Abstract
Checkpoint with FHA and RING finger domains (CHFR) was first recognized as an early mitotic checkpoint protein that delayed the cell cycle in response to microtubule-targeting drugs. It is an E3 ubiquitin ligase that ubiquitinates target proteins to direct them to the proteasome for degradation or to alter their activity. To date, however, the downstream target proteins critical to CHFR's normal cellular functions largely remain unidentified with the exception of the key mitosis regulators, and oncogenes, PLK1 and Aurora A kinases. Rapidly growing evidence in mice, primary human tumors, and mammalian cell culture models indicate that CHFR may also function as a potent tumor suppressor. Interestingly, studies reported to date suggest that CHFR both controls a novel prophase checkpoint early in mitosis and regulates chromosome segregation later in mitosis to maintain genomic stability. In addition, loss of CHFR sensitizes cancer cells to microtubule poisons, altering chemoresponsiveness to taxanes and making it a potential biomarker for chemotherapeutic response. Importantly, CHFR may be one of the few proteins that are required for regulating the cell cycle and maintaining genomic instability to inhibit tumorigenesis.
Introduction
Cancer is the second leading cause of death in the United States (http://www.cdc.gov/nchs/fastats/lcod.htm). Cancers are a molecularly complex set of diseases, often making them difficult to treat due to a great variation in sensitivity to radiation and chemotherapy drugs. Therefore, researchers and clinicians must identify the molecular and genetic characteristics of a tumor, or “biomarkers,” to determine the best course of treatment. In the past 8 years, Checkpoint with FHA and RING finger domains (CHFR) has gained attention as a mitotic checkpoint protein with tumor-suppressive function that has the potential to be a novel biomarker for chemotherapeutic response to microtubule-targeting drugs such as taxanes. Here, we discuss what is currently known about the structure and function of CHFR, as well as the growing amount of evidence supporting its role as a tumor-suppressor protein and biomarker for taxane treatment.
CHFR Protein Structure and Homologs
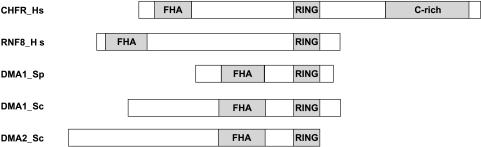
CHFR was initially identified through a screen to find novel mitotic checkpoint proteins that contained a forkhead-associated (FHA) domain [1]. In addition to the N-terminal FHA domain, CHFR was also found to have a central RING finger domain and a C-terminal cysteine-rich region (Figure 1) [1]. The functional relevance of the FHA domain in CHFR remains largely unknown. However, its deletion creates a dominant-negative form of the protein, suggesting that it is critical to its normal cellular function, and this domain was recently found to be responsible for the antiproliferative effects of CHFR [1–3]. The RING finger domain and the cysteine-rich region are better characterized. The zinc-binding RING finger domain has proven to be required for the early prophase checkpoint function of CHFR. It confers CHFR's E3 ubiquitin ligase activity to create ubiquitin chains on target proteins either through the amino acid residue lysine-48, thereby targeting proteins to the proteasome for degradation, or through lysine-63 linkages that may alter target protein function [4,5]. In addition to ubiquitinating target proteins, the RING finger domain was also found to be necessary for CHFR autoubiquitination [6]. The cysteine-rich region was identified as the region responsible for the interaction between CHFR and one of its target proteins, Aurora A [7]. Recently, a putative C2H2 zinc-finger motif, which was renamed a PAR-binding zinc-finger (PBZ) motif, was identified in the C-terminal cysteine-rich region of CHFR. This region was found to be poly(ADP-ribosyl)ated by PARP1. Although mutating the PBZ domain did not inhibit CHFR's ubiquitinating activity, it did impair the dominant-negative function of the FHA-domain deletion mutant. There was also inconclusive evidence that the PBZ motif may be required for CHFR's early mitotic checkpoint function [8]. Of interest, the targeting of proteins to portions of the mitotic apparatus is dependent on the recognition of poly(ADP-ribose) (PAR) by PAR-binding motifs.
Figure 1.
The domain architecture of CHFR is evolutionarily conserved. CHFR and its human homolog, RNF8, share the same domain architecture as the yeast orthologs DMA proteins in both Schizosaccharomyces pombe (Sp) and Saccharomyces cerevisiae (Sc). All of the proteins have an N-terminal FHA domain and a central or C-terminal RING domain that confers the proteins' ubiquitin ligase activities. CHFR is the only member of the family that has a C-terminal cysteine-rich region.
According to the domain architecture and organization, there are evolutionarily conserved protein orthologs of CHFR even in yeast, both Saccharomyces cerevisiae and Schizosaccharomyces pombe (Figure 1) [1,9]. These minimally described orthologs, defective in mitotic arrest 1 and 2 (DMA1 and DMA2), were found to be important in regulating the mitotic spindle checkpoint, mitotic spindle position, and cytokinesis through the septation-initiation network [9–11]. As will be described later, two studies have indicated that human and mouse CHFR may have similar functions as the yeast orthologs.
Interestingly, a human paralog of CHFR was also described recently. RING finger protein 8 (RNF8) shares the FHA and RING domains architecture of CHFR, although it seems to have diverged a bit in terms of cellular function (Figure 1). Initially, RNF8 was described as a mitotic protein important for spindle formation, cytokinesis, and mitotic exit, much like the yeast orthologs [12]. However, unlike CHFR, it was also characterized recently as an important DNA damage response protein to double-strand breaks. RNF8 interacts with phosphorylated MDC1, an interaction that is dependent on RNF8's FHA domain, to mediate double-strand break-associated ubiquitinations and facilitate the accumulation of 53BP1 and BRCA1 at the sites of DNA double-strand breaks [13,14]. In addition, both groups noted that RNF8 can ubiquitinate histones H2A and H2AX and that the loss of RNF8 expression by RNAi abrogated double-strand break retention of the ubiquitin-binding protein RAP80 and increased cellular sensitivity to ionizing radiation [13,14]. There is a growing body of evidence that suggests that CHFR, unlike RNF8, does not mediate a classic cellular response to DNA damage on the basis of the results from commonly used assays to assess DNA damage response.Whereas the role of CHFR in the cellular response to ionizing radiation has been inconclusive [15,16], it does not seem to have a role in the DNA damage response induced by cisplatin (CDDP), UV radiation, or the topoisomerase inhibitors etoposide (VP16) and topotecan [6,17,18].
Since its initial publication, studies on CHFR have primarily focused on its expression in cancer cells and potential role in oncogensis, but some progress has also been made in identifying its biochemical function and target proteins. This review will address the discoveries that have been published to date for CHFR, including its mitotic checkpoint functions, reported protein targets for its ubiquitin ligase activity, and evidence supporting its role as a tumor-suppressor protein and as a biomarker for chemotherapeutic response to taxanes.
CHFR Functions
The Microtubule Stress Prophase Checkpoint
As mentioned in the previous paragraphs, CHFR does not seem to participate in the DNA damage checkpoint or DNA repair pathways. On the contrary, CHFR regulates an early mitotic checkpoint, during prophase, in response to the disruption of normal microtubule formation or stabilization as assessed after treatment with microtubule poisons such as nocodazole, colcemid, and taxanes [1]. During mitotic stress, CHFR temporarily delays the cell cycle by approximately 3 hours, during which chromosome condensation and nuclear envelope breakdown are inhibited and cyclin B1/CDC2 is restricted to the cytoplasm where it is inactive [1,16,17]. CHFR also regulates the coordination of chromosome condensation and centrosome separation during prophase [1]. Together, these results identified CHFR as the first key member of a novel early mitotic checkpoint in prophase, referred to here as the CHFR-mediated prophase checkpoint.
The ability of CHFR to delay chromosome condensation has been confirmed both by visually identifying a lack of condensed chromosomes and by the absence of phosphorylation of histone H3 on residues Ser10 and Ser28 when CHFR is overexpressed in several cell lines [1,16,17,19]. In addition, the CHFR-mediated prophase checkpoint is typically monitored by calculating the mitotic index of cells treated with microtubule poisons; CHFR-expressing cells will have fewer mitotic cells, as evidenced by condensed chromosomes and no nuclear envelope, compared to nonexpressing cells [1,17,19,20].
Elegant studies performed by Kang et al. [5] in Xenopus laevis egg extracts have begun to elucidate how CHFR regulates entry into mitosis. As previously mentioned, CHFR delayed the onset of mitosis by retaining inactive cyclin B1/CDC2 in the cytoplasm in human cells. In X. laevis extracts, the addition of full-length CHFR, but not RING finger mutants, resulted in prolonged inhibitory phosphorylation of CDC2 on residue Tyr15. In addition, CHFR delayed the phosphorylation of the CDC2-regulatory proteins WEE1 and CDC25C at the G2-to-M transition. However, the steady state level of expression for these proteins, and for the CDC25C regulatory protein CHK1, remained unchanged in the presence of CHFR, indicating that they were not probable ubiquitination targets. Therefore, it is likely that the prolonged inhibitory phosphorylation of CDC2 at Tyr15 is due to the overactivation of WEE1 kinase, which phosphorylates CDC2 at Tyr15, and the inhibition of CDC25C phosphatase, which is normally responsible for activating cyclin B1/CDC2 by removing the inhibitory phosphorylation on Tyr15 of CDC2. However, as these experiments were performed in Xenopus extracts, they remain to be confirmed in mammalian cells.
One of the proteins that can regulate both CDC25C and WEE1 through phosphorylation is the polo-like kinase 1 (PLK1). Kang et al. [5] determined that CHFR could, in fact, ubiquitinate PLK1 in X. laevis extracts. In support of this model, the E3 ubiquitin ligase activity of CHFR through its RING finger domain is required for proper prophase checkpoint function, suggesting that CHFR must ubiquitinate a target protein for the cell cycle delay to occur. However, although the pathway described previously is a likely explanation for the series of events that occurs downstream of CHFR in response to mitotic stress, these results have not been easily replicated in mammalian cells. The ability of CHFR to ubiquitinate and/or regulate PLK1 has not been consistently reproduced, raising questions as to the legitimacy of this pathway in mammalian cells as discussed further in the next paragraphs.
Another potential pathway for the CHFR-regulated mitotic stress checkpoint during prophase is through the p38 stress-activated kinases in which CHFR acts upstream of p38 through an as-of-yet unknown mechanism [21]. There is also recent evidence that Sirtuin 2 (SIRT2), a tubulin and histone deacetylase, may also participate in the same microtubule stress-induced prophase checkpoint as CHFR does. Like CHFR, it was found that SIRT2 overexpression results in a decreased mitotic index and inhibits chromosome condensation in response to nocodazole or paclitaxel treatment and that the NAD-dependent tubulin deacetylase activity of SIRT2 was required for this response [22]. Finally, another potential player in the mitotic stress-induced prophase checkpoint is sensitivity to nitrogen mustard 1 (SNM1). Akhter et al. [23] found that mouse embryonic fibroblasts from Snm1 knockout mice behaved like cells that had lost CHFR expression. Snm1-null cells arrested during the spindle checkpoint with condensed chromosomes and separated (and duplicated) centrosomes after nocodazole exposure, whereas wild type cells from littermates arrested with decondensed chromosomes and unseparated centrosomes. Furthermore, Snm1 wild type cells delayed mitotic entry by approximately 4 hours and maintained cyclin A expression longer, indicating an arrest in prophase, when compared to null fibroblasts [23]. Much like CHFR-null cells, Snm1-/- mouse embryonic fibroblasts were less likely to survive nocodazole treatment when compared to wild type cells [23]. Of interest, because the CHFR homolog RNF8 can regulate 53BP1, Akhter et al. [23] also found that SNM1 could also interact with 53BP1 and the APC/cyclosome complex in HeLa cells [12]. Together, these observations suggest that a currently unrecognized pathway of protein interactions involving CHFR is critical for normal progression through the recently described prophase checkpoint.
The Mitotic Spindle Assembly Checkpoint
In addition to participating in an early mitotic checkpoint during prophase, a few reports indicate that CHFR may be required for the regulation of later events in mitosis, such as chromosome segregation to maintain genomic stability. Embryonic fibroblasts from Chfr knockout mice showed not only a prolonged time in prophase but also an extended amount of time in anaphase. The Chfr-null mouse embryonic fibroblasts, which became aneuploid in culture, also displayed lagging chromosomes during anaphase, failed nuclear segregation, and multinucleated cells indicating failed cytokinesis [7]. In addition, immortalized human mammary epithelial cells that had stably decreased CHFR expression by shRNA also became aneuploid [19]. When MCF10A immortalized human mammary epithelial cells were transiently transfected with siRNA targeting CHFR, they became aneuploid within 72 hours [24]. A role for CHFR in regulating chromosome segregation and the mitotic spindle assembly checkpoint was further supported by the finding that the key mitotic spindle checkpoint proteins BUBR1 and MAD2 were not properly localized to the kinetochores during metaphase in MCF10A cells lacking CHFR expression by RNAi [24]. This mislocalization was correlated with impaired MAD2/CDC20 complex formation during nocodazole treatment, which is required to prevent the anaphase-promoting complex (APC/C) from prematurely signaling the onset of anaphase before all of the sister chromatids are attached to the mitotic spindle [24]. This apparent mitotic spindle checkpoint defect was further supported by the characterization of an array of mitotic defects including misaligned chromosomes at the metaphase plate, lagging anaphase chromosomes, multipolar mitotic spindles, and tetraploid binucleated giant cells in MCF10A cells lacking CHFR by siRNA [24]. In support of these findings, recently published bioinformatics evidence indicated that CHFR may contain a KEN box motif, suggesting that CHFR may be targeted for proteasome-mediated degradation by the APC/C-CDH1 complex, which is a critical component of the mitotic spindle assembly checkpoint and the regulatory complex that controls mitotic exit [25]. Therefore, CHFR has multiple mitotic checkpoint functions and, contrary to initial thought, does not function only in response to mitotic stress due to microtubule-targeting drugs.
Regulation of Cellular Proliferation
Multiple cell culture models have indicated that CHFR can function as a negative regulator of proliferation. Decreasing CHFR expression by RNAi in MCF10A and HPV4-12 immortalized mammary epithelial cells caused a decrease in population growth rates, whereas overexpressing CHFR in the Hs578T breast cancer cell line decreased population growth rates. This was further supported by the finding that there was an increased frequency of phosphorylation of histone H3 on Ser28 in CHFR knockdown cells and a decreased frequency of histone H3 phosphorylation in CHFR overexpressing cells. This indicated that cells with altered CHFR expression entered into, or progressed through, mitosis at different rates compared to parental cells [19]. In addition, Fukuda et al. [3] discovered that overexpressing CHFR also inhibited cellular proliferation in HCT116 and RKO colon cancer cells, and this antiproliferative function was dependent on the FHA domain of CHFR. However, the mechanism(s) by which CHFR negatively regulates cellular proliferation remains to be elucidated.
CHFR Interacting Proteins
Polo-Like Kinase 1
As mentioned previously, there is evidence that CHFR can ubiquitinate PLK1 in Xenopus extracts, but tests to assess the ability of CHFR to regulate PLK1 activity or protein levels in mammalian cells have been inconclusive. Findings in support of CHFR controlling PLK1 include results indicating that overexpressed CHFR mutants, which mimic unphosphorylated CHFR, can decrease PLK1 expression and kinase activity in HeLa cells [15]. Of interest, mouse embryonic fibroblasts from Chfr knockout mice were found to overexpress PLK1 compared to cells from wild type and heterozygous littermates, suggesting that CHFR can ubiquitinate PLK1 to target it for degradation [7].
To the contrary, there are other reports that have not been able to find a correlation between CHFR expression and PLK1 expression or activity. For example, Matsusaka and Pines [21] were unable to find an association between PLK1 expression and induction of the CHFR-regulated prophase checkpoint due to colcemid treatment. In addition, other studies have not been able to find a relationship between CHFR expression and the amount of PLK1 protein, either as a trend among breast cell lines or after CHFR overexpression in HCT116 cells [16,20]. It is apparent that more work is required to determine whether PLK1 is a target for CHFR-mediated ubiquitination and regulation. Perhaps the interaction is specific for a particular species, tissue, or treatment.
Aurora A Kinase
A key protein that regulates the activity and translocation of cyclin B1 to the nucleus to initiate mitosis is Aurora A kinase [26]. Therefore, Aurora A has also been speculated to be a target for ubiquitination by CHFR. Summers et al. [16] analyzed Aurora A expression and activation in HCT116 cells overexpressing CHFR and found that, although there was no change in Aurora A expression or localization to the centrosomes, they did discover that the nocodazoleinduced CHFR-mediated mitotic delay was associated with the inactive Aurora A that was unphosphorylated at residue Thr288 at the centrosomes. Compelling evidence that CHFR ubiquitinates Aurora A was provided by Yu et al. [7] in which they found that Aurora A was overexpressed in Chfr-null mouse embryonic fibroblasts and tissues. In addition, using human cell lines, they determined that the C-terminal cysteine-rich region of CHFR interacts with the N-terminus of Aurora A by immunoprecipitation and that this interaction led to the ubiquitination of Aurora A [7]. In support of these findings, we have also reported that decreasing CHFR expression by siRNA in MCF10A-immortalized human mammary epithelial cells led to Aurora A overexpression, although its localization to the centrosomes was not perturbed, and that overexpressed Aurora A could interact with endogenous CHFR by immunoprecipitation [24].
Proteins Regulating Ubiquitination Activity
Because CHFR has been described as an E3 ubiquitin ligase due to the presence of its RING domain, there must be E2 enzymes that it interacts with for it to function. CHFR has been shown to use the E2 ubiquitin-conjugating (Ubc) proteins UBC4, UBC5A, and UBC5B to form Lys48-based polyubiquitin chains, but not E2 enzymes UBCH7, UBCH8, or UBCH10 [4,5]. CHFR has also been found to be able to interact with the E2 enzyme complex UBC13-MMS2 heterodimer to form lysine-63-linked polyubiquitin chains, which are associated with modifying protein function, not targeting them for degradation by the proteasome [4]. In addition to ubiquitin-conjugating E2 enzymes, CHFR has been shown to interact with the deubiquitinating protein ubiquitin-specific protease 7 (USP7) by immunoprecipitation [27]. USP7 was found to deubiquitinate CHFR and inhibit its auto-ubiquitinating activity, thereby preventing the degradation of CHFR [27]. Interestingly, USP7 and CHFR both localize to PML bodies in the nucleus; in particular, CHFR has been shown to be in PML bodies in interphase cells [2,27].
Additional Interacting Proteins
Due to the role of CHFR in initiating a cell cycle delay in response to microtubule poisons, it is feasible that it may interact with tubulin proteins. Recently, Privette et al. found that CHFR could interact with α-tubulin, by both immunoprecipitation and GST pull down experiments [24]. Importantly, CHFR was found to ubiquitinate α-tubulin when MCF10A cells were treated with nocodozaole to induce the checkpoint [24]. In the same work, CHFR and MAD2 were shown to interact by yeast two-hybrid analysis, coimmunoprecipitation, and colocalization, which further supported the hypothesis that CHFR may function in the mitotic spindle assembly checkpoint in addition to the prophase checkpoint [24]. Finally, it was determined that protein kinase B (PKB) could phosphorylate CHFR on residues Thr39 and Ser208, potentially inhibiting the ubiquitin ligase activity of CHFR; however, phosphorylation-defective mutants of CHFR did not alter the checkpoint response to paclitaxel [15]. Bothos et al. [4] also found that CHFR was phosphorylated during mitosis, although they did not determine which kinase was responsible.
CHFR and Cancer
Mutations, Alternative Transcripts, and Chromosomal Aberrations
CHFR has been implicated as a tumor suppressor in multiple cancers despite the fact that no heritable mutations in germline cells associated with a predisposition to cancer phenotypes have been identified and few mutations in somatic cancer cells have been described. Although many groups have not been able to identify coding mutations in the CHFR gene, particularly in breast and colon cancers, several single-nucleotide polymorphisms (SNPs) have been identified [20,28,29]. However, three missense mutations in CHFR were found in primary non-small cell lung cancers, that is, two mutations between the FHA and RING domains and one in the cysteine-rich region, all three of which were unable to rescue the CHFR checkpoint in DLD-1 cells during nocodazole treatment [29]. However, even these mutations were rare events, because they were only found in three patients, all of who were smokers, of 53 different patient samples tested [29]. Recent published data have indicated that one of these coding SNPs, V539M (Accession No. NM_018223; V580M for Accession No. AF_170724) found within the C-terminal cysteine-rich region, was significantly associated with a lower risk of colorectoral cancer if the patient had the methionine amino acid instead of the valine [30]. This SNP was also strongly associated with the absence of metastases, TNM stage, and microsatellite instability, all of which indicate a favorable prognosis. This variant was within the hap 10 (TGACTA) haplotype block that also contained the P138L SNP, which had correlated with the microsatellite instability phenotype [30]. Two additional deletion mutations have also been identified in which either residues 135 to 146 were deleted between the FHA and RING domains or residue Ala470 was deleted in the C-terminal cysteine-rich region [29].
Although they have not been studied extensively, alternative mRNA transcripts have been identified for CHFR. Toyota et al. [31] identified transcripts that were missing exons 2, 5, and/or 6 and the transcript missing exon 2 is believed to result in an isoform lacking the FHA domain [1]. This isoform was also found to be highly expressed in cancer cells when compared with matched normal tissues [31]. Although full-length CHFR was found to suppress cell growth when overexpressed, the FHA domain deletion mutant of CHFR was less effective in suppressing cell growth and had been previously identified as a dominant negative form of CHFR [1,31].
CHFR is located on human chromosome 12q24.33, which has been characterized as a site of allelic imbalance in many cancers. Deletion of 12q24 has been described in adenoid cystic carcinoma and as a prognostic indicator of cancer recurrence in Wilms tumor and pituitary adenomas [32–34]. Chromosome loci 12q24 has also been associated with the presence of a metastasis suppressor gene, because gain of 12q24 correlated with metastasis-free survival in breast cancer patients and decreased metastatic potential of prostate cancer cells [35–37]. Further characterization of 12q24 would be required to determine whether CHFR is the gene responsible for predicting cancer recurrence and metastasis in these association studies.
CHFR Expression and Correlations with Clinicopathologic Variables
Many studies have focused on studying the loss of CHFR mRNA expression due to promoter hypermethylation in cancers compared to normal cells and tissues, and they are summarized in Table 1 [17,18,20,28,31,38–62]. Although CHFR mRNA expression has been shown to be decreased or lost in many cancers, sometimes up to 50% of samples, promoter hypermethylation only accounts for a percentage of these instances and is often tissue-dependent [20]. In particular, 16% to 53% of cancers of the gastrointestinal tract have hypermethylated promoters of the CHFR gene; however, this is extremely rare in gynecologic cancers with the exception of HPV-positive cervical carcinomas [31,45,57,58,61]. As an example, 50% of breast cancer cell lines have decreased or lost CHFR expression compared to immortalized human mammary epithelial cells, but only 8% of the cell lines had a methylated promoter [20].
Table 1.
The Frequency of CHFR Promoter Hypermethylation and Correlations with Clinicopathological Variables.
| Cancer Tissue | Percent Hypermethylated | Correlations and Findings | Reference |
|---|---|---|---|
| Lung cancer | 19% (7/37) of primary cancers | Infrequent SNPs in coding region | [53]* |
| 19% (3/16) of cell lines | |||
| Esophageal cancer | 16.3% (7/43) of primary cancers | None found | [57]* |
| 26.7% (4/15) of cell lines | |||
| Colon adenocarcinomas | 37% (11/30) of primary cancers | Some methylation in normal colon | [42]* |
| Non-small cell lung | 10% (2/20) of primary cancers | ||
| Colorectal cancer | 40% (25/63) of primary cancers | Associated with increased mitotic index | [31]* |
| Colorectal adenomas | 53% (27/51) of primary cancers | ||
| Head and neck cancer | 30% (16/54) of primary cancers | ||
| Hepatocellular cancer | 0% (0/20) of primary cancers | ||
| Colon cancer | 36% (8/22) of primary cancers | None tested | [28]* |
| Colon cancer | 43% (9/21) of cell lines | ||
| Breast cancer | 0% (0/19) of cell lines | ||
| Breast cancer | 8% (2/24) of cell lines | Low expression associated with high mitotic index | [20]† |
| Gastric cancer | 39% (24/61) of primary cancers | None tested | [18]‡,§ |
| Gastric cancer | 20% (4/20) of cell lines | ||
| Gastric cancer | 35% (25/71) of primary cancers | None found | [47]* |
| Gastric cancer | 20% (2/10 cell lines) | ||
| Gastric cancer | 44% (19/43) of primary cancers | None tested | [49]*,§ |
| Gastric cancer | 67% (8/12) of cell lines | ||
| Biliary tract carcinomas | 16% (6/37) of primary cancers | None found | [59]* |
| Low-grade noninvasive gastric neoplasia | 10% (1/10) of primary cancers | Methylation correlated with advanced patient age and high tumor grade | [46]* |
| High-grade noninvasive gastric carcinoma | 45% (10/22) of primary cancers | ||
| Submucosal invasive gastric adenocarcinomas | 35% (7/20) of primary cancers | ||
| Colorectal cancer | 31% (19/62) of primary cancers | Associated with MLH1 promoter hypermethylation, microsatellite instability | [39]* |
| Cutaneous T-cell lymphoma | 19% (5/28) of primary cancers | None tested | [60]§,¶ |
| Nasopharyngeal carcinomas | 61% (22/36) of primary cancers | None found | [41]*,§ |
| 88% (7/8) of cell lines | |||
| Oral squamous cell carcinoma | 31% (4/13) of primary cancers | Low expression associated with high mitotic index and taxane sensitivity | [17]‡ |
| 22% (2/9) of cell lines | |||
| Acute myelocytic and lymphocytic leukemia | 39% (16/41) of primary cancers | None tested | [44]* |
| Hepatocellular cancer | 35% (22/62) of primary cancers | Correlated with infiltrative growth pattern and advanced stage | [56] |
| Gastric cancer | 37% (15/41) of primary cancers | No correlation with response to Taxol treatment | [62]‡ |
| Breast cancer | 0.9% (1/110) of primary cancers | None tested | [58]* |
| Gastric cancer | 52% (24/46) of primary cancers | Correlated with clinical response to Taxol treatment | [51]* |
| Esophageal cancer | 24% (9/38) of primary cancers | Correlated with female gender, but not with tumor stage | [55]* |
| Gastric cancer | 30% (16/53) of primary cancers | ||
| Colorectal cancer | 26% (25/98) of primary cancers | None tested | [54] |
| Endometrial cancer | 12% (6/50) of primary cancers | Correlated with poorly differentiated adenocarcinomas | [61]* |
| Ovarian cancer | 0% (0/48) of primary cancers | None tested | [52]* |
| Cervical adenocarcinomas | 12% (2/14) of primary cancers | Correlated with taxane sensitivity | [38]* |
| Cervical carcinoma | 33% (2/6) of cell lines | ||
| HPV+ cervical squamous cell carcinoma | 31% (5/16) of primary cancers | Methylation designated as a late event | [47]# |
| HPV+ cervical adenocarcinoma | 50% (4/8) of primary cancers | ||
| HPV+ cervical carcinoma | 56% (5/9) of cell lines | ||
| Head and neck squamous cell carcinoma | 25% (7/28) of primary cancers | Correlated with stage IV cancer | [40]# |
| Colorectal cancer | 41% (29/71) of primary cancers | hMLH1 promoter methylation, gain of chromosome 8q, BRAF mutations | [43]* |
| Gastric cancer | 48% (12/25) of primary cancers | None tested | [48]** |
| Colorectal cancer | 24% (217/888) of primary cancers | None tested | [50]** |
Methylation-specific polymerase chain reaction.
Treatment with 5-aza-dC.
Combined bisulfite restriction analysis.
Bisulfite sequencing.
CpG island microarray.
Methylation-specific multi-plex ligation-dependent probe amplification.
MethyLight quantitative polymerase chain reaction.
Altered mRNA expression of CHFR has been correlated with clinical and pathologic variables. In particular, low or lost CHFR was associated with a high mitotic index in colorectal and breast cancers [20,31]. A hypermethylated CHFR promoter was also associated with advanced patient age, high tumor grade, advanced stage, poor differentiation, female gender, and taxane sensitivity [38,40,45,46,51,54,56,61]. Of these correlations, high mitotic index, advanced tumor stage, and taxane sensitivity have been replicated among different research groups [20,31,38,40,51,56].
Only a few studies recently have reported on the frequency of altered CHFR protein expression in cancers versus normal tissue. In breast cancers, 41% of cultured cell lines were found to have decreased or lost CHFR expression by Western blot analysis, whereas 36% of primary invasive breast cancers were negative for CHFR expression by immunohistochemistry [19]. Of interest, the loss of CHFR expression in primary invasive breast cancers strongly correlated with larger tumor size and there was a trend of association between CHFR-negative and estrogen receptor (ER)-negative status [19]. Milne et al. [63] found that 33% of gastric cancers were negative for CHFR expression by immunohistochemistry, which correlated with a diffuse histologic diagnosis. Importantly, using immunohistochemistry, CHFR localization to the nucleus was altered in 66% of malignant peripheral nerve sheath tumors. In the same samples, decreased CHFR expression was associated with a multitude of clinical and pathologic variables including young age, site of tumor to the trunk, head, or neck, presentation in recurrent tumors, increased mitotic index, increased Ki67 staining for proliferation, abnormal karyotype, and poor patient prognosis [64].
CHFR Is a Tumor Suppressor and Regulator of Genomic Stability
The first indication that CHFR was a potent tumor suppressor was the creation of the Chfr knockout mouse, which was viable with no developmental defects [7]. A small percentage (9%) of these mice developed lymphomas by 40 weeks of age, and later, they were prone to developing spontaneous cancers of epithelial origin, primarily lung, liver, and gastrointestinal tumors. Fifty percent of the knockout mice also developed skin tumors after treatment with the chemical carcinogen dimethylbenz(a)anthracene at 4 months of age compared with none of their wild type littermates [7]. There was also an indication that the loss of CHFR led to cellular transformation because embryonic fibroblasts from the mice were able to form colonies in culture [7]. Interestingly, CHFR was found to be important for maintaining genomic stability in the embryonic fibroblasts from the Chfr-null mice. About 30% of the cells became aneuploid after four passages likely due to several mitotic defects that were observed, including lagging anaphase chromosomes, failed nuclear segregation, and failed cytokinesis, which resulted in 17% of the cells becoming multinucleated [7]. Of interest, they discovered that down-regulating Aurora A by RNAi rescued the genomic instability phenotype in Chfr-null mouse embryonic fibroblasts [7].
A multitude of cell culture assays using immortalized human mammary epithelial cell lines and the Hs578T breast cancer cell line provided further evidence that CHFR was a tumor suppressor in human cells. Two immortalized human mammary epithelial cell lines (HPV4-12 and MCF10A) that had decreased CHFR expression by RNAi developed many phenotypes reminiscent of malignant progression including increased population growth rates, higher mitotic index, enhanced invasion and motility, increased sensitivity to microtubule-targeting drugs, aneuploidy, and colony formation in soft agar [19]. MCF10A cells that had decreased CHFR expression by RNAi also displayed an epithelial-to-mesenchymal transition in morphology and an increase in the number of nucleoli. Interestingly, many of these tumorigenic phenotypes—particularly the growth rates, mitotic index, sensitivity to microtubule poisons, cellular invasion through Matrigel, and cellular motility—were reversed in a breast cancer cell line, Hs578T, which was transduced with a retrovirus to overexpress CHFR [19]. Further work found that siRNA against CHFR in MCF10A caused the cells to become aneuploid within 72 hours of transfection because of four mitotic defects: 1) misaligned chromosomes at the metaphase plate; 2) lagging anaphase chromosomes; 3) multipolar, poorly formed mitotic spindles indicating centrosome amplification; and 4) binucleated giant cells indicating cytokinesis defects [24]. The complementary data from the knockout mouse and human cell culture have provided strong evidence that CHFR, in addition to its checkpoint function, was involved in regulating growth rates, cellular invasion and motility, and genomic stability, which indicated that CHFR could be a potent tumor suppressor.
CHFR as a Potential Biomarker for Chemotherapeutic Response to Taxanes
As indicated in the previous paragraphs and in Table 1, CHFR promoter hypermethylation correlated with taxane sensitivity in gastric cancers and cervical carcinoma [38,51]. In support of these findings, cultured cells that were transfected to overexpress CHFR showed a decrease in apoptotic response to taxanes and enhanced cell survival, whereas decreasing CHFR by RNAi resulted in enhanced sensitivity (i.e., increased apoptotic response) to taxanes [1,17–19]. These reports have indicated that the CHFR expression status of cancers might be an excellent biomarker for tumor response to taxane treatment. However, this poses a very interesting dichotomy for CHFR expression. On one hand, losing the tumor-suppressive function of CHFR may lead to a larger, more advanced and aggressive tumor, conversely, an absence of CHFR expression might actually be favorable because it would be a positive indicator of chemotherapeutic response to taxane treatment. Future work studying the potential role of CHFR as a biomarker for chemotherapeutic response to taxanes in diverse tissues, and the mechanism of this response, would definitely be valuable both at the bench and at the clinic.
Conclusions
Since CHFR was first described in 2000, several investigators have been further characterizing its expression patterns in normal and malignant cells and trying to elucidate its functional roles in cell biology. At present, we are just beginning to understand how the CHFR-regulated prophase checkpoint might work in response to microtubule disruption. CHFR can regulate mitotic entry, particularly in the presence of microtubule poisons such as taxanes, by controlling cyclin B1/CDC2 activity and translocation to the nucleus. The mechanisms behind this regulation are unclear, although it likely involves CHFR-mediated ubiquitination and regulation of PLK1 and/or Aurora A kinases. More recently, CHFR has been implicated as a regulator of genomic stability. Cells that have lost or decreased expression quickly became aneuploid, a hallmark of many cancers, due to an impaired mitotic spindle checkpoint, which would result in chromosome mis-segregation. Interestingly, CHFR seems to have multiple checkpoint functions, because it participates in cellular response to microtubule poisons early in mitosis during prophase and it is also important for chromosome segregation, mitotic spindle formation, and the function of the mitotic spindle assembly checkpoint.
In addition, there is a rapidly growing amount of evidence to support the hypothesis that CHFR is also a potent tumor suppressor. CHFR has been shown to be a tumor suppressor in mice, which is further supported by studies in human cancer cells. Its expression often is lost or decreased in cancers when compared to normal tissues, sometimes due to promoter hypermethylation that varies by tissue. The pathways that regulate the transcriptional expression of CHFR are unknown, although the identification of these pathways will likely explain how/why expression is lost in cancers when promoter hypermethylation is not the cause. Surprisingly, CHFR expression alters cellular motility and invasion in cell culture models, which is supported by the presence of an unidentified “metastasis suppressor” at the chromosomal location where CHFR is found. This novel, poorly characterized role for CHFR certainly deserves more attention. Finally, CHFR is emerging as an important cancer biomarker for chemotherapeutic response to taxane treatment, although future research is required to verify this finding before translating these preliminary results for use in the clinic.
Footnotes
This work was supported by a Department of Defense Breast Cancer Research Program Fellowship, #BC050310, by the National Institutes of Health (NIH) National Research Service Award #5-T32-GM07544 from the National Institute of General Medicine Sciences to L.M.P., and by an NIH National Cancer Institute grant RO1CA072877 to E.M.P.
References
- 1.Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–435. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 2.Daniels MJ, Marson A, Venkitaraman AR. PML bodies control the nuclear dynamics and function of the CHFR mitotic checkpoint protein. Nat Struct Mol Biol. 2004;11:1114–1121. doi: 10.1038/nsmb837. [DOI] [PubMed] [Google Scholar]
- 3.Fukuda T, Kondo Y, Nakagama H. The anti-proliferative effects of the CHFR depend on the forkhead associated domain, but not E3 ligase activity mediated by RING finger domain. PLoS ONE. 2008;3:e1776. doi: 10.1371/journal.pone.0001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bothos J, Summers MK, Venere M, Scolnick DM, Halazonetis TD. The Chfr mitotic checkpoint protein functions with Ubc13-Mms2 to form Lys63-linked polyubiquitin chains. Oncogene. 2003;22:7101–7107. doi: 10.1038/sj.onc.1206831. [DOI] [PubMed] [Google Scholar]
- 5.Kang D, Chen J, Wong J, Fang G. The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J Cell Biol. 2002;156:249–259. doi: 10.1083/jcb.200108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaturvedi P, Sudakin V, Bobiak ML, Fisher PW, Mattern MR, Jablonski SA, Hurle MR, Zhu Y, Yen TJ, Zhou BB. Chfr regulates a mitotic stress pathway through its RING-finger domain with ubiquitin ligase activity. Cancer Res. 2002;62:1797–1801. [PubMed] [Google Scholar]
- 7.Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, Ward IM, Saya H, Fang G, van Deursen J, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 8.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 9.Fraschini R, Bilotta D, Lucchini G, Piatti S. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol Biol Cell. 2004;15:3796–3810. doi: 10.1091/mbc.E04-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guertin DA, Venkatram S, Gould KL, McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Dev Cell. 2002;3:779–790. doi: 10.1016/s1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- 11.Murone M, Simanis V. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 1996;15:6605–6616. [PMC free article] [PubMed] [Google Scholar]
- 12.Plans V, Guerra-Rebollo M, Thomson TM. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene. 2008;27:1355–1365. doi: 10.1038/sj.onc.1210782. [DOI] [PubMed] [Google Scholar]
- 13.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 15.Shtivelman E. Promotion of mitosis by activated protein kinase B after DNA damage involves polo-like kinase 1 and checkpoint protein CHFR. Mol Cancer Res. 2003;1:959–969. [PubMed] [Google Scholar]
- 16.Summers MK, Bothos J, Halazonetis TD. The CHFR mitotic checkpoint protein delays cell cycle progression by excluding cyclin B1 from the nucleus. Oncogene. 2005;24:2589–2598. doi: 10.1038/sj.onc.1208428. [DOI] [PubMed] [Google Scholar]
- 17.Ogi K, Toyota M, Mita H, Satoh A, Kashima L, Sasaki Y, Suzuki H, Akino K, Nishikawa N, Noguchi M, et al. Small interfering RNA-induced CHFR silencing sensitizes oral squamous cell cancer cells to microtubule inhibitors. Cancer Biol Ther. 2005;4:773–780. doi: 10.4161/cbt.4.7.1896. [DOI] [PubMed] [Google Scholar]
- 18.Satoh A, Toyota M, Itoh F, Sasaki Y, Suzuki H, Ogi K, Kikuchi T, Mita H, Yamashita T, Kojima T, et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63:8606–8613. [PubMed] [Google Scholar]
- 19.Privette LM, Gonzalez ME, Ding L, Kleer CG, Petty EM. Altered expression of the early mitotic checkpoint protein, CHFR, in breast cancers: implications for tumor suppression. Cancer Res. 2007;67:6064–6074. doi: 10.1158/0008-5472.CAN-06-4109. [DOI] [PubMed] [Google Scholar]
- 20.Erson AE, Petty EM. CHFR-associated early G2/M checkpoint defects in breast cancer cells. Mol Carcinog. 2004;39:26–33. doi: 10.1002/mc.10161. [DOI] [PubMed] [Google Scholar]
- 21.Matsusaka T, Pines J. Chfr acts with the p38 stress kinases to block entry to mitosis in mammalian cells. J Cell Biol. 2004;166:507–516. doi: 10.1083/jcb.200401139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26:945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 23.Akhter S, Richie CT, Deng JM, Brey E, Zhang X, Patrick C, Jr, Behringer RR, Legerski RJ. Deficiency in SNM1 abolishes an early mitotic checkpoint induced by spindle stress. Mol Cell Biol. 2004;24:10448–10455. doi: 10.1128/MCB.24.23.10448-10455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Privette LM, Weier JF, Nguyen HN, Yu X, Petty EM. Loss of CHFR expression in mammary epithelial cells causes genomic instability by disrupting the mitotic spindle assembly checkpoint. Neoplasia. 2008;10:643–652. doi: 10.1593/neo.08176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michael S, Trave G, Ramu C, Chica C, Gibson TJ. Discovery of candidate KEN box motifs using cell cycle keyword enrichment combined with native disorder prediction and motif conservation. Bioinformatics. 2008;24:453–457. doi: 10.1093/bioinformatics/btm624. [DOI] [PubMed] [Google Scholar]
- 26.Hirota T, Kunitoku N, Sasayama T, arumoto T, Zhang D, Nitta M, Hatakeyama K, Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/s0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 27.Oh YM, Yoo SJ, Seol JH. Deubiquitination of Chfr, a checkpoint protein, by USP7/HAUSP regulates its stability and activity. Biochem Biophys Res Commun. 2007;357:615–619. doi: 10.1016/j.bbrc.2007.03.193. [DOI] [PubMed] [Google Scholar]
- 28.Bertholon J, Wang Q, Falette N, Verny C, Auclair J, Chassot C, Navarro C, Saurin JC, Puisieux A. Chfr inactivation is not associated to chromosomal instability in colon cancers. Oncogene. 2003;22:8956–8960. doi: 10.1038/sj.onc.1207078. [DOI] [PubMed] [Google Scholar]
- 29.Mariatos G, Bothos J, Zacharatos P, Summers MK, Scolnick DM, Kittas C, Halazonetis TD, Gorgoulis VG. Inactivating mutations targeting the chfr mitotic checkpoint gene in human lung cancer. Cancer Res. 2003;63:7185–7189. [PubMed] [Google Scholar]
- 30.Kang HC, Kim IJ, Jang SG, Hong SH, Hwang JA, Shin HR, Park JG. Coding region polymorphisms in the CHFR mitotic stress checkpoint gene are associated with colorectal cancer risk. Cancer Lett. 2008;260:170–179. doi: 10.1016/j.canlet.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 31.Toyota M, Sasaki Y, Satoh A, Ogi K, Kikuchi T, Suzuki H, Mita H, Tanaka N, Itoh F, Issa JP, et al. Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci USA. 2003;100:7818–7823. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natrajan R, Williams RD, Hing SN, Mackay A, Reis-Filho JS, Fenwick K, Iravani M, Valgeirsson H, Grigoriadis A, Langford CF, et al. Array CGH profiling of favourable histology Wilms tumors reveals novel gains and losses associated with relapse. J Pathol. 2006;210:49–58. doi: 10.1002/path.2021. [DOI] [PubMed] [Google Scholar]
- 33.Rickert CH, Dockhorn-Dworniczak B, Busch G, Moskopp D, Albert FK, Rama B, Paulus W. Increased chromosomal imbalances in recurrent pituitary adenomas. Acta Neuropathol. 2001;102:615–620. doi: 10.1007/s004010100413. [DOI] [PubMed] [Google Scholar]
- 34.Rutherford S, Hampton GM, Frierson HF, Moskaluk CA. Mapping of candidate tumor suppressor genes on chromosome 12 in adenoid cystic carcinoma. Lab Invest. 2005;85:1076–1085. doi: 10.1038/labinvest.3700314. [DOI] [PubMed] [Google Scholar]
- 35.Aubele M, Auer G, Braselmann H, Nahrig J, Zitzelsberger H, Quintanilla-Martinez L, Smida J, Walch A, Hofler H, Werner M. Chromosomal imbalances are associated with metastasis-free survival in breast cancer patients. Anal Cell Pathol. 2002;24:77–87. doi: 10.1155/2002/820269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichikawa T, Hosoki S, Suzuki H, Akakura K, Igarashi T, Furuya Y, Oshimura M, Rinker-Schaeffer CW, Nihei N, Barrett JC, et al. Mapping of metastasis suppressor genes for prostate cancer by microcell-mediated chromosome transfer. Asian J Androl. 2000;2:167–171. [PubMed] [Google Scholar]
- 37.Jaeger EB, Chekmareva MA, Tennant TR, Luu HH, Hickson JA, Chen SL, Samant RS, Sokoloff MH, Rinker-Schaeffer CW. Inhibition of prostate cancer metastatic colonization by approximately 4.2 Mb of human chromosome 12. Int J Cancer. 2004;108:15–22. doi: 10.1002/ijc.11483. [DOI] [PubMed] [Google Scholar]
- 38.Banno K, Yanokura M, Kawaguchi M, Kuwabara Y, Akiyoshi J, Kobayashi Y, Iwata T, Hirasawa A, Fujii T, Susumu N, et al. Epigenetic inactivation of the CHFR gene in cervical cancer contributes to sensitivity to taxanes. Int J Oncol. 2007;31:713–720. [PubMed] [Google Scholar]
- 39.Brandes JC, van Engeland M, Wouters KA, Weijenberg MP, Herman JG. CHFR promoter hypermethylation in colon cancer correlates with the microsatellite instability phenotype. Carcinogenesis. 2005;26:1152–1156. doi: 10.1093/carcin/bgi058. [DOI] [PubMed] [Google Scholar]
- 40.Chen K, Sawhney R, Khan M, Benninger MS, Hou Z, Sethi S, Stephen JK, Worsham MJ. Methylation of multiple genes as diagnostic and therapeutic markers in primary head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2007;133:1131–1138. doi: 10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- 41.Cheung HW, Ching YP, Nicholls JM, Ling MT, Wong YC, Hui N, Cheung A, Tsao SW, Wang Q, Yeun PW, et al. Epigenetic inactivation of CHFR in nasopharyngeal carcinoma through promoter methylation. Mol Carcinog. 2005;43:237–245. doi: 10.1002/mc.20106. [DOI] [PubMed] [Google Scholar]
- 42.Corn PG, Summers MK, Fogt F, Virmani AK, Gazdar AF, Halazonetis TD, El-Deiry WS. Frequent hypermethylation of the 5′ CpG island of the mitotic stress checkpoint gene Chfr in colorectal and non-small cell lung cancer. Carcinogenesis. 2003;24:47–51. doi: 10.1093/carcin/24.1.47. [DOI] [PubMed] [Google Scholar]
- 43.Derks S, Postma C, Carvalho B, van den Bosch SM, Moerkerk PT, Herman JG, Weijenberg MP, de Bruine AP, Meijer GA, vanEngeland M. Integrated analysis of chromosomal, microsatellite and epigenetic instability in colorectal cancer identifies specific associations between promoter methylation of pivotal tumour suppressor and DNA repair genes and specific chromosomal alterations. Carcinogenesis. 2008;29:434–439. doi: 10.1093/carcin/bgm270. [DOI] [PubMed] [Google Scholar]
- 44.Gong H, Liu W, Zhou J, Xu H. Methylation of gene CHFR promoter in acute leukemia cells. J Huazhong Univ Sci Technolog Med Sci. 2005;25:240–242. doi: 10.1007/BF02828130. [DOI] [PubMed] [Google Scholar]
- 45.Henken FE, Wilting SM, Overmeer RM, van Rietschoten JG, Nygren AO, Errami A, Schouten JP, Meijer CJ, Snijders PJ, Steenbergen RD. Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br J Cancer. 2007;97:1457–1464. doi: 10.1038/sj.bjc.6604055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Homma N, Tamura G, Honda T, Jin Z, Ohmura K, Kawata S, Motoyama T. Hypermethylation of Chfr and hMLH1 in gastric noninvasive and early invasive neoplasias. Virchows Arch. 2005;446:120–126. doi: 10.1007/s00428-004-1146-6. [DOI] [PubMed] [Google Scholar]
- 47.Honda T, Tamura G, Waki T, Kawata S, Nishizuka S, Motoyama T. Promoter hypermethylation of the Chfr gene in neoplastic and non-neoplastic gastric epithelia. Br J Cancer. 2004;90:2013–2016. doi: 10.1038/sj.bjc.6601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, Campan M, Laird PW. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161–170. doi: 10.1038/labinvest.3700707. [DOI] [PubMed] [Google Scholar]
- 49.Kang HC, Kim IJ, Park JH, Shin Y, Park HW, Ku JL, Yang HK, Lee KU, Choe KJ, Park JG. Promoter hypermethylation and silencing of CHFR mitotic stress checkpoint gene in human gastric cancers. Oncol Rep. 2004;12:129–133. [PubMed] [Google Scholar]
- 50.Kawasaki T, Ohnishi M, Nosho K, Suemoto Y, Kirkner GJ, Meyerhardt JA, Fuchs CS, Ogino S. CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci. Mod Pathol. 2008;21:245–255. doi: 10.1038/modpathol.3800982. [DOI] [PubMed] [Google Scholar]
- 51.Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K. The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol. 2006;41:133–139. doi: 10.1007/s00535-005-1732-7. [DOI] [PubMed] [Google Scholar]
- 52.Ludwig AH, Bujko M, Bidzinski M, Kupryjanczyk J. CHFR gene is neither mutated nor hypermethylated in ovarian cancer. Cancer Detect Prev. 2007;31:257–261. doi: 10.1016/j.cdp.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 53.Mizuno K, Osada H, Konishi H, Tatematsu Y, Yatabe Y, Mitsudomi T, Fujii Y, Takahashi T. Aberrant hypermethylation of the CHFR prophase checkpoint gene in human lung cancers. Oncogene. 2002;21:2328–2333. doi: 10.1038/sj.onc.1205402. [DOI] [PubMed] [Google Scholar]
- 54.Morioka Y, Hibi K, Sakai M, Koike M, Fujiwara M, Kodera Y, Ito K, Nakao A. Aberrant methylation of the CHFR gene is frequently detected in noninvasive colorectal cancer. Anticancer Res. 2006;26:4267–4270. [PubMed] [Google Scholar]
- 55.Morioka Y, Hibi K, Sakai M, Koike M, Fujiwara M, Kodera Y, Ito K, Nakao A. Aberrant methylation of the CHFR gene in digestive tract cancer. Anticancer Res. 2006;26:1791–1795. [PubMed] [Google Scholar]
- 56.Sakai M, Hibi K, Kanazumi N, Nomoto S, Inoue S, Takeda S, Nakao A. Aberrant methylation of the CHFR gene in advanced hepatocellular carcinoma. Hepatogastroenterology. 2005;52:1854–1857. [PubMed] [Google Scholar]
- 57.Shibata Y, Haruki N, Kuwabara Y, Ishiguro H, Shinoda N, Sato A, Kimura M, Koyama H, Toyama T, Nishiwaki T, et al. Chfr expression is downregulated by CpG island hypermethylation in esophageal cancer. Carcinogenesis. 2002;23:1695–1699. doi: 10.1093/carcin/23.10.1695. [DOI] [PubMed] [Google Scholar]
- 58.Tokunaga E, Oki E, Nishida K, Koga T, Yoshida R, Ikeda K, Kojima A, Egashira A, Morita M, Kakeji Y, et al. Aberrant hypermethylation of the promoter region of the CHFR gene is rare in primary breast cancer. Breast Cancer Res Treat. 2006;97:199–203. doi: 10.1007/s10549-005-9112-9. [DOI] [PubMed] [Google Scholar]
- 59.Tozawa T, Tamura G, Honda T, Nawata S, Kimura W, Makino N, Kawata S, Sugai T, Suto T, Motoyama T. Promoter hypermethylation of DAP-kinase is associated with poor survival in primary biliary tract carcinoma patients. Cancer Sci. 2004;95:736–740. doi: 10.1111/j.1349-7006.2004.tb03254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Doorn R, Zoutman WH, Dijkman R, de Menezes RX, Commandeur S, Mulder AA, van der Velden PA, Vermeer MH, Willemze R, Yan PS, et al. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. Clin Oncol. 2005;23:3886–3896. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- 61.Yanokura M, Banno K, Kawaguchi M, Hirao N, Hirasawa A, Susumu N, Tsukazaki K, Aoki D. Relationship of aberrant DNA hypermethylation of CHFR with sensitivity to taxanes in endometrial cancer. Oncol Rep. 2007;17:41–48. [PubMed] [Google Scholar]
- 62.Yoshida K, Hamai Y, Suzuki T, Sanada Y, Oue N, Yasui W. DNA methylation of CHFR is not a predictor of the response to docetaxel and paclitaxel in advanced and recurrent gastric cancer. Anticancer Res. 2006;26:49–54. [PubMed] [Google Scholar]
- 63.Milne AN, Sitarz R, Carvalho R, Polak MM, Ligtenberg M, Pauwels P, Offerhaus GJ, Weterman MA. Molecular analysis of primary gastric cancer, corresponding xenografts, and 2 novel gastric carcinoma cell lines reveals novel alterations in gastric carcinogenesis. Hum Pathol. 2007;38:903–913. doi: 10.1016/j.humpath.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi C, Oda Y, Takahira T, Izumi T, Kawaguchi K, Yamamoto H, Tamiya S, Yamada T, Iwamoto Y, Tsuneyoshi M. Aberrant expression of CHFR in malignant peripheral nerve sheath tumors. Mod Pathol. 2006;19:524–532. doi: 10.1038/modpathol.3800548. [DOI] [PubMed] [Google Scholar]