Abstract
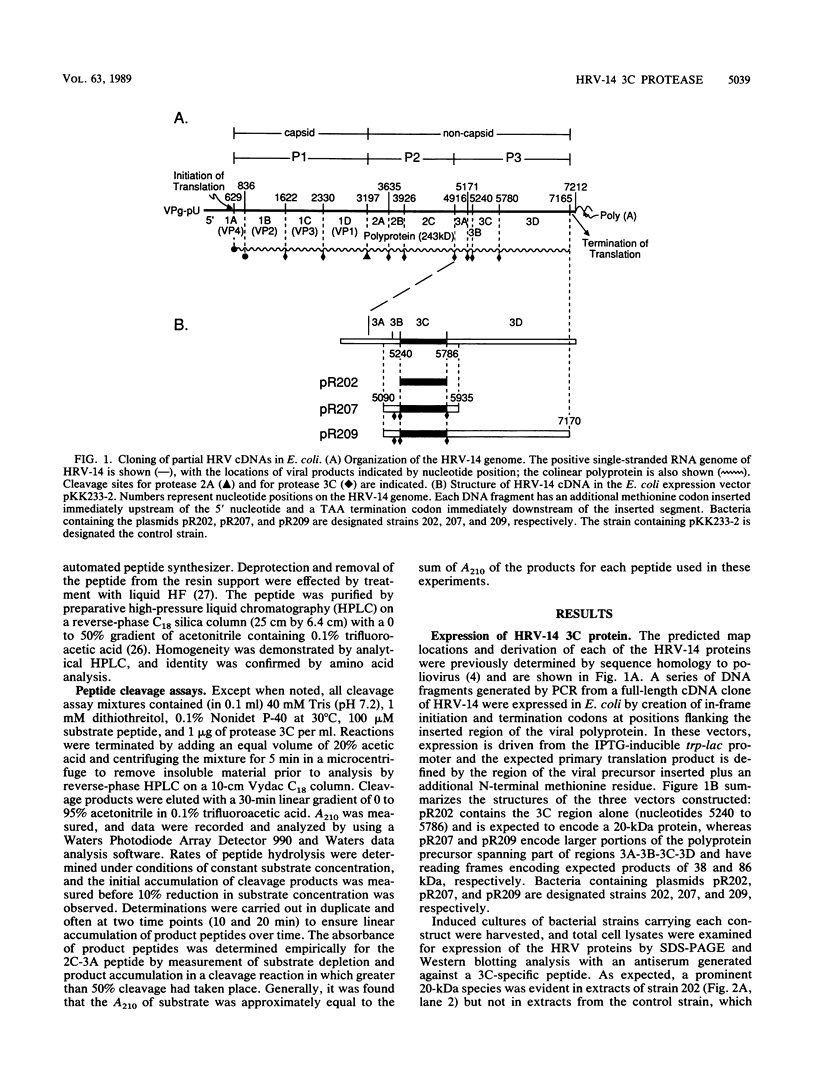
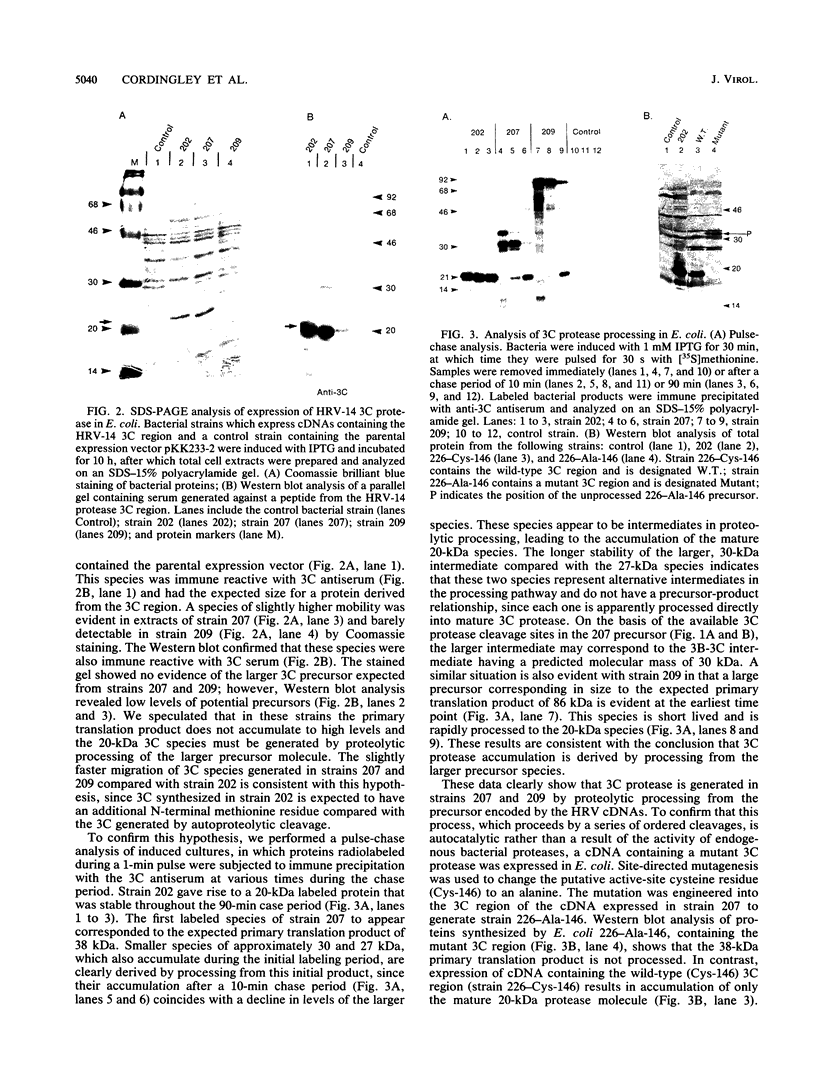
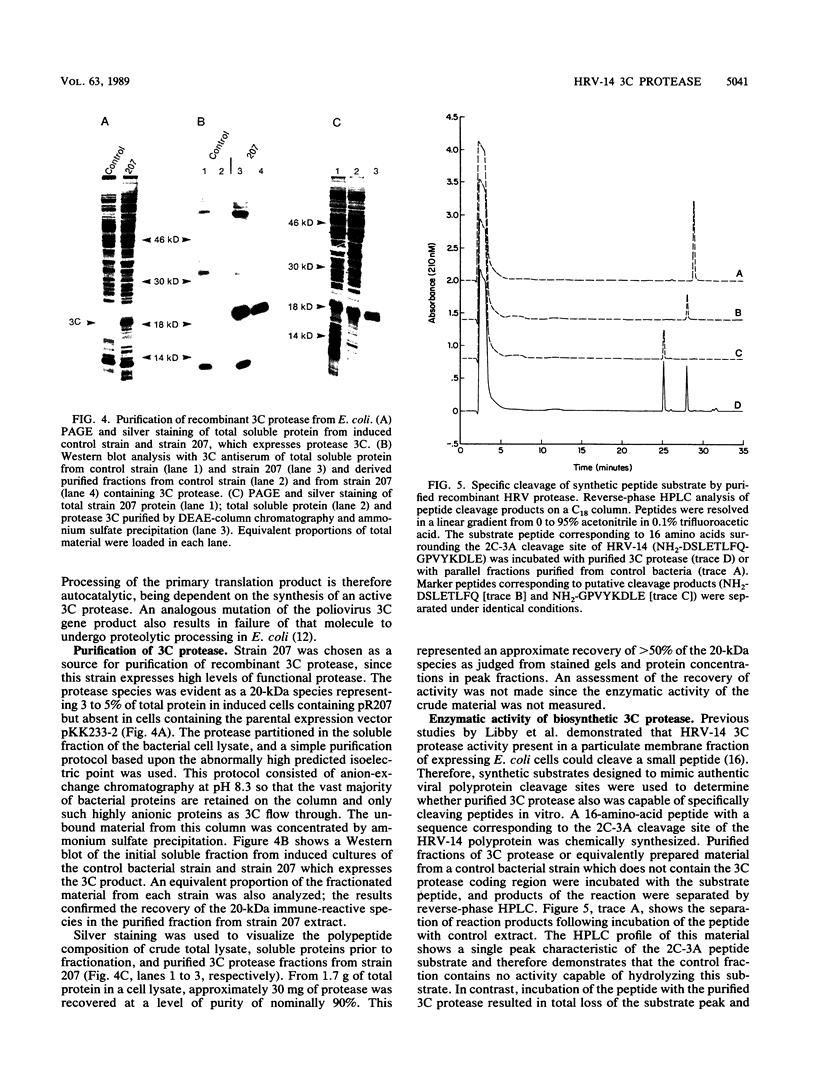
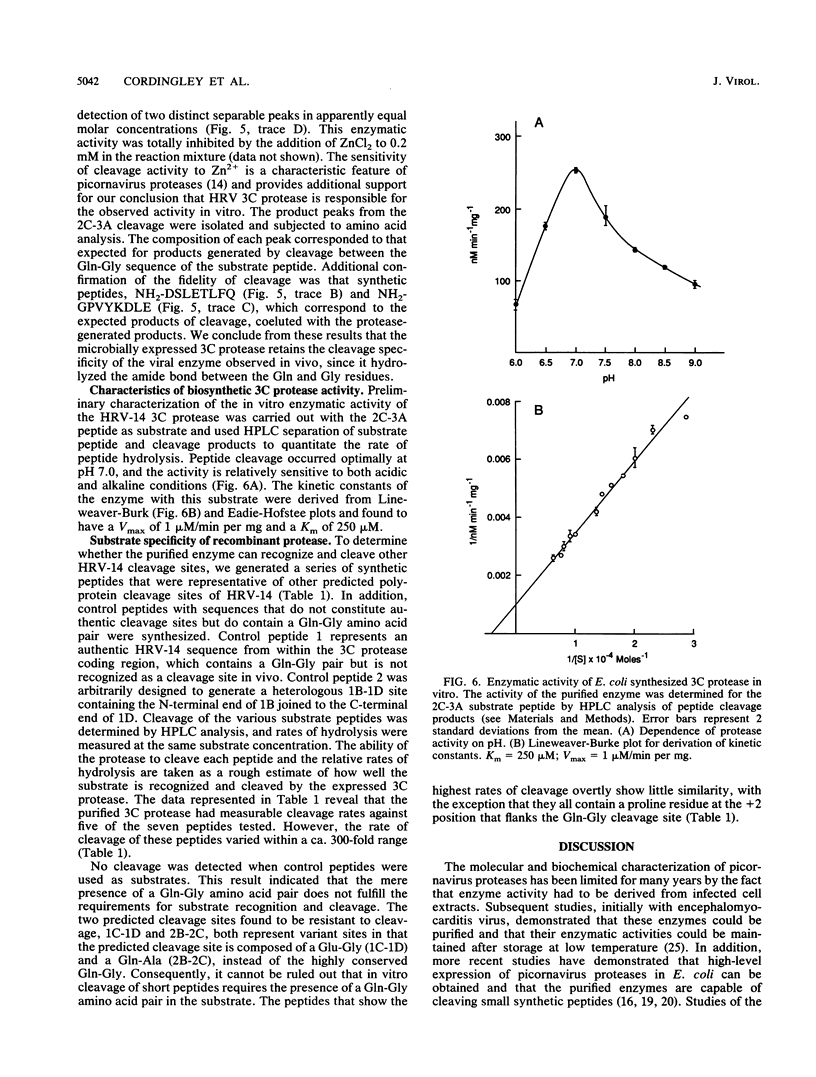
The 3C region of human rhinovirus 14 was expressed in Escherichia coli. The microbially synthesized protease was functional, since the expressed precursor underwent autoproteolytic processing to generate mature molecules of the expected molecular weight and antigenicity. Mutation of the putative active-site Cys-146 residue to an alanine resulted in the synthesis of unprocessed precursor molecules. Large quantities of the 20-kilodalton protease were purified by a simple purification protocol, and the resulting molecule was shown to be biologically active in vitro against synthetic peptides corresponding to the 2C-3A cleavage site. This site was cleaved with high efficiency and fidelity and was used to generate kinetic data on the 3C protease. The protease exhibited sensitivity to Zn2+, was capable of cleaving five of seven rhinovirus cleavage site peptides tested with variable efficiency, and could distinguish authentic substrate peptides from control peptides containing the dipeptide cleavage sequence pair Gln-Gly.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold E., Luo M., Vriend G., Rossmann M. G., Palmenberg A. C., Parks G. D., Nicklin M. J., Wimmer E. Implications of the picornavirus capsid structure for polyprotein processing. Proc Natl Acad Sci U S A. 1987 Jan;84(1):21–25. doi: 10.1073/pnas.84.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan J. F., Fletterick R. J. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H. D., Sarnow P., Baltimore D. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J Virol. 1986 Dec;60(3):1040–1049. doi: 10.1128/jvi.60.3.1040-1049.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan P. L., Mizutani S., Colonno R. J. Molecular cloning and complete sequence determination of RNA genome of human rhinovirus type 14. Proc Natl Acad Sci U S A. 1985 Feb;82(3):732–736. doi: 10.1073/pnas.82.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah K. C., Sankar S., Porter A. G. Expression and processing of human rhinovirus type 14 polypeptide precursors in Escherichia coli maxicells. Gene. 1988 Sep 30;69(2):265–274. doi: 10.1016/0378-1119(88)90437-4. [DOI] [PubMed] [Google Scholar]
- Dewalt P. G., Lawson M. A., Colonno R. J., Semler B. L. Chimeric picornavirus polyproteins demonstrate a common 3C proteinase substrate specificity. J Virol. 1989 Aug;63(8):3444–3452. doi: 10.1128/jvi.63.8.3444-3452.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Sigal I. S., Poe M., Scolnick E. M. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Blinov V. M., Donchenko A. P. Poliovirus-encoded proteinase 3C: a possible evolutionary link between cellular serine and cysteine proteinase families. FEBS Lett. 1986 Jan 6;194(2):253–257. doi: 10.1016/0014-5793(86)80095-3. [DOI] [PubMed] [Google Scholar]
- Hamparian V. V., Colonno R. J., Cooney M. K., Dick E. C., Gwaltney J. M., Jr, Hughes J. H., Jordan W. S., Jr, Kapikian A. Z., Mogabgab W. J., Monto A. A collaborative report: rhinoviruses--extension of the numbering system from 89 to 100. Virology. 1987 Jul;159(1):191–192. doi: 10.1016/0042-6822(87)90367-9. [DOI] [PubMed] [Google Scholar]
- Hanecak R., Semler B. L., Anderson C. W., Wimmer E. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc Natl Acad Sci U S A. 1982 Jul;79(13):3973–3977. doi: 10.1073/pnas.79.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanoff L. A., Towatari T., Ray J., Korant B. D., Petteway S. R., Jr Expression and site-specific mutagenesis of the poliovirus 3C protease in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5392–5396. doi: 10.1073/pnas.83.15.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore J., De Geus B., Jackson R. J., Pouwels P. H., Enger-Valk B. E. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J Gen Virol. 1988 Jul;69(Pt 7):1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Korant B. D., Butterworth B. E. Inhibition by zinc of rhinovirus protein cleavage: interaction of zinc with capsid polypeptides. J Virol. 1976 Apr;18(1):298–306. doi: 10.1128/jvi.18.1.298-306.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Libby R. T., Cosman D., Cooney M. K., Merriam J. E., March C. J., Hopp T. P. Human rhinovirus 3C protease: cloning and expression of an active form in Escherichia coli. Biochemistry. 1988 Aug 23;27(17):6262–6268. doi: 10.1021/bi00417a010. [DOI] [PubMed] [Google Scholar]
- Mizutani S., Colonno R. J. In vitro synthesis of an infectious RNA from cDNA clones of human rhinovirus type 14. J Virol. 1985 Nov;56(2):628–632. doi: 10.1128/jvi.56.2.628-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin M. J., Harris K. S., Pallai P. V., Wimmer E. Poliovirus proteinase 3C: large-scale expression, purification, and specific cleavage activity on natural and synthetic substrates in vitro. J Virol. 1988 Dec;62(12):4586–4593. doi: 10.1128/jvi.62.12.4586-4593.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallai P. V., Burkhardt F., Skoog M., Schreiner K., Bax P., Cohen K. A., Hansen G., Palladino D. E., Harris K. S., Nicklin M. J. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J Biol Chem. 1989 Jun 15;264(17):9738–9741. [PubMed] [Google Scholar]
- Pallansch M. A., Kew O. M., Semler B. L., Omilianowski D. R., Anderson C. W., Wimmer E., Rueckert R. R. Protein processing map of poliovirus. J Virol. 1984 Mar;49(3):873–880. doi: 10.1128/jvi.49.3.873-880.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A. C. Picornaviral processing: some new ideas. J Cell Biochem. 1987 Mar;33(3):191–198. doi: 10.1002/jcb.240330306. [DOI] [PubMed] [Google Scholar]
- Palmenberg A. C., Rueckert R. R. Evidence for intramolecular self-cleavage of picornaviral replicase precursors. J Virol. 1982 Jan;41(1):244–249. doi: 10.1128/jvi.41.1.244-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks G. D., Baker J. C., Palmenberg A. C. Proteolytic cleavage of encephalomyocarditis virus capsid region substrates by precursors to the 3C enzyme. J Virol. 1989 Mar;63(3):1054–1058. doi: 10.1128/jvi.63.3.1054-1058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks G. D., Duke G. M., Palmenberg A. C. Encephalomyocarditis virus 3C protease: efficient cell-free expression from clones which link viral 5' noncoding sequences to the P3 region. J Virol. 1986 Nov;60(2):376–384. doi: 10.1128/jvi.60.2.376-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier J., McClintock R., Galyean R., Anderson H. Reversed-phase high-performance liquid chromatography: preparative purification of synthetic peptides. J Chromatogr. 1984 Apr 24;288(2):303–328. doi: 10.1016/s0021-9673(01)93709-4. [DOI] [PubMed] [Google Scholar]
- Sakakibara S., Shimonishi Y., Kishida Y., Okada M., Sugihara H. Use of anhydrous hydrogen fluoride in peptide synthesis. I. Behavior of various protective groups in anhydrous hydrogen fluoride. Bull Chem Soc Jpn. 1967 Sep;40(9):2164–2167. doi: 10.1246/bcsj.40.2164. [DOI] [PubMed] [Google Scholar]
- Stanway G., Hughes P. J., Mountford R. C., Minor P. D., Almond J. W. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984 Oct 25;12(20):7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. F., Shaw E. N., Stewart M. L., Maizel J. V., Jr Inhibition of cleavage of large poliovirus-specific precursor proteins in infected HeLa cells by inhibitors of proteolytic enzymes. J Virol. 1972 Oct;10(4):880–884. doi: 10.1128/jvi.10.4.880-884.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H., Nicklin M. J., Murray M. G., Anderson C. W., Dunn J. J., Studier F. W., Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986 Jun 6;45(5):761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Dewalt P. G., Johnson V. H., Lamb J. G., Semler B. L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988 Sep;166(1):265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- Ypma-Wong M. F., Filman D. J., Hogle J. M., Semler B. L. Structural domains of the poliovirus polyprotein are major determinants for proteolytic cleavage at Gln-Gly pairs. J Biol Chem. 1988 Nov 25;263(33):17846–17856. [PubMed] [Google Scholar]