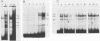Abstract
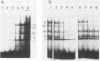
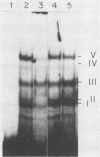
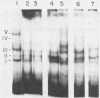
Long terminal repeat (LTR) sequences of murine leukemia viruses (MLVs) have been demonstrated to be mainly responsible for the pathogenic differences in these retroviruses. A region of the LTR which is downstream of the enhancer elements has been shown to contribute both to enhancer activity as well as to disease specificity of MLVs. We have identified protein-DNA complexes generated by this region of a lymphomagenic MLV (MCF13) and one which is nonpathogenic (Akv). One protein-DNA complex we have observed for this region is unique to MCF13 DNA sequences. Detection of protein involved in this unique MCF13 complex in different cell lines revealed that it was ubiquitous.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg P. E., Popovic Z., Anderson W. F. Promoter dependence of enhancer activity. Mol Cell Biol. 1984 Aug;4(8):1664–1668. doi: 10.1128/mcb.4.8.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösze Z., Thiesen H. J., Charnay P. A transcriptional enhancer with specificity for erythroid cells is located in the long terminal repeat of the Friend murine leukemia virus. EMBO J. 1986 Jul;5(7):1615–1623. doi: 10.1002/j.1460-2075.1986.tb04404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran T., Franza B. R., Jr Fos and Jun: the AP-1 connection. Cell. 1988 Nov 4;55(3):395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984 Nov;52(2):448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura F. K. Nuclear activity from F9 embryonal carcinoma cells binding specifically to the enhancers of wild-type polyoma virus and PyEC mutant DNAs. Nucleic Acids Res. 1986 Apr 11;14(7):2845–2861. doi: 10.1093/nar/14.7.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E., Li Y., Fredrickson T. N., Hartley J. W., Hopkins N. Distinct segments within the enhancer region collaborate to specify the type of leukemia induced by nondefective Friend and Moloney viruses. J Virol. 1989 Jan;63(1):328–337. doi: 10.1128/jvi.63.1.328-337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Anklesaria P., Sakakeeny M. A., Greenberger J. S. Enhancer sequences of a retroviral vector determine expression of a gene in multipotent hematopoietic progenitors and committed erythroid cells. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8662–8666. doi: 10.1073/pnas.84.23.8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland C. A., Wozney J., Chatis P. A., Hopkins N., Hartley J. W. Construction of recombinants between molecular clones of murine retrovirus MCF 247 and Akv: determinant of an in vitro host range property that maps in the long terminal repeat. J Virol. 1985 Jan;53(1):152–157. doi: 10.1128/jvi.53.1.152-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollon T., Yoshimura F. K. Mapping of functional regions of murine retrovirus long terminal repeat enhancers: enhancer domains interact and are not independent in their contributions to enhancer activity. J Virol. 1989 Aug;63(8):3353–3361. doi: 10.1128/jvi.63.8.3353-3361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C., Albert V. R., Elsholtz H. P., Lu L. I., Rosenfeld M. G. Activation of cell-specific expression of rat growth hormone and prolactin genes by a common transcription factor. Science. 1988 Mar 18;239(4846):1400–1405. doi: 10.1126/science.2831625. [DOI] [PubMed] [Google Scholar]
- Oliff A., Signorelli K., Collins L. The envelope gene and long terminal repeat sequences contribute to the pathogenic phenotype of helper-independent Friend viruses. J Virol. 1984 Sep;51(3):788–794. doi: 10.1128/jvi.51.3.788-794.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Haseltine W. A., Lenz J., Ruprecht R., Cloyd M. W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985 Sep;55(3):862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C., Mermod N., Andrews P. C., Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988 Jul 21;334(6179):218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- Short M. K., Okenquist S. A., Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987 Apr;61(4):1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]