Abstract
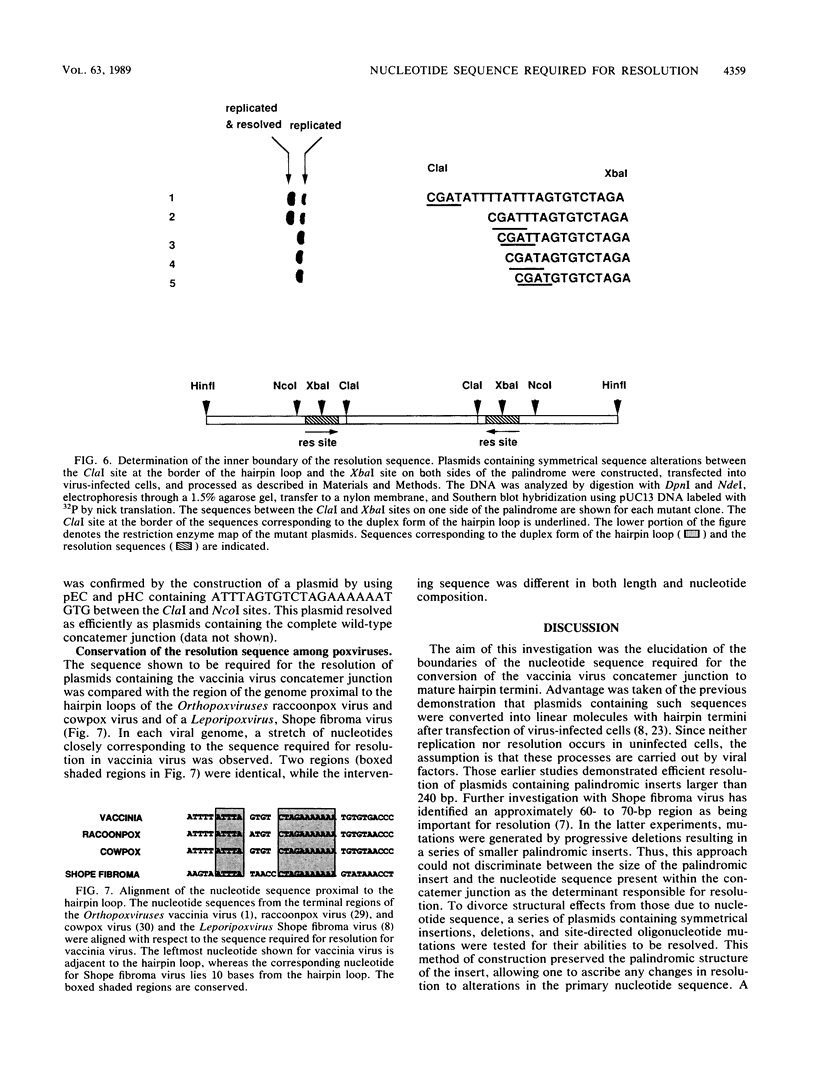
The mature form of the vaccinia virus genome consists of a linear, 185,000-base-pair (bp) DNA molecule with a 10,000-bp inverted terminal repetition and incompletely base-paired 104-nucleotide hairpin loops connecting the two strands at each end. In concatemeric forms of intracellular vaccinia virus DNA, the inverted terminal repetitions of adjacent genomes form an imperfect palindrome. The apex of this palindrome corresponds in sequence to the double-stranded form of the hairpin loop. Circular plasmids containing palindromic concatemer junction fragments of 250 bp or longer are converted into linear minichromosomes with hairpin ends when they are transfected into vaccinia virus-infected cells, providing a model system with which to study the resolution process. To distinguish between sequence-specific and structural requirements for resolution, plasmids with symmetrical insertions, deletions, and oligonucleotide-directed mutations within the concatemer junction were constructed. A sequence (ATTTAGTGTCTAGAAAAAAA) located on both sides of the apex segment was found to be critical for resolution. Resolution was more efficient when additional nucleotides, TGTG, followed the run of A residues. Both the location and sequence of the proposed resolution signal are highly conserved among poxviruses.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baroudy B. M., Venkatesan S., Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982 Feb;28(2):315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- Baroudy B. M., Venkatesan S., Moss B. Structure and replication of vaccinia virus telomeres. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):723–729. doi: 10.1101/sqb.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. I., Binns M. M., Tomley F. M., Boursnell M. E. Tandem repeated sequences within the terminal region of the fowlpox virus genome. J Gen Virol. 1989 Jan;70(Pt 1):145–154. doi: 10.1099/0022-1317-70-1-145. [DOI] [PubMed] [Google Scholar]
- Craig N. L. The mechanism of conservative site-specific recombination. Annu Rev Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- DeLange A. M., McFadden G. Efficient resolution of replicated poxvirus telomeres to native hairpin structures requires two inverted symmetrical copies of a core target DNA sequence. J Virol. 1987 Jun;61(6):1957–1963. doi: 10.1128/jvi.61.6.1957-1963.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange A. M., McFadden G. Sequence-nonspecific replication of transfected plasmid DNA in poxvirus-infected cells. Proc Natl Acad Sci U S A. 1986 Feb;83(3):614–618. doi: 10.1073/pnas.83.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange A. M., Reddy M., Scraba D., Upton C., McFadden G. Replication and resolution of cloned poxvirus telomeres in vivo generates linear minichromosomes with intact viral hairpin termini. J Virol. 1986 Aug;59(2):249–259. doi: 10.1128/jvi.59.2.249-259.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie P., McFadden G., Morgan A. R. The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J Biol Chem. 1987 Oct 25;262(30):14826–14836. [PubMed] [Google Scholar]
- Dickie P., Morgan A. R., McFadden G. Cruciform extrusion in plasmids bearing the replicative intermediate configuration of a poxvirus telomere. J Mol Biol. 1987 Aug 5;196(3):541–558. doi: 10.1016/0022-2836(87)90031-3. [DOI] [PubMed] [Google Scholar]
- Geshelin P., Berns K. I. Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J Mol Biol. 1974 Oct 5;88(4):785–796. doi: 10.1016/0022-2836(74)90399-4. [DOI] [PubMed] [Google Scholar]
- González A., Talavera A., Almendral J. M., Viñuela E. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 1986 Sep 11;14(17):6835–6844. doi: 10.1093/nar/14.17.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Lakritz N., Foglesong P. D., Reddy M., Baum S., Hurwitz J., Bauer W. R. A vaccinia virus DNase preparation which cross-links superhelical DNA. J Virol. 1985 Mar;53(3):935–943. doi: 10.1128/jvi.53.3.935-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M., Garon C. F., Moss B. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J Mol Biol. 1988 Feb 5;199(3):399–413. doi: 10.1016/0022-2836(88)90613-4. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M. Intramolecular homologous recombination in cells infected with temperature-sensitive mutants of vaccinia virus. J Virol. 1989 May;63(5):2030–2035. doi: 10.1128/jvi.63.5.2030-2035.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M., Moss B. Resolution of linear minichromosomes with hairpin ends from circular plasmids containing vaccinia virus concatemer junctions. Cell. 1986 Jun 20;45(6):879–884. doi: 10.1016/0092-8674(86)90562-3. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M., Moss B. Resolution of vaccinia virus DNA concatemer junctions requires late-gene expression. J Virol. 1989 Apr;63(4):1595–1603. doi: 10.1128/jvi.63.4.1595-1603.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981 Dec;27(2 Pt 1):391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Parsons B. L., Pickup D. J. Tandemly repeated sequences are present at the ends of the DNA of raccoonpox virus. Virology. 1987 Nov;161(1):45–53. doi: 10.1016/0042-6822(87)90169-3. [DOI] [PubMed] [Google Scholar]
- Pickup D. J., Bastia D., Stone H. O., Joklik W. K. Sequence of terminal regions of cowpox virus DNA: arrangement of repeated and unique sequence elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7112–7116. doi: 10.1073/pnas.79.23.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. K., Bauer W. R. Activation of the vaccinia virus nicking-joining enzyme by trypsinization. J Biol Chem. 1989 Jan 5;264(1):443–449. [PubMed] [Google Scholar]
- Wittek R., Menna A., Müller H. K., Schümperli D., Boseley P. G., Wyler R. Inverted terminal repeats in rabbit poxvirus and vaccinia virus DNA. J Virol. 1978 Oct;28(1):171–181. doi: 10.1128/jvi.28.1.171-181.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]






