Abstract
We have isolated a novel cDNA, that appears to represent a new class of ion channels, by using the yeast two-hybrid system and the SH3 domain of the neural form of Src (N-src) as a bait. The encoded polypeptide, BCNG-1, is distantly related to cyclic nucleotide-gated channels and the voltage-gated channels, Eag and H-erg. BCNG-1 is expressed exclusively in the brain, as a glycosylated protein of ≈132 kDa. Immunohistochemical analysis indicates that BCNG-1 is preferentially expressed in specific subsets of neurons in the neocortex, hippocampus, and cerebellum, in particular pyramidal neurons and basket cells. Within individual neurons, the BCNG-1 protein is localized to either the dendrites or the axon terminals depending on the cell type. Southern blot analysis shows that several other BCNG-related sequences are present in the mouse genome, indicating the emergence of an entire subfamily of ion channel coding genes. These findings suggest the existence of a new type of ion channel, which is potentially able to modulate membrane excitability in the brain and could respond to regulation by cyclic nucleotides.
Ion channels are a diverse group of proteins that regulate the flow of ions across cellular membranes. In the nervous system, ion channel activity has evolved into a rapid and accurate system for intercellular communication. The electrical excitability characteristic of each neuron is in part determined by the set of channels it expresses. However, cells are also able to regulate the activity of individual channels in response to physiological or developmental events, and there is growing evidence that ion channels can be the site of integration of multiple electrical and biochemical pathways.
In vivo, ion channels appear to be multimeric proteins with the potential for heterogeneity arising from the combinatorial assembly of different pore-forming and auxiliary subunits (1). In addition to auxiliary (β) subunits, pore-forming subunits can interact with a variety of intracellular proteins, including G-proteins, cytoskeleton-associated proteins, and protein kinases (2).
Several classes of ion channels bind directly, and are regulated by, second messenger molecules such as cyclic nucleotides (3–5) or Ca2+ (6, 7). Channels with this property can be key elements in the control of neuronal signaling, as they couple biochemical cascades with electrical activity. Cyclic nucleotide-gated (CNG) channels play a distinct role both in visual and olfactory signal transduction; their recent identification in the hippocampus and other regions of the brain, where cAMP and cGMP are known to mediate different forms of synaptic plasticity (8–10), suggests that CNG channels may also contribute to the regulation of excitability in central neurons (11, 12).
We are interested in defining signal transduction pathways that contribute to the control of synaptic strength in the brain. In an effort to identify the biochemical targets of Src-family tyrosine kinases in the central nervous system, we used the yeast two-hybrid system to clone proteins that could interact with the neural specific form of Src (13, 14). As a result of this screening we have isolated a new protein, which we have named BCNG-1 (for brain cyclic nucleotide gated 1).
BCNG-1 has been identified as an ion channel protein based on its sequence homology to voltage-gated potassium channels, CNG channels, and plant inward rectifiers. Southern blot analysis suggests that this is the first member of a new family of proteins. BCNG-1 is expressed exclusively in the brain and is preferentially localized to the processes of subsets of neurons in the neocortex, cerebellar cortex, and hippocampus. The specific localization pattern of BCNG-1 and the potential for a direct interaction with cyclic nucleotides suggest that it may represent an important component in the expression of inter- and intracellular signaling systems.
MATERIALS AND METHODS
Two-Hybrid Interaction Screen.
Standard manipulations of Escherichia coli, Saccharomyces cerevisiae, proteins, and nucleic acids, including recombinant DNA procedures, were performed essentially as described (15, 16).
Two-hybrid screens were performed according to Zervos et al. (17). Plasmids (pEG202, pJG4–5, pJK103) and S. cerevisiae EGY48 (MATa trp1 ura3 his3 LEU2::pLexAop6-LEU2) were generously provided by R. Brent. The N-src fusion in plasmid pEG202 contains amino acids 83–147 from the mouse N-src sequence (14). The cDNA fusion library was constructed in plasmid pJG4–5, using poly(A)+ RNA from an adult mouse brain (C57BL/6J) and the GIBCO/BRL SuperScript II synthesis kit (random hexamers priming).
Colonies that were visible after 3 days of growth on Trp−-Ura−-His−-Leu−-galactose+ medium were assayed for β-galactosidase activity using a filter lift assay (18). The positively reacting fusion products were isolated and tested for specificity following retransformation into an independent yeast strain.
λgt10 Library Screening and Northern/Southern Blot Hybridization.
For the isolation of the 5′ end of the BCNG-1 cDNA, we first performed nested PCR reactions on the pJG4–5 library, using oligonucleotides derived from the pJG-d5 sequence. The longest amplification product, containing amino acids 207–430 from the BCNG-1 sequence (Fig. 1A), was used as a probe to screen a mouse brain cDNA library in λgt10 (CLONTECH, ML3000a). Positively reacting clones were screened by PCR, subcloned, and sequenced. The longest extension contained amino acids 1–463 of the BCNG-1 sequence. The overlapping region of this insert with the insert contained in clone pJG-d5 (amino acids 404–463) includes a BglII site, which was used to join the 5′ and 3′ fragments of the BCNG-1 cDNA in plasmid pSD64TF for in vitro transcription (MessageMachine, Ambion, Austin, TX) and translation (In Vitro Express, Stratagene).
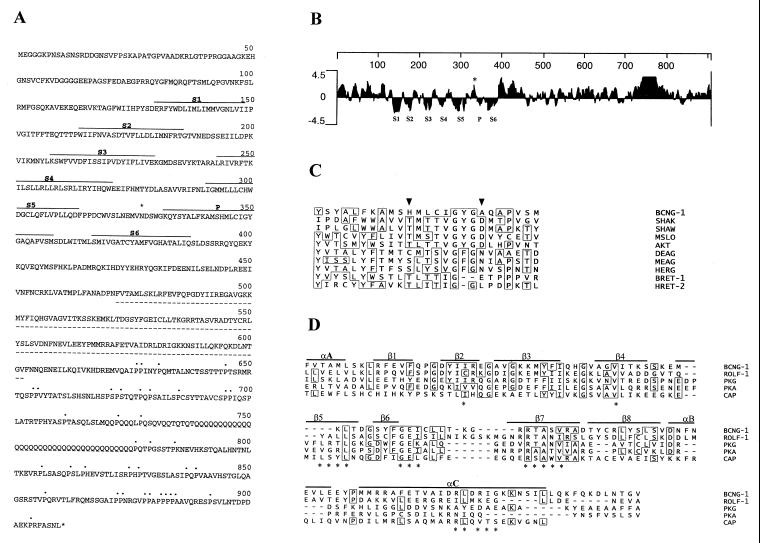
Figure 1.
Primary structure of BCNG-1. (A) Deduced amino acid sequence encoded by the BCNG-1 cDNA. Hydrophobic domains homologous to the six transmembrane domains (S1–S6) and pore (P) of K+ channels are indicated (———). The putative CNBs (- - - - -), C-terminal prolines (⋅⋅⋅), and the consensus N-glycosylation site with presumptive extracellular localization (∗) are also marked. (B) Kyte and Doolittle hydropathy plot of the predicted amino acid sequence of BCNG-1. Hydrophobic regions corresponding to S1 through S6 and the P region lie below the zero line while the N-glycosylation site (∗) is in a hydrophilic region between S5 and P. Numbering (top line) indicates position in the BCNG-1 sequence. Profile generated with a window size of seven residues. (C) Multiple alignment of the putative P region of BCNG-1 with the P regions of Drosophila Shaker (SHAK, refs. 32 and 33), Drosophila Shaw (SHAW, ref. 31), mouse Slo (MSLO, ref. 46), Arabidopsis AKT1 (AKT, ref. 26), Drosophila Eag (DEAG, ref. 34), mouse Eag (MEAG, ref. 26), human Erg (HERG, ref. 26), α-subunit of bovine retinal CNG channel (BRET-1, 36), and β-subunit of human retinal CNG channel (HRET-2, 27). Arrowheads mark residues 344 and 352 (see Results). (D) Alignment of the CNBs of BCNG-1 with the corresponding site in the rat olfactory CNG channel (ROLF-1, ref. 47), bovine cGMP-dependent protein kinase (PKG, ref. 25), bovine cAMP-dependent protein kinase (PKA, ref. 25), and catabolite activator protein of Escherichia coli (CAP, ref. 25). Continuous lines mark α-helical (α) and β-strand (β) elements of the secondary structure of catabolite gene activator protein, while asterisks indicate specific amino acids that appear to lie close to the cAMP molecule in the catabolite gene activator protein crystal structure.
PCR-generated cDNA fragments corresponding to amino acids 6–131 (λgt10-derived 5′ sequence) and 594–720 (pJG4–5 derived 3′ sequence) were used to probe a Multiple Tissue Northern Blot (CLONTECH, 7762–1) (15).
For Southern blots, a Mouse Geno-Blot (CLONTECH, 7650–1) was probed using a PCR generated cDNA fragment (B1-T) corresponding to amino acids 270–463 (15). Blots were hybridized at 65°C (5× standard saline citrate (SSC; 1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) buffer in aqueous solution) and washed as described in figure legends. The melting temperature for the B1-T probe was calculated according to melting temperature = 81.5°C + 16.6(logM) + 0.41(%GC) − (675/L), where M is the cation concentration and L is the probe length in bp.
Immunochemistry and in Situ Hybridization.
Glutathione S-transferase (GST)-fusion proteins were created by subcloning fragments q1, q2, or d5 (see Results) in plasmid pGEX-1λT (Pharmacia), followed by induction and purification essentially as described (19). Fusion proteins were eluted in PBS and injected into rabbits as a 1:1 suspension with Freund adjuvant (Pierce). Antisera were prepared and tested essentially as described (20).
For Western Blot analysis, mouse brain extracts (21) were separated on a 10% SDS/PAGE and electroblotted to polyvinylidene difluoride membranes (Immobilon-P, Millipore) (21) after deglycosylation with N-glycosidase F (20 units/ml, Boehringer Mannheim) where indicated. Blocking and antibody incubations were done in TBST (10 mM Tris, pH 7.5/150 mM NaCl/0.1% Tween-20) + 2% BSA. The αq1 and αq2 antisera were used at a 1:1000 dilution. Secondary anti-rabbit antibodies coupled to alkaline phosphatase (Bio-Rad) were used at a 1:5000 dilution, and the bands were visualized by incubation in nitroblue tetrazolium/5-bromo-4-chloro-3-indoyl phosphate p-toluidine salt (Boehringer Mannheim).
For immunohistochemistry, 20 μm cryostat sections of mouse brain (fixed in 4% paraformaldehyde/PBS, quenched in 50 mM NH4Cl/PBS), were blocked (10% goat serum/0.1% saponin in PBS), and then exposed to αq1 or αq2 antisera (diluted 1:400 in blocking solution). After washing in PBS + 0.1% saponin, sections were incubated with Cy3-conjugated goat anti-rabbit F(ab′)2 fragments (Jackson ImmunoResearch) diluted 1:200 in blocking solution.
In situ hybridization was performed essentially as described (22) using oligonucleotide probes labeled by 3′ tailing with (α-35S)thio-dATP and terminal transferase (Boehringer Mannheim) to a specific activity of 5 × 108 cpm/μg. Hybridizations were carried out at 37°C. Slides were washed at 60°C in 0.2× SSC and exposed to film for 2 weeks.
RESULTS
Isolation of BCNG-1 by Interactive Cloning with the N-src SH3 Domain in a Yeast Two-Hybrid Screen.
The src gene expresses an alternatively spliced form (N-src or pp60c-src(+)), which is specific for neuronal cells and has an increased kinase activity (13). The N-src protein differs from the nonneuronal form (c-src or pp60c-src) by an insertion of six amino acids in the region corresponding to the Src homology 3 (SH3) domain of the protein (14). As SH3 domains are considered modules for protein-protein interaction (23), we used the yeast two-hybrid screen (24, 17) to identify brain specific proteins that would selectively interact with the N-src SH3 domain.
The screening of 5 × 105 independent clones with the N-src SH3 bait resulted in the isolation of a single positively reacting fusion product (pJG-d5). This clone encoded a protein that showed a strong interaction with the N-src SH3 domain, but no significant interaction with the c-src, fyn, or abl SH3 domains, indicating a specific recognition of the N-src SH3 domain in the yeast two-hybrid system. The sequence analysis of pJG-d5 indicated that it encodes the C-terminal portion of a larger protein. We therefore isolated overlapping cDNA clones from a λgt10 library and identified an ORF that encodes a 910 amino acids long polypeptide with a predicted molecular mass of 104 kDa (Fig. 1A). The pJG-d5 insert corresponds to its C-terminal amino acids 404–910.
The N-terminal part of the predicted protein contains an hydrophobic core comprising seven hydrophobic domains (Fig. 1B). These domains show significant homology to the six transmembrane domains (S1–S6) and the pore region (P) of voltage activated K+ channels (Fig. 1C). In addition to the hydrophobic core, there is a putative cyclic nucleotide binding site (CNBs) in the C-terminal half of the protein (amino acids 472–602, Fig. 1 A and D). This CNBs is most closely related to the corresponding region in cyclic nucleotide-gated channels (30% similarity). The amino acids that lie close to the bound cyclic nucleotide in the bacterial catabolite gene activator protein are conserved in the N-src interacting protein, suggesting that the CNBs is functional (25). On the basis of these features, we have named the newly identified protein BCNG-1.
Among all the known K+ channel superfamily genes (26), the core region of BCNG-1 displays the highest similarity (22%) to the corresponding region in the mouse Eag protein, whereas the similarity to CNG channels is only 17% in this region (distances were determined by the MegAlign program of dnastar). The S4 domain of BCNG-1 has a total of eight positively charged residues (two groups of four, separated by a serine), which again makes it more similar to voltage activated K+ channels (Sh and eag families) than to CNG channels.
The putative pore forming region of the BCNG-1 protein (Fig. 1C) is also most closely related to the corresponding region in Shaker and Eag-related channels (30% similarity in either case). However, it contains significant substitutions in two positions that are otherwise highly conserved in voltage activated K+ channels: the aspartate residue that follows the GYG triplet is replaced with alanine (position 352) and the serine/threonine residue at −8 from that position is replaced with histidine (position 344). Similar substitutions are found in the β-subunit of the retinal CNG channel, where the position corresponding to the aspartate is occupied by a leucine and a lysine is found at −8 from that position (27). This suggests that the BCNG-1 protein might be incapable of conducting current per se, but may act in combination with a second not yet identified polypeptide to form a functional heteromultimeric ion channel.
In conclusion, the sequence of the BCNG-1 cDNA indicates that BCNG-1 may be a distinct member of the K+ channel superfamily and could be regulated by cyclic nucleotide binding.
BCNG-1 Is a 132-kDa Glycoprotein.
To characterize the protein encoded by the BCNG-1 cDNA, we generated antibodies against two separate domains in the predicted cytoplasmic tail: amino acids 594–720 (fusion protein GST-q1; antiserum αq1) and 777–910 (fusion protein GST-q2; antiserum αq2). Both antisera specifically immunoprecipitate the in vitro translation product of the cloned BCNG-1 sequence (not shown).
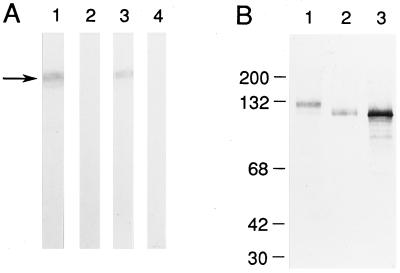
In Western blots of mouse brain extracts, both the αq1 and αq2 antisera recognize a diffuse band with an apparent molecular mass of 132 kDa (Fig. 2A). Complete abolition of the labeling by preadsorbing the antisera with a GST-fusion protein incorporating both the antigenic domains (GST-d5, amino acids 404–910) indicates it represents the native BCNG-1 subunit. Following treatment of the brain extract with N-glycosidase F, the deglycosylated native protein comigrated with the in vitro translated BCNG-1 product (Fig. 2B).
Figure 2.
BCNG-1 is a 132 kDa N-glycosylated protein. (A) Western blot analysis of BCNG-1 protein in a mouse brain extract. Ten micrograms of a total brain SDS-extract was loaded per strip then probed with αq1 (1) or αq2 (3) antiserum or αq1 (2) or αq2 (4) antiserum preadsorbed with the GST-d5 fusion protein. The arrow marks the position of native BCNG-1 protein. (B) Western blot using the αq1 antiserum against total brain extract (1), total brain extract pretreated with N-glycosidase F (2), and in vitro translated BCNG-1 protein (3). Positions of molecular mass standards are shown on the left.
Sequence analysis indicates that three N-glycosylation consensus sites are present in the BCNG-1 protein. Among these, Asn-327 is predicted to lie between transmembrane domain S5 and the pore (P) on the extracellular side of the plasma membrane (Fig. 1 A and B). This site corresponds to Asn-327 of the cGMP-gated channel from bovine rod photoreceptors, where it has been demonstrated to be the sole site of glycosylation (28). Together, these data suggest that the cloned cDNA sequence encodes the full length product of the BCNG-1 gene and that BCNG-1 is a N-linked glycoprotein.
BCNG-1 Is Expressed in Neurons.
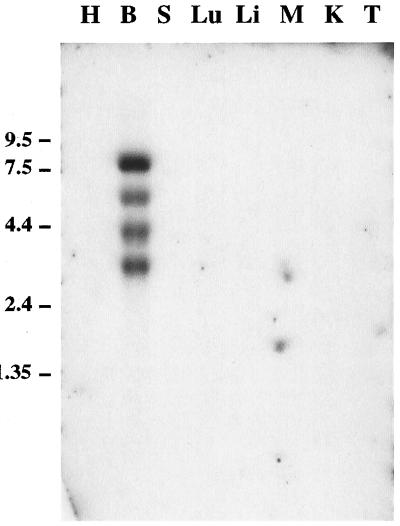
Northern blot analysis reveals the presence of multiple BCNG-1 transcripts in poly(A)+ RNA from the brain, the most abundant species being 3.4, 4.4, 5.8, and 8.2 kb long (Fig. 3). The 3.4 kb transcript corresponds in size to the cloned cDNA. No expression is detected in the heart, spleen, lung, liver, skeletal muscle, kidney, or testis. The specific expression of the BCNG-1 protein in brain was confirmed by Western blot analysis (not shown).
Figure 3.
Northern blot analysis of BCNG-1 expression in different mouse tissues. Two micrograms of poly(A)+ RNA from each of heart (H), brain (B), spleen (S), lung (Lu), liver (Li), skeletal muscle (M), kidney (K), and testis (T) were loaded. The filter was probed with a DNA fragment encoding amino acids 6–131 of the BCNG-1 sequence. A probe corresponding to amino acids 594–720 recognized the same bands (not shown), confirming that the cDNA fragments isolated from the λgt10 and pJG4–5 libraries are from a contiguous mRNA sequence. Positions of molecular mass standards are shown on the left.
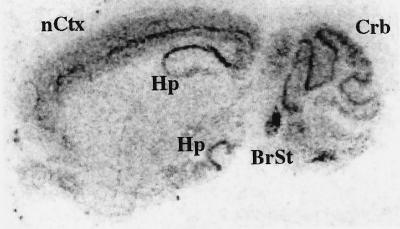
The cellular localization of BCNG-1 within the brain was examined by in situ hybridization (Fig. 4) and by immunohistochemical staining (Fig. 5). In both cases, the highest levels of BCNG-1 expression were detected in the cerebral cortex, in the hippocampus, and in the cerebellum.
Figure 4.
In situ hybridization analysis of BCNG-1 expression in the brain. Parasagittal section of a mouse brain probed with an antisense oligonucleotide directed to the mRNA region corresponding to amino acids 648–657 of the BCNG-1 sequence. nCtx, Neocortex; Hp, hippocampus; Crb, cerebellum; BrSt, brainstem.
Figure 5.
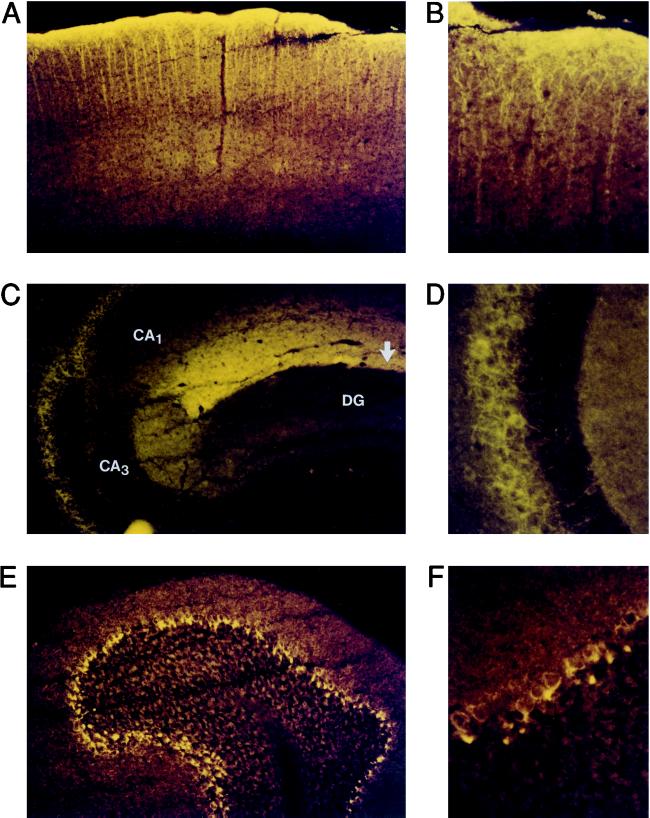
Immunohistochemical analysis of BCNG-1 expression in the brain. Parasagittal sections of a mouse brain were stained with αq1 and αq2 antisera. The patterns of BCNG-1 expression detected with the two different antisera were identical, and in both cases the staining was entirely abolished by preadsorbing the sera with the GST-d5 fusion protein (not shown). (A, B) BCNG-1 immunoreactivity in the cerebral cortex. (C, D) BCNG-1 immunoreactivity in the hippocampus. In C, arrow shows position of hippocampal fissure; areas CA1, CA3, and dentate gyrus (DG) are labeled. D shows a detail of the stratum pyramidale of area CA3. (E, F) BCNG-1 immunoreactivity in the cerebellum. Magnification: ×100 (A,C,E); ×400 (B,D,F).
In the cerebral cortex, in situ hybridization shows a strong expression of the BCNG-1 mRNA in layer V pyramidal neuron cell bodies that are distributed in a continuous line along the neocortex (Fig. 4). Immunohistochemical analysis reveals a strict subcellular localization of the BCNG-1 protein within these cells. Staining of the apical dendrites (Fig. 5A) extends into the terminal branches of these fibers and is particularly intense in layer I, which contains the terminal dendritic plexus of the pyramidal neurons (Fig. 5B).
A similar expression pattern can be recognized in the hippocampus. Here, in situ hybridization shows strong BCNG-1 mRNA expression in the pyramidal cell body layer of areas CA1 and CA3 (Fig. 4). The labeling in area CA3 is somewhat less prominent than the labeling in area CA1. At the protein level, the most intense BCNG-1 immunostaining is observed along the hippocampal fissure, in the layer corresponding to the stratum lacunosum-moleculare (Fig. 5C). This layer contains the terminal branches of the apical dendrites of the pyramidal neurons in area CA1 (29). Further BCNG-1 immunoreactivity is detected within the stratum pyramidale of areas CA1 and CA3; the staining, however, is absent from the pyramidal cell bodies but rather is present in the fibers surrounding them (Fig. 5D). These fibers most likely represent the basket cell plexus associated to pyramidal neurons.
The immunostaining in the cerebellum also shows a pattern characteristic of basket cell expression. In the cerebellar cortex, basket cell nerve endings branch and contact the initial segment of the Purkinje cell axon in a distinct structure known as “pinceau” (30). As shown in Figs. 5 E and F, these structures are intensely labeled by the αq1 and αq2 antisera, while the staining excludes the Purkinje cell bodies. Thus, in basket cells, the BCNG-1 protein appears to be selectively localized to axons and is particularly enriched in the nerve terminals.
An intense labeling of some brainstem nuclei is observed by in situ hybridization (Fig. 4) and areas of immunoreactivity were detected in other brain regions, including the olfactory bulb (not shown).
BCNG-1 Defines a New Subfamily of K+ Channel Genes.
Most of the ion channel sequences characterized so far are members of evolutionarily related multigene families. To investigate whether more sequences related to BCNG-1 exist, we analyzed mouse genomic DNA Southern blots under variable stringency conditions.
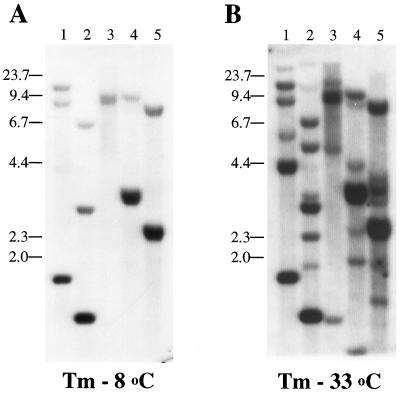
The probe (B1-T) was designed in the hydrophobic core region of BCNG-1, including transmembrane domains S5, P, and S6; the repeat region in the C-terminal portion of the protein was excluded. Reducing the stringency of the hybridization conditions from 8°C below melting temperature (Fig. 6A) to 33°C below melting temperature (Fig. 6B) resulted in the detection of additional signals in every lane of the blot. None of the known sequences in the K+ channel superfamily has sufficient homology to BCNG-1 to hybridize under these conditions.
Figure 6.
Southern blot analysis of mouse genomic DNA. Four μg of mouse genomic DNA were loaded onto each lane following digestion with EcoRI (1), HindIII (2), BamHI (3), PstI (4), or BglII (5). The filter was probed with a DNA fragment encoding amino acids 270–463 of the BCNG-1 sequence at high (A) and low (B) stringency. Positions of molecular mass standards are shown on the left.
This result suggests that BCNG-1 is the first member of a larger group of related genes, which represent a new branch in the voltage-gated potassium channel superfamily.
DISCUSSION
Voltage-gated potassium channels constitute a large and still expanding superfamily of related genes (31, 26). The most widely used strategy for cloning new genes in the voltage-gated potassium channel family has been by homology to a small number of initial members (Sh, eag, and slo from Drosophila, refs. 32–35; cGMP-channel from bovine retina, ref. 36). Unfortunately, this approach is not well suited for identifying more divergent sequences. Expression cloning in Xenopus oocytes can circumvent this problem, however, this implies a pre-existing or readily detectable physiological characterization of the channel.
An alternative cloning strategy that requires no a priori knowledge of the structure or activity of the target protein is to screen for K+ channels by means of protein-protein interactions. Using the SH3 domain of N-src as a bait we have obtained a protein that displays the motifs of a voltage-gated K+ channel (six transmembrane spanning domains, a highly basic S4, and a P region) (refs. 31 and 26 and Fig. 1). However, BCNG-1 shows considerable divergence from all other known sequences. Although the CNBs site of BCNG-1 is most similar to the site present in CNG channels (30%), the S4 and P regions are most closely related to the corresponding regions in Shaker and Eag. Overall, the highest similarity in the hydrophobic core region is to mouse Eag (22%). Thus, BCNG-1 appears to constitute a new branch of the K+ channel superfamily.
The fusion between an ancestral K+ channel and an ancestral CNBs is likely to have occurred prior to the evolutionary separation between plants and animals (26). Divergence from this common ancestor would have led on one hand to Eag-related channels and plant inward rectifiers (which maintained more of the features of voltage-activated K+ channels, while showing a progressive deviation from the original CNBs sequence) and on the other hand to CNG channels (which show a higher evolutionary constraint on the cyclic nucleotide binding site, while they have lost voltage activation and K+ selectivity). The features of BCNG-1 suggest that it may have remained closer to the ancestral molecule that represents the evolutionary link between voltage-gated K+ channels and cyclic nucleotide-gated channels.
Our attempts to detect currents following BCNG-1 expression in Xenopus oocytes have so far proven unsuccessful. However, by analogy with the emerging pattern for retinal and olfactory CNG channels, the non-consensus sequence of the putative pore forming region of BCNG-1 suggests that this protein may represent a β-subunit of a heteromultimeric channel (27, 37, 38). Indeed our data show the existence of a number of BCNG-related sequences in the mouse genome, and one or more of these genes could encode additional subunits required for the formation of an active channel.
The BCNG-1 protein is expressed only in the brain and in particular in two of the principal classes of neurons within the cerebral, hippocampal, and cerebellar cortices: pyramidal neurons and basket cells. This distribution would be consistent with an in vivo interaction of BCNG-1 with N-src, which is also expressed in cerebral and hippocampal pyramidal neurons (39). The observed interaction between BCNG-1 and the N-src SH3 domain is intriguing; its physiological relevance and the role of the proline-rich region of BCNG-1 remain to be investigated. The possibility that other factors may target the proline-rich region of BCNG-1 has also to be considered, particularly in view of the recent discovery of WW domains (40, 41).
The varied subcellular localization of BCNG-1—dendritic in pyramidal cells and axonal in basket cells—suggests that BCNG-1 could play different roles in different populations of neurons, perhaps by regulating presynaptic or postsynaptic membrane excitability depending on the cell type. A similar distribution has been demonstrated for the K+ channel subunit Kv 1.2 (42, 43). Kv 1.2 forms heteromultimeric K+ channels with several other Shaker type subunits, which have an overlapping yet differential pattern of expression, giving rise to a range of conductances with diversified functional characteristics.
Can we envision any physiological function for BCNG-1, based on its sequence and distribution? The presence of BCNG-1 in the dendrites of hippocampal pyramidal cells is particularly intriguing, as cAMP has been shown to be important for the establishment of some forms of long-term synaptic potentiation in these cells (8, 9, 44). The structural features of BCNG-1 would predict a K+ conducting activity, directly modulated by cyclic nucleotide binding. Interestingly, a current with similar characteristics has been described in the hippocampal pyramidal neurons of area CA1 (45), where BCNG-1 is highly expressed. This current (IQ) is believed to contribute to the noradrenergic modulation of hippocampal activity, by regulating neuronal excitability in response to cAMP levels. It remains to be determined whether BCNG-1 could participate in the formation of the channels responsible for this type of current.
Acknowledgments
We thank Paul Skehel, Francois Rassendren, Gareth Tibbs, and Steve Siegelbaum for their valuable contributions throughout the course of this study. We are grateful to Roger Brent and Ruibao Ren for providing us with the yeast, bacterial strains, and plasmids. Ke Huang and Feng Ye provided us with excellent technical assistance. We also thank Harriet Ayers and Chuck Lam for help in preparation of the manuscript. This research was supported by the Howard Hughes Medical Institute and a fellowship from Istituto Pasteur/Fondazione Cenci-Bolognetti to B.S.
ABBREVIATIONS
- CNG
cyclic nucleotide-gated
- CNBs
cyclic nucleotide binding site
- GST
glutathione S-transferase
- SH3
Src homology 3
- P
pore region
- S1–S6
six transmembrane domains
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF028737).
References
- 1.Greene W N, Millar N S. Trends Neurosci. 1995;18:280–287. [PubMed] [Google Scholar]
- 2.Adelman J P. Curr Opin Neurobiol. 1995;5:286–295. doi: 10.1016/0959-4388(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 3.Zagotta W N, Siegelbaum S A. Annu Rev Neurosci. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
- 4.Brüggemann A, Pardo L, Stühmer W, Pongs O. Nature (London) 1993;365:445–448. doi: 10.1038/365445a0. [DOI] [PubMed] [Google Scholar]
- 5.Hoshi T. J Gen Physiol. 1995;105:309–328. doi: 10.1085/jgp.105.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adelman J P, Shen K-Z, Kavanaugh M P, Warren R A, Wu Y-N, Lagrutta A, Bond C T, North R A. Neuron. 1992;9:209–216. doi: 10.1016/0896-6273(92)90160-f. [DOI] [PubMed] [Google Scholar]
- 7.Kohler M, Hirschberg B, Bond C T, Kinzie J M, Marrion N V, Maylie J, Adelman J P. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 8.Frey U, Huang Y-Y, Kandel E R. Science. 1993;260:1661–1664. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- 9.Bolshakov V Y, Golan H, Kandel E R, Siegelbaum S A. Neuron. 1997;19:635–651. doi: 10.1016/s0896-6273(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 10.Arancio O, Kandel E R, Hawkins R D. Nature (London) 1995;376:74–80. doi: 10.1038/376074a0. [DOI] [PubMed] [Google Scholar]
- 11.Kingston P A, Zufall F, Barnstable C J. Proc Natl Acad Sci USA. 1996;93:10440–10445. doi: 10.1073/pnas.93.19.10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradley J, Zhang Y, Bakin R, Lester H A, Ronnett G V, Zinn K. J Neurosci. 1997;17:1993–2005. doi: 10.1523/JNEUROSCI.17-06-01993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brugge J S, Cotton P C, Querel A E, Barrett J N, Nonner D, Keane R W. Nature (London) 1985;316:554–557. doi: 10.1038/316554a0. [DOI] [PubMed] [Google Scholar]
- 14.Martinez R, Mathey-Prevot B, Bernards A, Baltimore D. Science. 1987;237:411–415. doi: 10.1126/science.2440106. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 17.Zervos A S, Gyuris J, Brent R. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 18.Breede L, Nasmyth K. Cold Spring Harbor Symp Q Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 19.Frangioni J V, Neel B G. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 21.Grant S G N, Karl K A, Kiebler M, Kandel E R. Genes Dev. 1995;9:1909–1921. doi: 10.1101/gad.9.15.1909. [DOI] [PubMed] [Google Scholar]
- 22.Mayford M, Wang J, Kandel E R, O’Dell T J. Cell. 1995;81:891–904. doi: 10.1016/0092-8674(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 23.Pawson T. Nature (London) 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 24.Fields S, Song O K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 25.Weber I T, Shabb J B, Corbin J D. Biochemistry. 1989;28:6122–6127. doi: 10.1021/bi00440a059. [DOI] [PubMed] [Google Scholar]
- 26.Warmke J W, Ganetzky B. Proc Natl Acad Sci USA. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen T Y, Peng Y W, Dhallan R S, Ahamed B, Reed R R, Yau K W. Nature (London) 1993;362:764–767. doi: 10.1038/362764a0. [DOI] [PubMed] [Google Scholar]
- 28.Wohlfart P, Haase W, Molday R S, Cook N J. J Biol Chem. 1992;267:644–648. [PubMed] [Google Scholar]
- 29.Raisman G, Cowan W M, Powell T P S. Brain. 1965;88:963–996. [Google Scholar]
- 30.Palay S L, Chan-Palay V. Cerebellar Cortex: Cytology and Organization. New York: Springer; 1974. [Google Scholar]
- 31.Strong M, Chandy G, Gutman G. Mol Biol Evol. 1993;10:221–242. doi: 10.1093/oxfordjournals.molbev.a039986. [DOI] [PubMed] [Google Scholar]
- 32.Papazian D M, Schwartz T L, Tempel B L, Jan Y N, Jan L Y. Science. 1987;237:749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- 33.Kamb A, Iverson L E, Tanouye M A. Cell. 1987;50:405–413. doi: 10.1016/0092-8674(87)90494-6. [DOI] [PubMed] [Google Scholar]
- 34.Warmke J, Drysdale R, Ganetzky B. Science. 1991;252:1560–1562. doi: 10.1126/science.1840699. [DOI] [PubMed] [Google Scholar]
- 35.Atkinson N S, Robertson G A, Ganetzky B. Science. 1991;253:551–555. doi: 10.1126/science.1857984. [DOI] [PubMed] [Google Scholar]
- 36.Kaupp U B, Niidome T, Tanabe T, Terada S, Bönigk W, Stühmer W, Cook N J, Kangawa K, Matsuo H, Hirose T, Miyata T, Numa S. Nature (London) 1989;342:762–766. doi: 10.1038/342762a0. [DOI] [PubMed] [Google Scholar]
- 37.Liman E R, Buck L B. Neuron. 1994;13:1–20. doi: 10.1016/0896-6273(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 38.Bradley J, Li J, Davidson N, Lester H A, Zinn K. Proc Natl Acad Sci USA. 1994;91:8890–8894. doi: 10.1073/pnas.91.19.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugrue M M, Brugge J S, Marshak D R, Greengard P, Gustafson E L. J Neurosci. 1990;10:2513–2527. doi: 10.1523/JNEUROSCI.10-08-02513.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudol M. Trends Biol Sci. 1996;21:161–163. [PubMed] [Google Scholar]
- 41.Staub O, Dho S, Henry P C, Correa J, Ishikawa T, McGlade J, Rotin D. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 42.Sheng M, Tsaur M-L, Jan Y N, Jan L Y. J Neurosci. 1994;14:2408–2417. doi: 10.1523/JNEUROSCI.14-04-02408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Kunkel D D, Schwartzkroin P A, Tempel B L. J Neurosci. 1994;14:4588–4599. doi: 10.1523/JNEUROSCI.14-08-04588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas M J, Moody T D, Makhinson M, O’Dell T J. Neuron. 1996;17:475–482. doi: 10.1016/s0896-6273(00)80179-8. [DOI] [PubMed] [Google Scholar]
- 45.Pedarzani P, Storm J F. Proc Natl Acad Sci USA. 1995;92:11716–11720. doi: 10.1073/pnas.92.25.11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pallanck L, Ganetzky B. Hum Mol Genet. 1994;3:1239–1243. doi: 10.1093/hmg/3.8.1239. [DOI] [PubMed] [Google Scholar]
- 47.Dhallan R S, Yau K W, Schrader K A, Reed R R. Nature (London) 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]