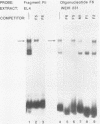
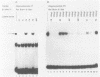
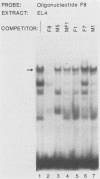
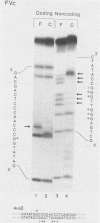
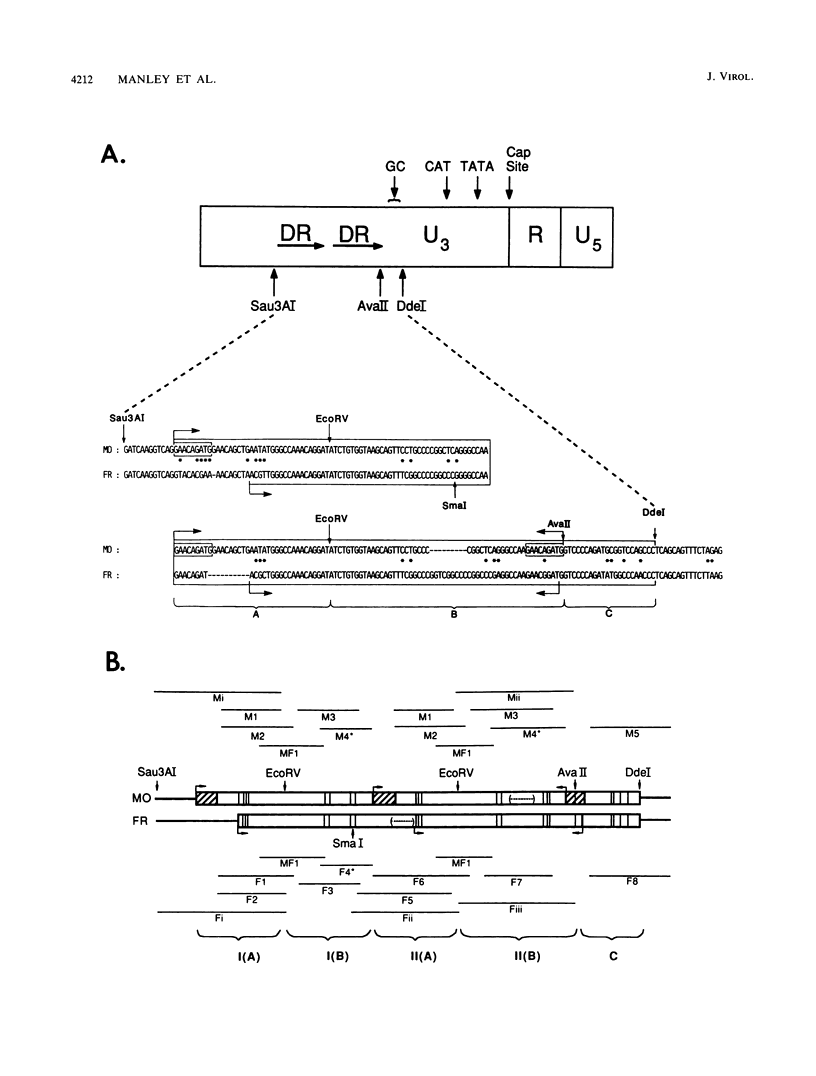
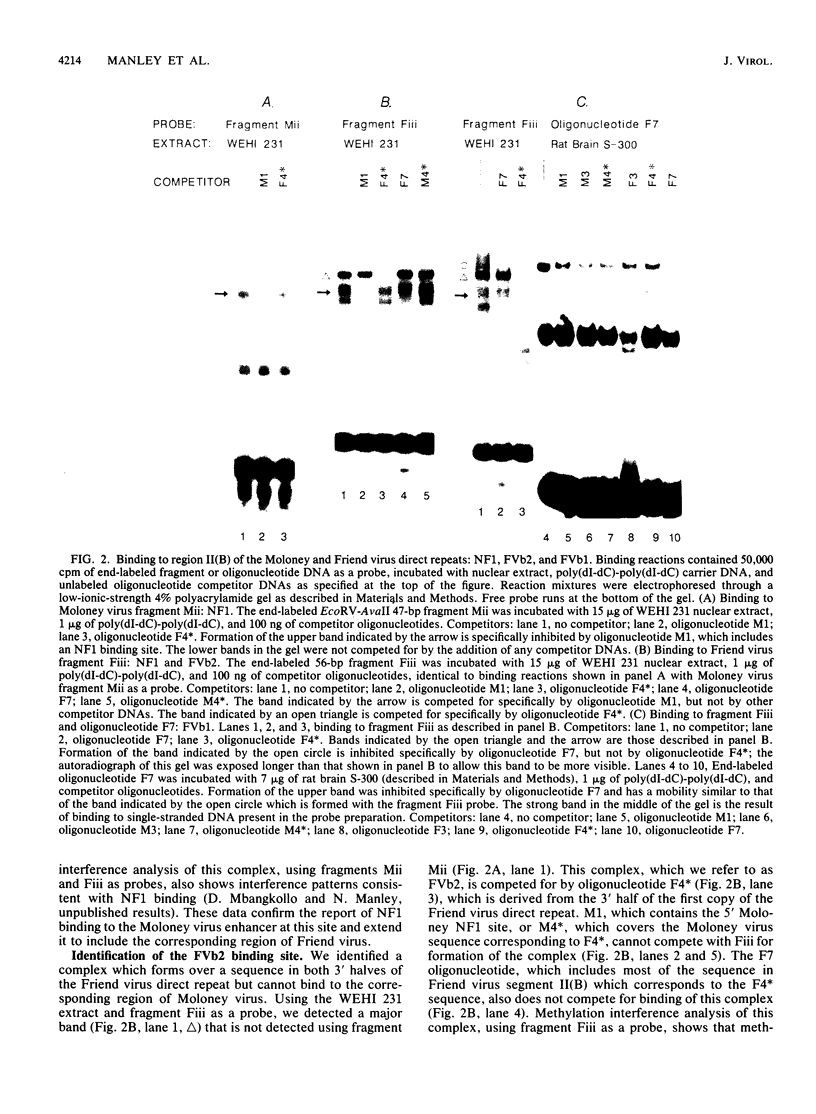
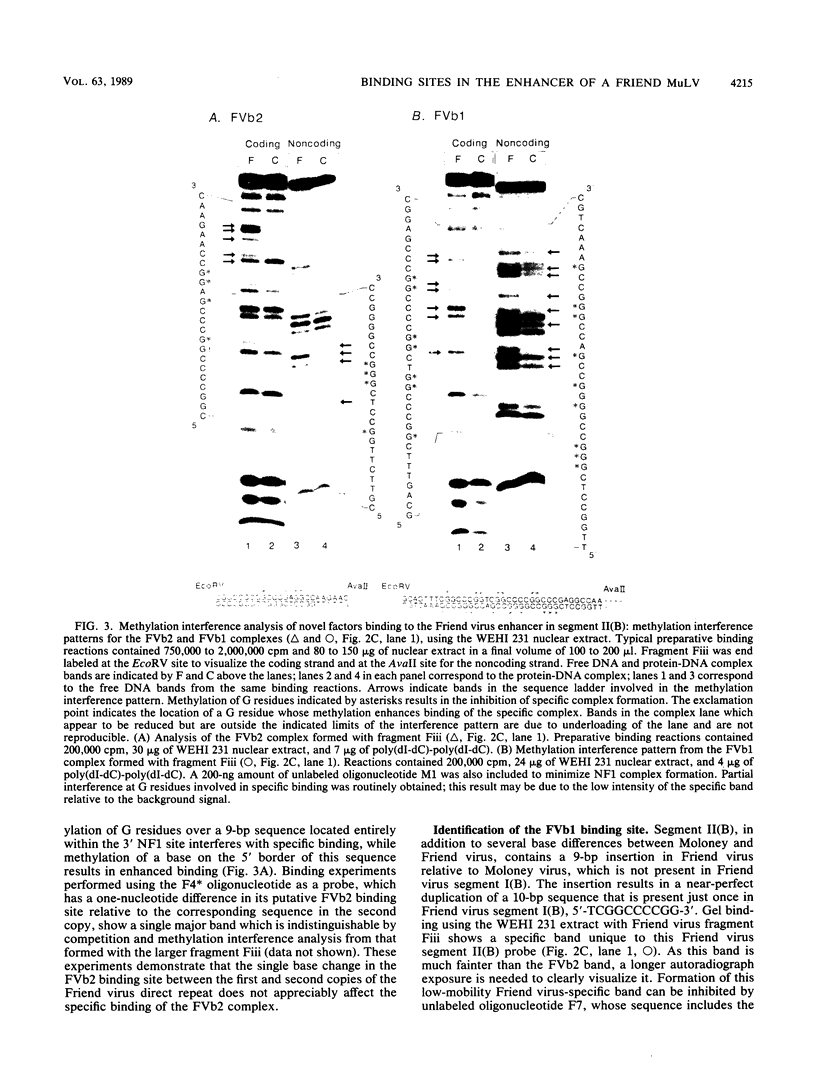
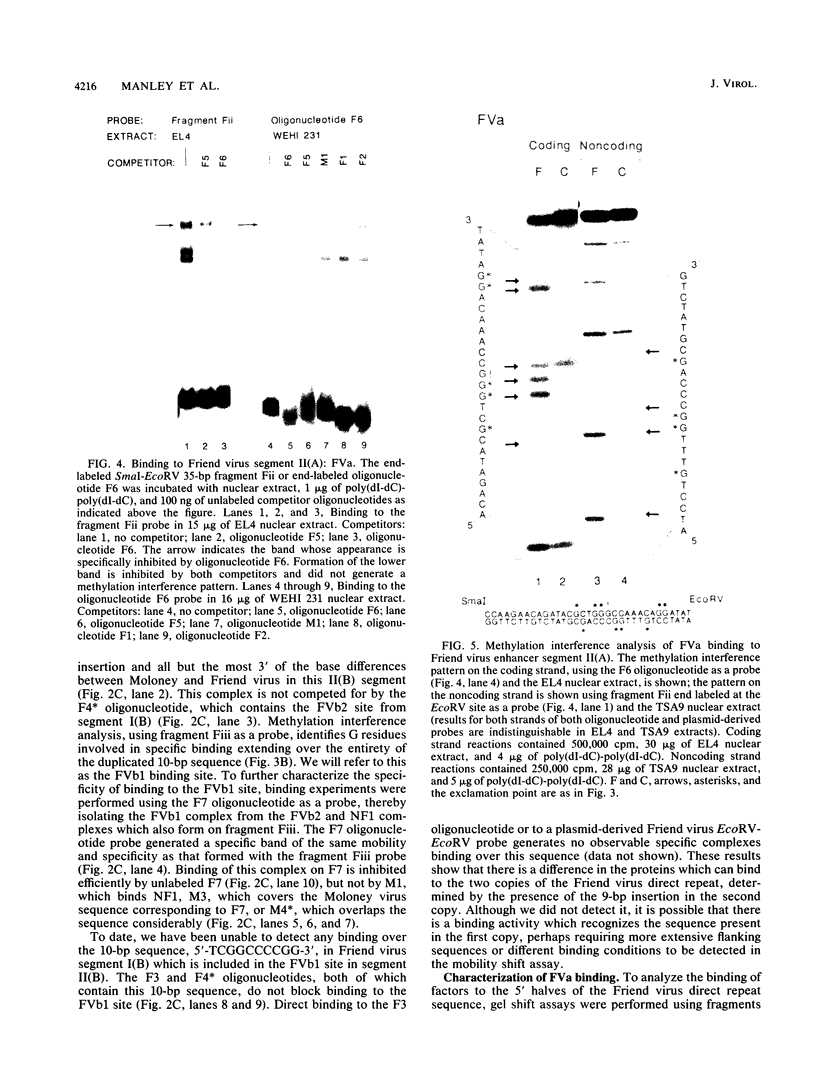
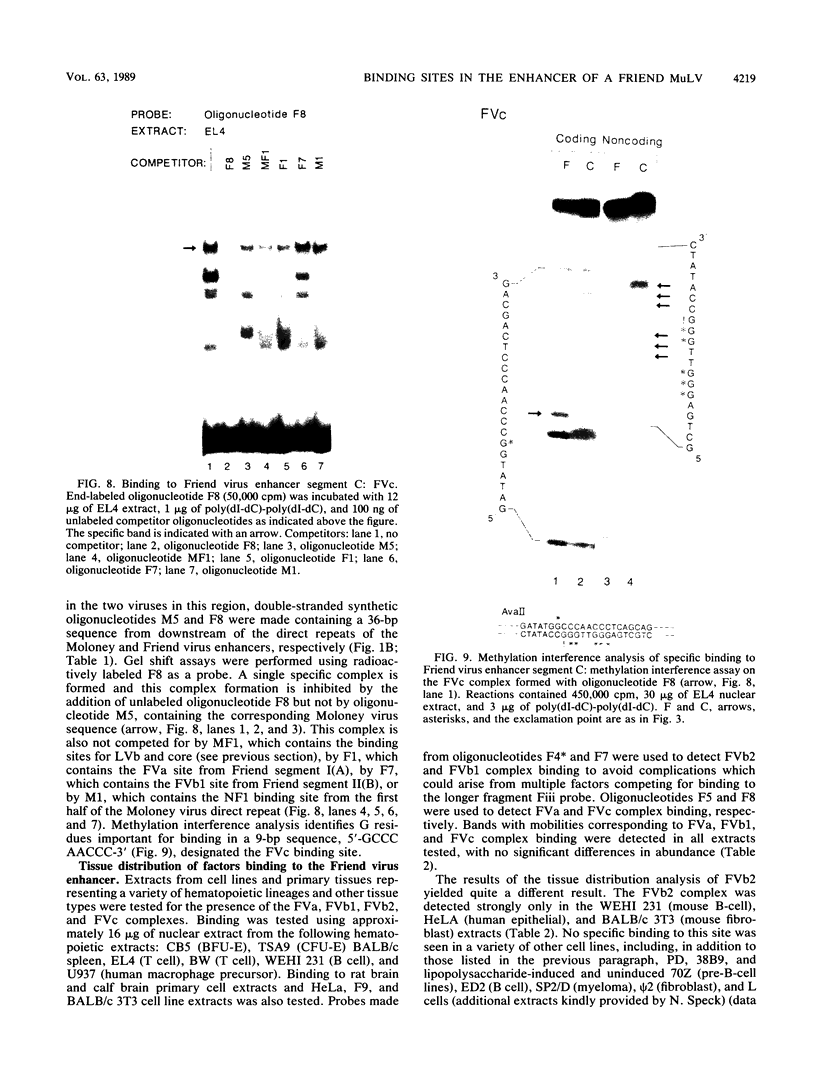
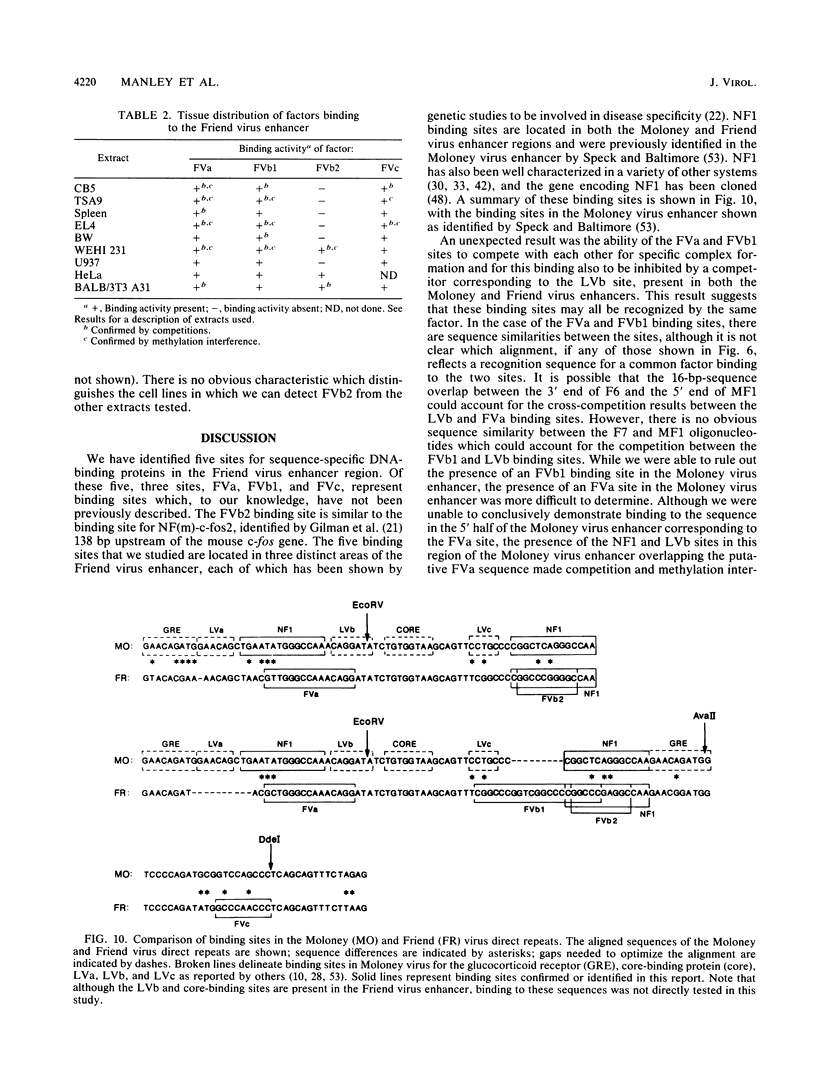
Nondefective Friend murine leukemia virus (MuLV) causes erythroleukemia when injected into newborn NFS mice, while Moloney MuLV causes T-cell lymphoma. Exchange of the Friend virus enhancer region, a sequence of about 180 nucleotides including the direct repeat and a short 3'-adjacent segment, for the corresponding region in Moloney MuLV confers the ability to cause erythroid disease on Moloney MuLV. We have used the electrophoretic mobility shift assay and methylation interference analysis to identify cellular factors which bind to the Friend virus enhancer region and compared these with factors, previously identified, that bind to the Moloney virus direct repeat (N. A. Speck and D. Baltimore, Mol. Cell. Biol. 7:1101-1110, 1987). We identified five binding sites for sequence-specific DNA-binding proteins in the Friend virus enhancer region. While some binding sites are present in both the Moloney and Friend virus enhancers, both viruses contain unique sites not present in the other. Although none of the factors identified in this report which bind to these unique sites are present exclusively in T cells or erythroid cells, they bind to three regions of the enhancer shown by genetic analysis to encode disease specificity and thus are candidates to mediate the tissue-specific expression and distinct disease specificities encoded by these virus enhancer elements.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baumruker T., Sturm R., Herr W. OBP100 binds remarkably degenerate octamer motifs through specific interactions with flanking sequences. Genes Dev. 1988 Nov;2(11):1400–1413. doi: 10.1101/gad.2.11.1400. [DOI] [PubMed] [Google Scholar]
- Bender A., Sprague G. F., Jr MAT alpha 1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell. 1987 Aug 28;50(5):681–691. doi: 10.1016/0092-8674(87)90326-6. [DOI] [PubMed] [Google Scholar]
- Bösze Z., Thiesen H. J., Charnay P. A transcriptional enhancer with specificity for erythroid cells is located in the long terminal repeat of the Friend murine leukemia virus. EMBO J. 1986 Jul;5(7):1615–1623. doi: 10.1002/j.1460-2075.1986.tb04404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Baldwin A. S., Carthew R. W., Sharp P. A. Human CCAAT-binding proteins have heterologous subunits. Cell. 1988 Apr 8;53(1):11–24. doi: 10.1016/0092-8674(88)90483-7. [DOI] [PubMed] [Google Scholar]
- Clark S. P., Kaufhold R., Chan A., Mak T. W. Comparison of the transcriptional properties of the Friend and Moloney retrovirus long terminal repeats: importance of tandem duplications and of the core enhancer sequence. Virology. 1985 Jul 30;144(2):481–494. doi: 10.1016/0042-6822(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Davidson I., Xiao J. H., Rosales R., Staub A., Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988 Sep 23;54(7):931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Yamamoto K. R. Two different factors act separately or together to specify functionally distinct activities at a single transcriptional enhancer. Mol Cell Biol. 1986 Apr;6(4):993–1001. doi: 10.1128/mcb.6.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984 Nov;52(2):448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan C., Theis J., O'Farrell P. H. The sequence specificity of homeodomain-DNA interaction. Cell. 1988 Sep 23;54(7):1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. H., Morrey J. D. Tissue-specific replication of Friend and Moloney murine leukemia viruses in infected mice. J Virol. 1987 May;61(5):1350–1357. doi: 10.1128/jvi.61.5.1350-1357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. M. The steroid and thyroid hormone receptor superfamily. Science. 1988 May 13;240(4854):889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Mittal S., Chute H., Chao E., Pattengale P. K. Rearrangements and insertions in the Moloney murine leukemia virus long terminal repeat alter biological properties in vivo and in vitro. J Virol. 1986 Oct;60(1):204–214. doi: 10.1128/jvi.60.1.204-214.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Simian virus 40 early- and late-region promoter functions are enhanced by the 72-base-pair repeat inserted at distant locations and inverted orientations. Mol Cell Biol. 1983 Jun;3(6):991–999. doi: 10.1128/mcb.3.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Wilson R. N., Weinberg R. A. Multiple protein-binding sites in the 5'-flanking region regulate c-fos expression. Mol Cell Biol. 1986 Dec;6(12):4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E., Li Y., Fredrickson T. N., Hartley J. W., Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989 Jan;63(1):328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C., Johnson A. D. a1 protein alters the DNA binding specificity of alpha 2 repressor. Cell. 1988 Mar 25;52(6):875–882. doi: 10.1016/0092-8674(88)90429-1. [DOI] [PubMed] [Google Scholar]
- Holland C. A., Anklesaria P., Sakakeeny M. A., Greenberger J. S. Enhancer sequences of a retroviral vector determine expression of a gene in multipotent hematopoietic progenitors and committed erythroid cells. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8662–8666. doi: 10.1073/pnas.84.23.8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. At least four viral genes contribute to the leukemogenicity of murine retrovirus MCF 247 in AKR mice. J Virol. 1985 Jan;53(1):158–165. doi: 10.1128/jvi.53.1.158-165.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Wozney J., Chatis P. A., Hopkins N., Hartley J. W. Construction of recombinants between molecular clones of murine retrovirus MCF 247 and Akv: determinant of an in vitro host range property that maps in the long terminal repeat. J Virol. 1985 Jan;53(1):152–157. doi: 10.1128/jvi.53.1.152-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Jolicoeur P., Nicolaiew N., DesGroseillers L., Rassart E. Molecular cloning of infectious viral DNA from ecotropic neurotropic wild mouse retrovirus. J Virol. 1983 Mar;45(3):1159–1163. doi: 10.1128/jvi.45.3.1159-1163.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Rosenfeld P. J., Kelly T. J., Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987 Jan 16;48(1):79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater P. A., van der Vliet P. C., Rupp R. A., Nowock J., Sippel A. E. Functional homology between the sequence-specific DNA-binding proteins nuclear factor I from HeLa cells and the TGGCA protein from chicken liver. EMBO J. 1986 Feb;5(2):381–386. doi: 10.1002/j.1460-2075.1986.tb04223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Levinson B., Khoury G., Vande Woude G., Gruss P. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature. 1982 Feb 18;295(5850):568–572. doi: 10.1038/295568a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoSardo J. E., Cupelli L. A., Short M. K., Berman J. W., Lenz J. Differences in activities of murine retroviral long terminal repeats in cytotoxic T lymphocytes and T-lymphoma cells. J Virol. 1989 Mar;63(3):1087–1094. doi: 10.1128/jvi.63.3.1087-1094.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung M. L., Hartley J. W., Rowe W. P., Hopkins N. H. Large RNase T1-resistant oligonucleotides encoding p15E and the U3 region of the long terminal repeat distinguish two biological classes of mink cell focus-forming type C viruses of inbred mice. J Virol. 1983 Jan;45(1):275–290. doi: 10.1128/jvi.45.1.275-290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Pargellis C. A., Hasan N. M., Bushman E. W., Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988 Sep 23;54(7):923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowock J., Borgmeyer U., Püschel A. W., Rupp R. A., Sippel A. E. The TGGCA protein binds to the MMTV-LTR, the adenovirus origin of replication, and the BK virus enhancer. Nucleic Acids Res. 1985 Mar 25;13(6):2045–2061. doi: 10.1093/nar/13.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., McKinney M. D., Agranovsky O. Contribution of the gag and pol sequences to the leukemogenicity of Friend murine leukemia virus. J Virol. 1985 Jun;54(3):864–868. doi: 10.1128/jvi.54.3.864-868.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Ruscetti S. A 2.4-kilobase-pair fragment of the Friend murine leukemia virus genome contains the sequences responsible for friend murine leukemia virus-induced erythroleukemia. J Virol. 1983 Jun;46(3):718–725. doi: 10.1128/jvi.46.3.718-725.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Signorelli K., Collins L. The envelope gene and long terminal repeat sequences contribute to the pathogenic phenotype of helper-independent Friend viruses. J Virol. 1984 Sep;51(3):788–794. doi: 10.1128/jvi.51.3.788-794.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Haseltine W. A., Lenz J., Ruprecht R., Cloyd M. W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985 Sep;55(3):862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C., Mermod N., Andrews P. C., Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988 Jul 21;334(6179):218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Shibuya T., Mak T. W. Isolation and induction of erythroleukemic cell lines with properties of erythroid progenitor burst-forming cell (BFU-E) and erythroid precursor cell (CFU-E). Proc Natl Acad Sci U S A. 1983 Jun;80(12):3721–3725. doi: 10.1073/pnas.80.12.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Short M. K., Okenquist S. A., Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987 Apr;61(4):1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon M., Sola B., Evans L., Nishio J., Hayes S. F., Nathanson K., Garon C. F., Chesebro B. Hemolytic anemia and erythroleukemia, two distinct pathogenic effects of Friend MuLV: mapping of the effects to different regions of the viral genome. Cell. 1986 Dec 26;47(6):851–859. doi: 10.1016/0092-8674(86)90800-7. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen H. J., Bösze Z., Henry L., Charnay P. A DNA element responsible for the different tissue specificities of Friend and Moloney retroviral enhancers. J Virol. 1988 Feb;62(2):614–618. doi: 10.1128/jvi.62.2.614-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M. Virus cloned from the Rauscher virus complex induces erythroblastosis and thymic lymphoma. Virology. 1982 Apr 15;118(1):225–228. doi: 10.1016/0042-6822(82)90336-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 1985;19:209–252. doi: 10.1146/annurev.ge.19.120185.001233. [DOI] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Ahe D., Janich S., Scheidereit C., Renkawitz R., Schütz G., Beato M. Glucocorticoid and progesterone receptors bind to the same sites in two hormonally regulated promoters. Nature. 1985 Feb 21;313(6004):706–709. doi: 10.1038/313706a0. [DOI] [PubMed] [Google Scholar]