Abstract
p53 tumor suppressor protein negatively regulates cell growth, mainly through the transactivation of its downstream target genes. As a sequence-specific DNA binding transcription factor, p53 specifically binds to a 20-bp consensus motif 5′-PuPuPuC(A/T) (T/A)GPyPyPyPuPuPuC(A/T)(T/A)GPyPyPy-3′. We have now identified, partially purified, and characterized an additional ≈40-kDa nuclear protein, p53CP (p53 competing protein), that specifically binds to the consensus p53 binding sites found in several p53 downstream target genes, including Waf-1, Gadd45, Mdm2, Bax, and RGC. The minimal sequence requirement for binding is a 14-bp motif, 5′-CTTGCTTGAACAGG-3′ [5′-C(A/T)(T/A)GPyPyPyPuPuPuC(A/T)(T/A)G-3′], which includes the central nucleotides of the typical p53 binding site with one mismatch. p53CP and p53 (complexed with antibody) showed a similar binding specificity to Waf-1 site but differences in Gadd45 and T3SF binding. Like p53, p53CP also binds both double- and single-stranded DNA oligonucleotides. Important to note, cell cycle blockers and DNA damaging reagents, which induce p53 binding activity, were found to inhibit p53CP binding in p53-positive, but not in p53-negative, cells. This finding suggested a p53-dependent coordinate regulation of p53 and p53CP in response to external stimuli. p53CP therefore could be a third member of the p53 family, in addition to p53 and p73, a newly identified p53 homolog. p53CP, if sequestering p53 from its DNA binding sites through competitive binding, may provide a novel mechanism of p53 inactivation. Alternatively, p53CP may have p53-like functions by binding and transactivating p53 downstream target genes. Cloning of the p53CP gene ultimately will resolve this issue.
p53, a 53-kDa nuclear protein, is one of the most fascinating molecules in the field of cancer research. The multiple biochemical and biological functions of p53 can be mainly defined from its protein primary structure (1). Structurally, the p53 protein consists mainly of three distinct domains: a transactivation domain at the N terminal, a central specific DNA binding domain, and the oligomerization domain at the C terminal of the molecule (ref. 2 and references therein). As a transcription factor, p53 either transactivates or transrepresses gene expression. Other p53 biochemical activities include inhibition of DNA helicase (3), binding to single-stranded DNA (ssDNA) and stimulating their annealing (refs. 4 and 5 and references therein), and action as exonuclease (6). As a typical tumor suppressor, p53 has been shown to inhibit tumor cell growth and suppress transformation by either inducing G1 arrest or apoptosis (refs. 2 and 7 and references therein). As a “genome guard,” p53 is induced on DNA damage to prevent gene amplification and preserve genetic stability (2, 7–9). In addition, p53 may play a role in differentiation, senescence, and angiogenesis (10–12).
Many of p53 functions were mediated by its transactivation activity. As a transcription factor, p53 binds specifically to its consensus DNA sequence consisting of two copies of the 10-bp motif 5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′, separated by 0–13 bp (13). This sequence has been found in many p53 regulatory genes, including Waf-1/p21 (14), Mdm-2 (15), Bax (16), Gadd45 (17), PCNA (18), thromspodin (19), and type IV collagenase (20) among others. Those are the genes involved in regulation of cell growth and differentiation, apoptosis, DNA damage/replication, and angiogenesis. Because of its biological significance in cell growth control, p53 becomes inactivated by many ways during human carcinogenesis (ref. 21 and references therein). The most common way to inactivate p53 in cells is by point mutations in its DNA binding domain, which were detected in about 50% of all human cancers. Mutant p53 proteins often lose DNA binding and transactivation activity whereas some of them gain oncogenic activity (ref. 22 and references therein). p53 also can be inactivated by binding to and being inhibited by several viral proteins such as simian virus 40 large T antigen, E1B, and human papillomavirus E6 (refs. 2 and 7 and references therein). In some sarcomas, oncoprotein Mdm2, which binds to and inactivates p53 (refs. 2 and 7 and references therein), was overexpressed (23). The third way to inactivate p53, as seen in some breast cancers (24), is through the nuclear exclusion by which p53 is excluded from the nucleus where it normally functions as a transcription factor. Recently, p53 was found to be subjected to redox regulation both in vitro and in vivo (ref. 25 and references therein; ref. 26).
We recently have cloned the gene encoding mouse tissue inhibitor of metalloproteinases-3 (TIMP-3) and its promoter (27, 28), and identified in the promoter a putative p53 binding site (28). We have conducted extensive studies to determine whether TIMP-3 is a p53 downstream target gene and have concluded that TIMP-3 is not subjected to p53 regulation (29). During the course of that study, we identified a nuclear protein that also specifically bound to the p53 sites. We named this protein p53CP (p53 competing protein) for its potential competition with p53 for p53 DNA binding sites. We report here the identification, partial purification, and characterization of p53CP and propose a novel mechanism for p53 inactivation. Identification of such a protein may open a new avenue in study of p53 inactivation and regulation during human carcinogenesis.
MATERIALS AND METHODS
Cell Culture, Nuclear Extract Preparation, and Gel Retardation Assay.
The mouse H-Tx liver tumor cells (30) were grown in 10% DMEM with 1 mM sodium pyruvate. The human Saos-2 cells and p53−/− mouse embryonic fibroblast cells both were grown in 10% DMEM. The human PC-3 cells were grown in 10% RPMI. Nuclear extract from confluent cells was prepared, and 1–5 μg of nuclear protein was subjected to gel retardation assay as detailed previously (26). For determination of binding specificity, 50× excess of unlabeled oligonucleotides was included in some reactions.
Southwestern Analysis.
Each strand (10 pmol) of T3SF oligonucleotides (T3SF.01: 5′-AGGGCTTGCTTGAACAGGGTCT-3′ and T3SF.12: 5′-CCCTAGACCCTGTTCAA GCAAG-3′) was labeled separately with 32P, annealed, ligated, and purified. Nuclear extracts of H-Tx and PC-3 cells (20 μg of protein) were run on 10–20% SDS tricine gel (Novax) and transferred to nitrocellulose membrane. The membrane was incubated sequentially with cold buffer A (50 mM KCl/10 mM Tris⋅HCl, pH 7.5/2 mM EDTA/0.1 mM DTT/1 mM phenylmethylsulfonyl fluoride) containing 4 M urea for 1 hr, buffer A with 2 M urea for 1 hr, and then buffer A without urea for 15 min at 4°C. The membrane was blocked with 5% dry milk in buffer A for 1 hr, washed with the binding buffer used in gel retardation assay, and incubated with labeled T3SF concatemer probe (5 × 106 cpm) in 1 ml of gel retardation buffer for 4 hr with gentle rocking. The membranes were washed with the gel retardation buffer three times (5 min per wash) and exposed to a Kodak film.
RESULTS AND DISCUSSION
Identification of a Nuclear Protein (p53CP) Other Than p53 that Binds to p53 Consensus Sequence.
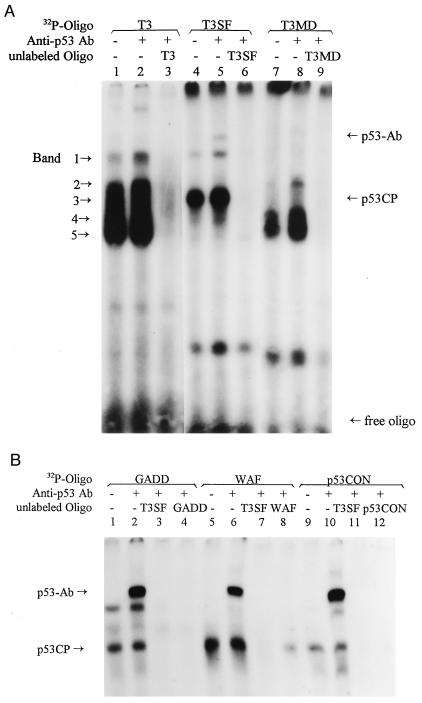
We previously have identified a putative p53 binding site in the promoter of the gene encoding mouse TIMP-3. This site, named as T3 (5′-AGGGCTTGCTTGACGTCCAGAACAGGGTCT-3′) consists of two copies of the p53 binding motif (bold) separated by 8-bp spacer (italics) with two mismatches (underlined) in the second motif (28). To test if p53 binds to this sequence, we performed a gel retardation assay by using the nuclear extract prepared from a mouse liver tumor line, H-Tx. This line is a spontaneously transformed line established by conventional subcultivation of an immortalized liver line (30). H-Tx cells contain a high level of wild-type p53 (31). As shown in Fig. 1A, T3 did not bind to p53 because p53 antibody, pAb421 [a mAb against C terminus of the p53 molecule (amino acids 370–378) that activates p53 DNA binding (32)] did not induce a supershift (lanes 1 and 2). It, however, binds to five nuclear proteins as indicated by arrows. The binding is sequence-specific because the proteins can be largely blocked by unlabeled T3 oligo (lane 3). A closer examination of T3 sequence revealed within the spacer a consensus sequence (TGACGT) for a cellular transcription factor ATF, a cAMP-responsive element binding protein (33). Some of these multiple bands therefore may result from the binding of cAMP-responsive element binding proteins. To test this hypothesis, we deleted the spacer sequence and made the oligo T3SF (spacer free), 5′-AGGGCTTGCTTGAACAGGGTCT-3′ and performed the same gel retardation assay. T3SF is a typical p53 binding site, without the spacer, but with two mismatches (underlined). As shown in Fig. 1A, lanes 4 and 5, T3SF binds to band 3 strongly, as well as to p53 as evident by a supershift induced by pAb421. Band 3, however, cannot be supershifted by pAb421. Again, the binding of the T3SF to band 3 and p53 is sequence-specific, because it can be blocked completely by cold T3SF (lane 6) but not at all by a mutant T3SF (5′-AGGGGTTCCTTGAAGAGCGTCT-3′) that contained a substitution in both motifs of the C to the G and the G to the C at conserved positions 4 and 7 (underlined), respectively (data not shown). The disappearance of bands 2, 4, and 5 indicates that they bind to 8-bp spacer sequence. The identity of band 1 is not clear. It seems to have similar migration to p53 but should not be p53 because (i) T3 did not bind to purified p53 as described previously (29), and (ii) it cannot be supershifted by pAb421 (lane 2). To further test the binding specificity of band 3 to p53 consensus sequence, we made another oligo (T3MD, 5′-AGGGCTTGCTTGACGTCCAGGTCT-3′) for gel retardation assay, which retained the spacer sequence (cAMP-responsive element site) but had the first six nucleotides deleted in the second p53 binding motif. As shown in Fig. 1A, lanes 7–9, T3MD binds specifically to bands 2, 4, and 5, but not to band 3 and p53, further confirming that (i) bands 2, 4, and 5 belong to cAMP-responsive element binding proteins, and (ii) two 10-bp motifs are needed for both p53 and band 3 to bind. These experiments indicate that a nuclear protein, present in H-Tx cells, specifically binds to an artificial p53 binding site, and the binding is dependent on the integrity of the p53 binding motif.
Figure 1.
Identification of a nuclear protein that binds to p53 binding sites. (A) Binding of p53CP to T3SF and (B) binding of p53CP to Gadd45, Waf1, and p53CON. The nuclear extract was prepared from H-Tx mouse liver tumor cells and subjected to gel retardation assay with a poly(I/dC) concentration of 10 μg/ml. In some reactions, p53 antibody, pAb421 was included. The oligo nucleotides used were the following: T3 5′-AGGGCTTGCTTGACGTCCAGAACAGGGTCT-3′, the sequence found in the promoter of mouse TIMP-3; T3SF 5′-AGGGCTTGCTTGAACAGGGTCT-3′; T3MD 5′-AGGGCTTGCTTGACGTCCAGGTCT-3′; GADD 5′-GAACATGTCTAAGCATGCTG-3′; WAF 5′-GAACATGTCCCAACATGTTG-3′; and p53CON 5′-AGACATGCCTAGACATGCCT-3′. The protein complexes bound to T3 oligonucleotide are indicated by arrows. Also indicated are p53-Ab complex, p53CP (band 3), and free probe.
We next examined if the nuclear protein (revealed as band 3 after being complexed with T3SF) also binds to the p53 consensus binding sequences found in genes encoding Gadd45 and Waf-1. Again the nuclear extract from H-Tx cells was used. As shown in Fig. 1B, indeed, both p53 and band 3 bind Gadd45 and Waf-1 (lanes 1 and 5). The binding to p53, but not to band 3, was enhanced and supershifted by pAb421 (lanes 2 and 6). The binding to both p53 and band 3 is sequence-specific. It can be blocked by cold Gadd45 (lane 4) or Waf (lane 8), respectively, as well as by cold T3SF (lanes 3 and 7). We have included another artificial p53 consense sequence (p53CON, without mismatch) and found that it binds to both p53 and band 3 (lanes 9 and 10) with a stronger binding to p53 in the presence of pAb421 (lane 10). Again, binding is specific, it can be specifically blocked by cold p53CON and T3SF (lanes 11 and 12). Band 3 also binds to the p53 site found in the genes encoding Mdm-2, Bax, and RGC (data not shown), and the binding was detected only in nuclear, not cytoplasmic, fractions (data not shown). Because this nuclear protein binds to all tested p53 consensus DNA binding sequences found in p53-regulated genes, it may compete with p53 for specific DNA binding in vivo, we have named it p53CP (p53 competing protein).
p53CP Is Not Another Form of p53.
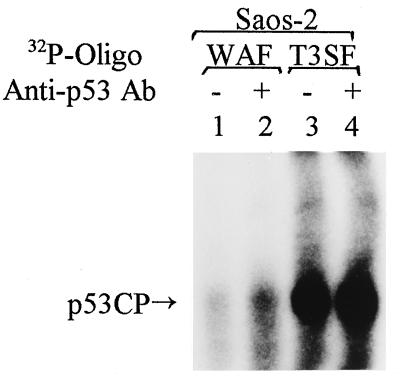
To exclude the possibility that p53CP observed in the gel retardation assay is an alternate form of p53, we used antibody against alternatively spliced p53 (34) and found that the antibody neither blocked nor supershifted p53CP (data not shown). We next used nuclear extract from human Saos-2 cells, a sarcoma cell line with the p53 gene deleted (35). If p53CP is another form of p53, it should not be detected in Saos-2 cells. As shown in Fig. 2, p53CP, but not p53, can be detected with both the Waf or T3SF DNA binding oligonucleotides, and again, the presence of pAb421 did not induce a supershift of p53CP. We further confirmed this result by using nuclear extract prepared from mouse embryonic fibroblast cells derived from p53 knockout mice and from PC-3 human prostate carcinoma cells, having one nucleotide deletion in the p53 coding region (36, 37). Only p53CP, not p53, showed binding in both lines (data not shown). All of these experiments confirmed that p53CP is not p53, but specifically binds to p53 consensus sequences. In addition to Saos-2 and PC-3 cells, p53CP can be detected in multiple human tumor or transformed cell lines originating from prostate (Du145, LNCap), bone (HT1080), cervix (HeLa), and foreskin (Rhek-ras), although the expression level varies (data not shown). p53CP therefore is present in both mouse and human cells.
Figure 2.
p53CP is not another form of p53. Nuclear extract was prepared from p53 negative human Saos-2 cells and subjected to gel retardation assay. The oligo probes used were WAF and T3SF. The position of p53CP is indicated.
p53CP and p53 Have Differential Binding Specificity in a p53 Binding Site-Dependent Manner.
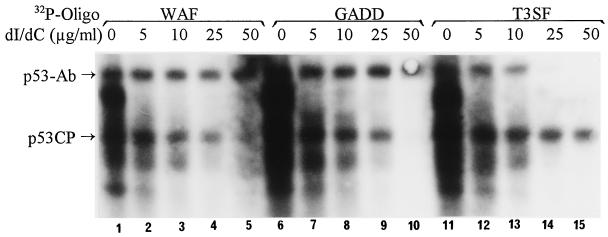
For p53CP to compete with p53 in vivo efficiently, p53CP should have a comparable binding specificity to the p53 DNA binding sites as p53 does. We therefore examined binding specificity of p53CP and p53 by using several p53 consensus sequences. We used oligonucleotides of p53 binding sites found in the genes encoding Waf1 and Gadd45 as well as of T3SF by which we originally identified p53CP. The nuclear extract from H-Tx cells, which contains high levels of both p53CP and p53, was used as a protein source. Because p53 binds weakly to these oligonucleotides in the absence of pAb421 and because pAb421 did not influence the binding of p53CP (see Fig. 1), we included it in the assay. To determine specificity, we used increasing amounts of poly(dI/dC) as nonspecific competitor. As shown in Fig. 3, binding of all three oligonucleotides to both p53CP and p53-Ab complex decreases as the amount of poly(dI/dC) increases. No significant binding difference between p53CP and p53/Ab complex can be seen for Waf binding site (lanes 1–5). However, some differential binding specificity revealed when Gadd and T3SF were used. Compared with p53CP, p53/Ab complex requires more poly(dI/dC) to compete its binding with GADD, suggesting that p53/Ab complex binds to the Gadd site more specifically (lanes 6–10). On the other hand, when the T3SF site was used, p53CP requires more poly(dI/dC) for competition, suggesting a tighter binding (lanes 11–15). These results demonstrated a sequence-dependent difference in binding specificity between p53CP and p53. It implies that some in vivo p53 target genes (such as Gadd45) may be subjected to differential regulation by p53 and p53CP, whereas others (such as Waf-1) may be regulated by the two proteins in a competitive way.
Figure 3.
p53CP and p53-Ab complex showed differential binding specificity to the p53 binding consensus sequences. Nuclear extract was prepared from mouse H-Tx cells and subjected to gel retardation assay with increasing amounts of nonspecific competitor, poly(dI/dC). All gel shift reactions contained p53 antibody, pAb421 to enhance p53 binding. The oligonucleotides used were WAF, GADD, and T3SF. The positions of p53-Ab complex and p53CP are indicated.
p53CP Binds to Both Double-Stranded DNA (dsDNA) and ssDNA But with a Higher Preference for dsDNA.
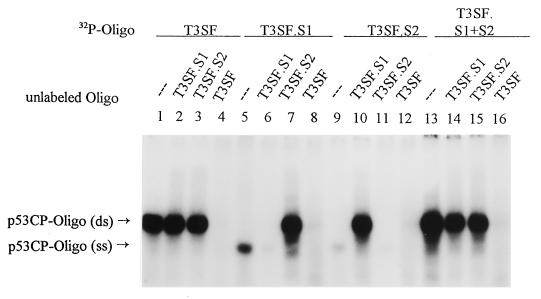
p53 has been shown previously to bind to both dsDNA and ssDNA (refs. 4 and 5 and references therein). To determine if p53CP also binds to ssDNA, we performed a gel retardation assay by using a single-stranded oligo T3SF.S1 (top strand) and T3SF.S2 (complementary/bottom strand) separately or in combination. To avoid interference with p53 binding, we increased the poly(dI/dC) concentration to 50 μg/ml and excluded addition of pAb421. As shown in Fig. 4, T3SF binds to p53CP (lane 1), which cannot be blocked by either single-stranded T3SF.S1 or T3SF.S2 (lanes 2 and 3), but can be blocked by dsT3SF (lane 4). When T3SF.S1 was used as a labeled probe, p53CP binding was visualized as a fast-migrating band (lane 5), which can be blocked by cold T3SF.S1 (lane 6) or dsT3SF (lane 8). Interestingly, inclusion of cold T3SF.S2 caused the formation of dsT3SF, and a typical p53CP band was observed (lane 7). Similarly, when T3SF.S2 was used as a labeled probe, it binds to p53CP but a little weaker (lane 9). The binding could be blocked by cold T3SF.S2 or T3SF (lanes 11 and 12). Again, addition of cold T3SF.S1 formed dsT3SF, and a strong p53CP band was revealed (lane 10). Moreover, we have labeled T3SF.S1 and T3SF.S2 separately and added them to reaction mixture without pre-annealing. It was found that both strands annealed rapidly and bound to p53CP in a double-stranded form (lane 13). The binding can be blocked only by dsT3SF (lane 16), not by its single-stranded oligo (lanes 14 and 15). The results clearly demonstrated that like p53, p53CP can bind both ssDNA and dsDNA with a high preference for dsDNA binding. These data point out an additional interesting feature of p53CP. If it functions like p53, it could be involved in both transactivation (dsDNA binding) and DNA repair (ssDNA binding). On the other hand, if it competes with p53, it may compete with p53 for both of these functions.
Figure 4.
p53CP also binds to ssDNA. Two complementary strands of T3SF (T3SF.S1 and T3SF.S2) were labeled separately and included individually or in combination in the gel retardation reaction [containing 50 μg/ml poly(dI/dC)] with H-Tx cell nuclear extract. The p53CP-oligo (ds) and p53CP-oligo (ss) are indicated.
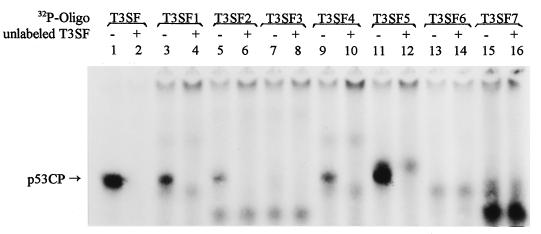
Mapping of the p53CP Binding Core Sequence: 14 Nucleotides Residing at the Center of the p53 Binding Site.
The minimal sequence required for p53 binding is a 20-mer consisting of PuPuPuC(A/T)(T/A)GPyPyPyPuPuPuC(A/T)(T/A)GPyPyPy (13). To further define the biochemical features of p53CP, we performed a deletion/mutation mapping analysis to identify the minimal sequence requirement for p53CP binding. The oligonucleotides used in this assay are listed in Table 1. To keep the oligonucleotides at a length of 20 bases, each deletion was compensated by the addition of a T, either in the 5′ end or the 3′ end. To increase the binding specificity and avoid possible interference from p53 binding, we have increased poly(dI/dC) concentration to 50 μg/ml. Under these conditions, p53 will not bind to T3SF (data not shown). Again, nuclear extract from H-Tx was used. As shown in Fig. 5, deletion of two purine residues from the 5′ end of the p53 site decreased p53CP binding (lane 3, T3SF1), and one additional purine deletion further decreased the binding (lane 5, T3SF2, and lane 9, T3SF4). Deletion of the C at position 4 completely abolished binding, indicating its importance (lane 7, T3SF3). Strong binding restored by the replacement of the first three purine residues by three pyrimidines (lane 11, T3SF5), indicating that the first three purine residues in the p53 DNA binding site are not required for p53CP binding. Deletion of the G at the position 7 or deletion of both the G at position 7 and the C at position 4 from the 3′ end completely abolished p53CP binding (lanes 13 or 15, T3SF6 & 7). All of the p53CP binding can be blocked by cold T3SF (lanes 2, 4, 6, 10, and 12), indicating that the binding is specific, and the band does represent p53CP as we identified previously. We further examined the potential blockage of p53CP binding to T3SF by these deletion mutants. As summarized in Table 1, the stronger the binding, the better is the competition. The binding can be completely blocked by cold T3SF, partially blocked by T3SF5, slightly blocked by T3SF1, T3SF2, and T3SF4, and not at all by T3SF3, T3SF6, and T3SF7 (data not shown). From these data, particularly T3SF5, we deduced that the minimal sequence requirement for the p53CP binding is a 14-bp oligonucleotide 5′-CTTGCTTGAACAGG-3′. We have named this core sequence T3SF-Core. The nucleotides C and G at positions 1, 4, 11, and 14 (underlined) are critical for the binding. Substitution of the C to the G or vice versa at these positions will abolish the binding (data not shown). This 14-bp oligonucleotide may represent a consensus sequence of 5′-C(A/T)(T/A)GPyPyPyPuPuPuC (A/T)(T/A)G-3′, which is the typical p53 DNA binding sequence with the deletions at the first three purines and the last three pyrimidines. It is worth noting that three nonspecific nucleotides are needed in the 5′ end of this core sequence (perhaps at 3′-end also, not tested) to ensure a strong p53CP binding. The T3SF5 (3 nts) has a stronger binding than T3SF1 or T3SF4 (2 nts) than T3SF2 (1 nt) (Fig. 5, compare lane 11 to lanes 3 and 9 and to lane 5). As expected, the binding of p53 to T3SF5 (the first three purines was replaced by three pyrimidines) is reduced significantly even in the presence of pAb421 (data not shown).
Table 1.
Oligonucleotides used to define the core p53CP binding site
| Names | Oligo sequences | p53CP binding | T3SF competition |
|---|---|---|---|
| 147147 | |||
| T3SF | AGGGCTTGCTTGAACAGGGTCT | +++ | +++ |
| T3SF1 | TGCTTGCTTGAACAGGGTCT | ++ | ++ |
| T3SF2 | TCTTGCTTGAACAGGGTCTT | + | + |
| T3SF3 | TTTTGCTTGAACAGGGTTTT | − | − |
| T3SF4 | TTCTTGCTTGAACAGGGTTT | + | + |
| T3SF5 | TTTCTTGCTTGAACAGGTTT | +++ | +++ |
| T3SF6 | TTTTCTTGCTTGAACAGTTTT | − | − |
| T3SF7 | TTTTCTTGCTTGAATTTTT | − | − |
Arabic numerals labeled on the top of sequence indicate the position of each nucleotide in p53 consensus sequence. The two 10-nucleotide motifs are underlined, and the critical C and G at positions 4 and 7 are in bold.
Figure 5.
Mapping the minimal sequence required for p53CP binding. A series of deletion oligonucleotides based on T3SF sequence were synthesized and labeled to define the minimal p53CP binding site. The sequences of these oligonucleotides were listed in Table 1. The gel retardation assay was performed with a poly(dI/dC) concentration of 50 μg/ml. The position of p53CP-DNA complex is indicated.
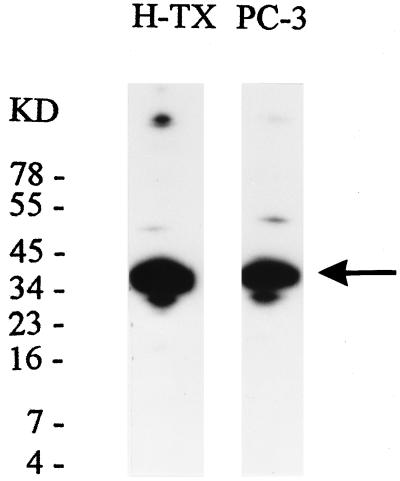
A 40-kDa Nuclear Protein Is p53CP or its DNA Binding Component and Partial Purification of p53CP from Nuclear Extract.
We next determined the actual molecular mass of p53CP by using three different approaches. First, we performed Southwestern analysis. The nuclear extracts from mouse H-Tx cells or human PC-3 (a prostate carcinoma line) was run in a SDS denaturing gel and transferred onto a nitrocellulose membrane. Proteins were refolded and incubated with 32P-labeled T3SF. As shown in Fig. 6, a strong T3SF binding band with a molecular mass ≈40 kDa was observed in both cell lines. Second, we labeled mouse embryonic fibroblast cells from p53−/− mice with [35S]methionine, prepared nuclear extract, and ran a gel retardation assay using 32P-T3SF as the probe. We then cut out the band corresponding to p53CP-DNA complex and resolved the p53CP in a SDS gel. Again, a major 40-kDa band was observed (data not shown). Third, we performed a UV crosslinking assay using H-Tx nuclear extract and 32P-T3SF oligonucleotide, and detected a 46-kDa p53CP-DNA complex. As a positive control, partial purified p53 protein was included in the assay, and a 59-kDa protein-DNA complex was observed (data not shown). These three independent determinations suggest that p53CP has a denatured molecular mass of about 40 kDa. We, however, cannot exclude the possibility that p53CP is a multicomponent proteins with the 40-kDa protein being its DNA binding subunit.
Figure 6.
Determination of the molecular mass of p53CP. Southwestern analysis was performed to determine molecular mass of both mouse and human p53CP, by using nuclear extracts from mouse H-Tx or human PC-3 cells and 32P-T3SF as a hybridization probe. A major band with a size of ≈40 kDa was revealed.
We have partially purified p53CP by a sequence-specific DNA affinity column by using oligo concatemer (made of either T3SF or T3SF-Core oligo)-conjugated Sepharose 4B as described (38, 39). DNA affinity chromatography of H-Tx cell nuclear extracts resulted in elution of p53CP in the fractions of 0.3 M and 0.4 M salt. Similar elution pattern was seen when p53−/− mouse embryonic fibroblast cells were used as source of nuclear extract, and T3SF-Core sequence was prepared for DNA affinity column (data not shown).
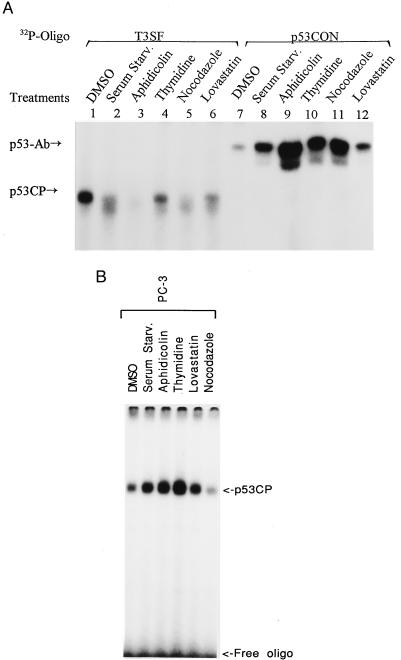
p53 Activation and p53CP Inactivation by Cell Cycle Blocking Reagents.
Finally, we determined the potential coregulation of the activities of p53 and p53CP. If crosstalk exists between these two proteins, they may be regulated in a coordinate way. Because p53 is actively involved in regulation of the various phases of the cell cycle (ref. 2 and references therein), we examined the effect of cell cycle blockers on DNA binding activity of p53 and p53CP. H-Tx cells were synchronized by incubating with the following cell cycle blockers for 24 hr: (i) serum starvation for quiescence G0 phase; (ii) lovastatin for G1 arrest; (iii) aphidicolin and thymidine for S phase arrest; and (iv) nocodazole for G2/M arrest. Nuclear extract then was prepared and subjected to gel retardation assay. We used p53CON and T3SF as binding oligonucleotides because our previous experiments showed that they bound the best (among the oligonucleotides tested) to p53 and p53CP, respectively (Fig. 1 and ref. 29). As shown in Fig. 7A, treatment of H-Tx cells with cell cycle blockers induced p53 DNA binding activity (lanes 7–12). The highest activation was seen with aphidicolin treatment (lane 9). In contrast, cell cycle blocker treatment inactivated p53CP binding (lanes 1–6). A nearly complete inactivation was observed in aphidicolin-treated cells (lane 3). We also have treated cells with DNA damaging reagents (camptothecin and etoposide) and found activation of p53 binding and inactivation of p53CP binding (data not shown). To further examine whether p53CP inactivation by cell cycle blockers was p53-dependent, we performed the same treatment with PC-3 cells, a human prostate carcinoma line with a deletion mutation in the p53 gene (38, 39). As shown in Fig. 7B, in this p53 nonfunctional cell line, cell cycle blockers did not induce an inactivation of p53CP DNA binding activity, they (except nocodazole), in contrast, activate p53CP binding to some extent. The results demonstrated that p53CP inactivation is dependent on p53 activation. Overall, the data presented here highly suggest a p53-dependent coordinate regulation of p53 and p53CP in response to external stimuli. It will be of great interest to identify the reagents that could activate p53CP activity and to see if they would, at the same time, inactivate p53 activity.
Figure 7.
Coordinate regulation of p53 and p53CP in response to cell cycle blockers. Mouse H-Tx cells (A) and human PC-3 cells (B) were subjected to treatment with cell cycle blockers for 24 hr. The concentrations used were dimethyl sulfoxide (for control, 0.1%), aphidicolin (10 μg/ml), thymidine (2.5 mM), nocodazole (4 μg/ml), and lovastatin (1 μg/ml). Nuclear extracts were prepared after treatment and subjected (2 μg) to gel retardation assay. To measure p53CP binding activity, T3SF was used with a poly(dI/dC) concentration of 50 μg/ml (A, lanes 1–6 and B). To determine p53 binding, p53CON oligo was used with a poly(dI/dC) concentration of 10 μg/ml in the presence of p53Ab (A, lanes 7–12). The p53CP and p53-Ab complexed with oligonucleotides are indicated.
What Is the Function of p53CP?
We have identified a nuclear protein (p53CP) that is not p53, but has p53-like DNA binding activity. There are two alternative mechanisms by which p53CP may effect p53 functions. First, p53CP has p53-like functions and regulates a group of p53 target genes to which it has high binding specificity. This hypothesis is supported by: (i) p53 and p53CP showed differential binding specificity in a target sequence-dependent manner; (ii) both proteins bind dsDNA and ssDNA; and (iii) for sequence specific binding, p53CP only requires a 14-bp motif, 5′-CTTGCTTGAACAGG-3′, which is located in the center of the p53 consensus sequence. This finding suggested that p53CP may regulate both p53 responsive and nonresponsive genes. Alternatively, p53CP may compete with p53 for specific sequence DNA binding. This hypothesis is supported by: (i) the minimal sequence requirement for p53CP binding is a 14-bp motif, which centered in the typical p53 binding site, providing a physical basis for its potential competition; (ii) there is a coordinate regulation of DNA binding between p53CP and p53 on external stimuli in a p53-dependent manner; and (iii) it is noteworthy that determinations of the binding specificity were made based on p53CP vs. p53-antibody complex rather than p53 alone. That p53 has very weak binding in the absence of p53 antibody may suggest that it can be easily competed away by p53CP in vivo.
Competition between two transcription factors for specific DNA binding site with biological consequence has been reported previously (40). The binding site of Sp1 and Egr-1 (early-growth-response gene) overlaps in the platelet-derived growth factor B-chain (PDGF-B) promoter. In unstimulated cells, Sp1 occupies the binding site. During acute mechanical injury, Egr-1 expression increased, which displaced Sp-1 from the binding site, and induced expression of PDGF-B (40). The overlapping binding site of Sp-1 and Egr-1 also was found in interleukin 2 receptor-β chain (IL-2Rβ) promoter. In this promoter, however, Sp1 (responsible mainly for constitutive expression) and Egr-1 (for induced expression) cooperate physically and functionally to mediate maximal IL-2Rβ expression (41). Another case of transcription factor competition recently was reported in hepatitis B virus X promoter (42). The binding site for activating transcription factor 2 (ATF2) and activating protein 1 (AP-1) overlaps in the hepatitis B virus E element. The basal transcription mediated by AP-1 was inhibited by ATF2 through the competition for the AP-1 binding site as well as the formation of the ATF2-jun heterodimer (42).
p53 has been involved in many cellular functions, including growth arrest and apoptosis, tumor cell growth inhibition, genome guardian, differentiation, senescence, and angiogenesis (refs. 2, 7, and 10–12 and references therein). Inactivation of p53 was found to be the most frequent genetic alterations in human cancers. About 50% of human cancers contain p53 mutations (22). Competition with p53 for its specific DNA binding by p53CP, as proposed here, may provide a novel mechanism of p53 inactivation. Alteration or overexpression of p53CP may play an important role in the other 50% of human cancers harboring a wild-type p53. Inactivation of p53 by p53CP through the competition for DNA binding can render p53 mutation unnecessary in these cancers. The true biochemical activities and biological functions of p53CP as well as its effect on p53 must, however, await the cloning of the encoding gene.
Subsequent to our submission of this manuscript, Kaghad et al. (43) reported the cloning of p73, a p53 homolog with 63% identity to p53 in DNA binding domain. Although DNA binding of p73 to p53 consensus DNA binding sites was not examined, p73 did transactivate luciferase/chloramphenicol acetyltransferase reporters driven by the p53 site-containing promoters as well as induce expression of endogenous p21, a well-known p53 target gene (44). Cloning of p73 indicates that like other tumor suppressor genes, there exists a family gene for p53 (45). Apparent size difference between p73 and p53CP (a 40-kDa protein) suggests that p53CP could be the third member of p53 family. Cloning of the p53CP encoding gene ultimately will resolve this issue.
Acknowledgments
We thank Dr. M. Kulesz-Martin at the Roswell Park Cancer Institute for the antibody against alternatively spliced p53. We also thank Drs. H. Lu at Oregon Health Science University for helpful discussion and J. Menetski at Parke-Davis for discussion, densitometrical quantitation, and critical reading of the manuscript.
ABBREVIATIONS
- p53CP
p53 competing protein
- TIMP-3
tissue inhibitor of metalloproteinases-3
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
References
- 1.Arrowsmith C H, Morin P. Oncogene. 1996;12:1379–1385. [PubMed] [Google Scholar]
- 2.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 3.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J-M, Wang Z, Friedberg E C, Evans M K, Taffe B G, Bohr V A, Weeda G, Hoeimakers J H J, Forrester K, Harris C C. Nat Genet. 1995;10:188–193. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 4.Bakalkin G, Yakovleva T, Selivanova G, Magnusson K P, Szekely L, Kiseleva E, Klein G, Terenius L, Wiman K G. Proc Natl Acad Sci USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selivanova G, Wiman K G. Adv Cancer Res. 1995;66:143–180. doi: 10.1016/s0065-230x(08)60253-5. [DOI] [PubMed] [Google Scholar]
- 6.Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 7.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 8.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 10.Rotter V, Aloni-Grinstein R, Schwartz D, Elkind N B, Simons A, Wolkowicz R, Lavigne M, Beserman P, Kapon A, Goldfinger N. Semin Cancer Biol. 1994;5:229–236. [PubMed] [Google Scholar]
- 11.Vojta P J, Barrett J C. Biochim Biophys Acta. 1995;1242:29–41. doi: 10.1016/0304-419x(95)00002-w. [DOI] [PubMed] [Google Scholar]
- 12.Bouck N. Biochim Biophys Acta. 1996;1287:63–66. doi: 10.1016/0304-419x(96)00005-4. [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W S, Kern S, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 14.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin E, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 15.Barak Y, Juven T, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 17.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 18.Morris G F, Bischoff J R, Mathews M B. Proc Natl Acad Sci USA. 1996;93:895–899. doi: 10.1073/pnas.93.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dameron K M, Volpert O V, Tainsky M A, Bouck N. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 20.Bian J, Sun Y. Mol Cell Biol. 1997;17:6330–6338. doi: 10.1128/mcb.17.11.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang F, Syrjanen S, Syrjanen K. J Clin Oncol. 1995;13:1009–1022. doi: 10.1200/JCO.1995.13.4.1009. [DOI] [PubMed] [Google Scholar]
- 22.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 23.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Nature (London) 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 24.Moll U M, Riou G, Levin A J. Proc Natl Acad Sci USA. 1992;89:7262–7266. doi: 10.1073/pnas.89.15.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Oberley L W. Free Rad Biol Med. 1996;21:335–348. doi: 10.1016/0891-5849(96)00109-8. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, Bian J, Wang Y, Jacobs C. Oncogene. 1997;14:385–393. doi: 10.1038/sj.onc.1200834. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Hegamyer G, Colburn N H. Cancer Res. 1994;54:1139–1144. [PubMed] [Google Scholar]
- 28.Sun Y, Hegamyer G, Kim H, Sithanandam K, Li H, Watts R, Colburn N H. J Biol Chem. 1995;270:19312–19319. doi: 10.1074/jbc.270.33.19312. [DOI] [PubMed] [Google Scholar]
- 29.Bian J, Jacobs C, Wang Y, Sun Y. Carcinogenesis. 1996;17:2559–2562. doi: 10.1093/carcin/17.12.2559. [DOI] [PubMed] [Google Scholar]
- 30.Sun Y, Oberley L W, Oberley T D, Elwell J H, Sierra-Rivera E. Carcinogenesis. 1993;14:1457–1463. doi: 10.1093/carcin/14.7.1457. [DOI] [PubMed] [Google Scholar]
- 31.Sun Y, Hegamyer G, Nakamura K, Kim H, Oberley L W, Colburn N H. Int J Cancer. 1993;55:952–956. doi: 10.1002/ijc.2910550613. [DOI] [PubMed] [Google Scholar]
- 32.Hupp T R, Meek D W, Midgley C A, Lane D P. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 33.Deutsch P J, Hoeffler J P, Jameson J L, Lin J, Habener J F. J Biol Chem. 1988;263:18466–18472. [PubMed] [Google Scholar]
- 34.Kulesz-Martin M, Lisafeld B, Huang H, Kisiel N D, Lee L. Mol Cell Biol. 1994;14:1698–1708. doi: 10.1128/mcb.14.3.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuda H, Miller C, Koeffler H P, Battifora H, Cline M J. Proc Natl Acad Sci USA. 1987;84:7716–7719. doi: 10.1073/pnas.84.21.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaacs W B, Carter B S, Ewing C M. Cancer Res. 1991;51:4716–4720. [PubMed] [Google Scholar]
- 37.Borner M B, Myers C E, Sartor O, Sei Y, Toko T, Trepel J B, Schneider E. Cancer Res. 1995;55:2122–2128. [PubMed] [Google Scholar]
- 38.Kadonaga J T, Tjian R. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 40.Khachigian L M, Lindner V, Williams A J, Collins T. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 41.Lin J-X, Leonard W J. Mol Cell Biol. 1997;17:3714–3722. doi: 10.1128/mcb.17.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi C Y, Choi B H, Park G T, Rho H M. J Biol Chem. 1997;272:16934–16939. doi: 10.1074/jbc.272.27.16934. [DOI] [PubMed] [Google Scholar]
- 43.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, Ferrara P, McKeon F, Caput D. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 44.Jost C A, Marin M C, Kaelin W G., Jr Nature (London) 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 45.Oren M. Cell. 1997;90:829–832. doi: 10.1016/s0092-8674(00)80347-5. [DOI] [PubMed] [Google Scholar]