Protein interactions are a central mechanism for the generation of biological regulatory specificity. Complexes formed by different combinations of proteins can perform a much larger number of biological functions than would be possible if each protein were to act independently. By interacting with different partners in different cell types and in response to different extracellular stimuli, a single protein can contribute to divers cellular responses.
Many methods for the investigation of protein interactions have been developed. Methods that enable direct detection of interactions (i.e. co-precipitation) generally require removal of the proteins from their normal cellular environment. Conversely, methods that allow interactions to be studied with minimal perturbation of their environment (i.e. genetic suppressor analysis) generally rely on indirect detection of their consequences. This trade-off is not absolute; protein-fragment complementation methods based on the fusion of complementary fragments of a reporter protein (Fig. 1) to two putative interacting proteins combine nearly direct detection of protein interactions with limited cellular perturbation. Functional complementation between the reporter protein fragments, mediated by the interaction of the two fusion proteins, results in a quantifiable signal (Table 1) that can be used to investigate the localization and regulation of protein interactions in their normal environment.
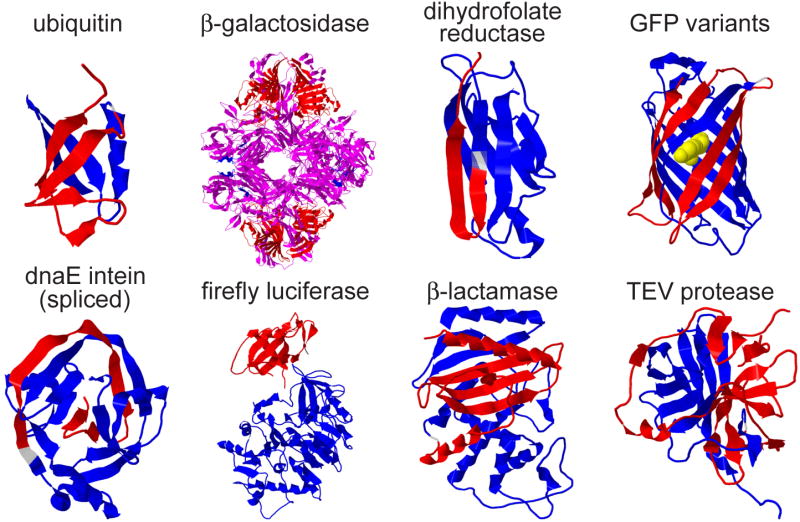
Figure 1.
Examples of protein fragments that support complementation are shown in red and blue using models based on the X-ray crystal structures of the intact proteins. In β-galactosidase, the overlap between the fragments is shown in magenta. The discontinuity in the polypeptide backbone is shown in translucent grey. The images were generated using jmol.
Table 1.
Characteristics of complementation methods using fragments of different proteins.
| Complementation: | Detection: | Spatial resolution*: | Time resolution*: | Experimental systems*: | Reference: |
|---|---|---|---|---|---|
| Ubiquitin | Ub-protease coupled reporters | Cell population | Day | Yeast | 10 |
| β-galactosidase | FDG hydrolysis | Cellular | Hours | Cultured cells, D. melanogaster | 11 |
| Dihydrofolate reductase | Fl-MTX binding | Sub-cellular | Minutes | Cultured cells, plants | 12 |
| GFP variants | Intrinsic fluorescence | Sub-cellular | Minutes-Hours | Cultured cells, plants, fungi | 5 13 |
| Synechocystis dnaE intein | Reporter ligation | Cell population | Hours | Cultured, implanted cells | 14 |
| β-lactamase | CCF2/AM hydrolysis | Cellular | Minutes | Cultured cells, primary neurons | 6, 15, 16 |
| Firefly luciferase | Luciferin luminescence | Cell population | Hours | Cultured, implanted cells | 4 |
| Renilla luciferase | Coelenterazine luminescence | Cell population | Minutes-Hours | Cultured, implanted cells | 3 |
| Gaussia luciferase | Coelenterazine luminescence | Cell population | Minutes | Cultured cells | 1 |
| TEV protease | Coupled reporters | Cellular | Minutes | Cultured cells | 2 |
The spatial and temporal resolutions as well as the experimental systems listed are those reported in publications using these complementation methods and are not intended to represent the limits of these approaches
Protein-fragment complementation methods share several characteristics that set them apart from other methods for the detection of protein interactions in cells. Among the latter, fluorescence resonance energy transfer (FRET) analysis is overwhelmingly the most widely used. The principal trade-off between these methods is one of sensitivity versus dynamics. Protein-fragment complementation can, in principle, be more sensitive since complementation produces novel functions whereas FRET produces changes in an existing fluorescence signal. Conversely, detection of protein fragment association can involve a significant time lag and the association between the fragments can stabilize the complex, whereas FRET ideally reflects the real-time dynamics of complex formation and dissociation. The relative significance of sensitivity versus dynamics depends on the specific application, which should guide the choice of method.
Although the choice between alternative methods is ideally based on the performance of each assay, often the issue is decided by available instrumentation and technical expertise. Complementation methods are generally technically straightforward, require only common equipment and require little data processing for interpretation. All methods for the detection of protein interactions in cells, however, require numerous controls to determine if a signal results from a specific interaction.
Fragments of many proteins have been identified that can associate with each other to form a functional complex when they are brought together by an interaction between proteins fused to the fragments (Fig. 1 and Table 1). In this issue of Nature Methods, two groups report the development of complementation methods based on Gaussia luciferase and tobacco etch virus (TEV) protease, respectively1, 2. Since no general strategy has been defined for prediction of protein fragments that support complementation, in each of these cases the complementary fragments were identified by screening for positions that tolerate a discontinuity in the protein backbone.
Gaussia luciferase complementation makes use of smaller protein fragments than the previously described firefly and Renilla luciferase complementation methods3, 4. The rapid association and moderately fast dissociation of the fragments in conjunction with the high sensitivity of the luciferase assay make it possible to detect reversible associations between model proteins within minutes in a cell population1, but require an exogenous substrate and have limited spatial resolution.
TEV protease complementation, on the other hand, provides flexibility in detection since it can be coupled to several different reporter systems that use either fluorescent proteins or luciferase to detect protease activity2. The multiple reporter systems allow selection of different reporters for different purposes (i.e fluorescent proteins for single cells and luciferase for a cell population), but introduce an additional step between the interaction and the measurement.
Complementation methods based on fragments of different proteins have distinct advantages and disadvantages (Table 1). An ideal method would provide maximal spatial and temporal resolution as well as maximal sensitivity and specificity for the detection of all protein interactions in every cell type and organism with minimal perturbation. At present, the highest spatial resolution can be obtained by bimolecular fluorescence complementation (BiFC) between fragments of fluorescent proteins5. The fastest response times in living cells have been reported for β-lactamase and Gaussia luciferase fragment complementation6 1. Conversely, in many cases, the BiFC approach does not reflect the real-time dynamics of complex formation or dissociation. For most complementation methods, the effects of fragment association on the dynamics of complex formation and dissociation have been examined for a single interaction, which may not be representative of the majority of interactions. Ubiquitin and dihydrofolate reductase (DHFR) complementation require respectively, the presence or absence of endogenous activities. Many complementation methods (β-galactosidase, DHFR, β-lactamase, luciferases) require the use of exogenous substrates or ligands for detection of complementation.
The sensitivities of the different methods have not been directly compared. It is likely that they depend on the identities of the interaction partners. The specificities of complementation methods must be tested by examining the effects of mutations that eliminate the interaction but do not alter other properties of the proteins. Different complementation methods are likely to be optimal for detection of interactions between different proteins and for studies of different aspects of the interaction.
Finally, all complementation methods and most other approaches for the study of protein interactions inside living cells require the use of fusion proteins. Thus, these methods can't be used to study interactions between endogenous proteins and it is critical to compare the fusions with the corresponding endogenous proteins under the conditions used to study the interactions. In this issue of Nature Methods, a proximity ligation in situ assay (P-LISA) is described that -enables determination of the locations of endogenous protein complexes in fixed samples7. Clearly, a combination of experimental approaches, including analysis of both fusion proteins in living cells and endogenous proteins in fixed or lysed samples provides the most comprehensive understanding of a protein interaction.
The ultimate impact of complementation methods on the study of protein interactions depends on their general applicability. BiFC has been used to visualize more than two hundred protein interactions8 (see http://sitemaker.umich.edu/kerppola.bifc for an updated list), and the number of interactions examined using other complementation methods is steadily increasing. Most of the complementation methods have been used in intact multicellular organisms and in cells from a variety of species. BiFC has been used to screen for interaction partners and for semi-high-throughput analysis of small molecule modulators of protein complexes. Given the widespread use of luciferase assays in high-throughput screens, the development of complementation methods that either directly or indirectly produce a luciferase reporter should expand the opportunities for high-throughput analysis of protein interactions in living cells. The many complementation methods that have been developed expand the applicability of this class of methods to a larger range of problems than can be investigated using any single approach.
Most complementation methods are based on naturally occurring protein sequences. It is likely that the characteristics of these assays could be improved by searching for fragments with favorable characteristics. Spectral variants of fluorescent proteins inspired multicolor BiFC analysis, and allow simultaneous visualization of multiple protein complexes9. Likewise, complementation by luciferases, proteases or other enzymes with different substrate specificities could be used for multiplex analysis. Such multiplex analysis is especially valuable for the incorporation of same-cell controls into the experiments. Future studies of protein interactions in living cells are likely to involve an expanding array of complementation methods that will enable interrogation of detailed molecular mechanisms in the normal cellular environment.
Footnotes
Two new protein-fragment complementation methods expand the toolkit available for the detection of protein interactions in living cells.
References
- 1.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on the enzyme Gaussia Luciferase. Nature Methods. 2006 doi: 10.1038/nmeth979. this issue. [DOI] [PubMed] [Google Scholar]
- 2.Wehr MC, et al. Monitoring Regulated Protein-Protein Interactions Using Split-TEV. Nature Methods. 2006 doi: 10.1038/nmeth967. this issue. [DOI] [PubMed] [Google Scholar]
- 3.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–9. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci U S A. 2002;99:15608–13. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–98. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 6.Wehrman T, Kleaveland B, Her JH, Balint RF, Blau HM. Protein-protein interactions monitored in mammalian cells via complementation of beta -lactamase enzyme fragments. Proc Natl Acad Sci U S A. 2002;99:3469–74. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Söderberg O, et al. Direct observation in situ of individual endogenous protein complexes by proximity ligation. Nature Methods. 2006 doi: 10.1038/nmeth947. this issue. [DOI] [PubMed] [Google Scholar]
- 8.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nature Reviews Molecular Cell Biology. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–45. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc Natl Acad Sci U S A. 1997;94:8405–10. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10340–4. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelletier JN, Campbell-Valois FX, Michnick SW. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc Natl Acad Sci U S A. 1998;95:12141–6. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- 14.Ozawa T, Kaihara A, Sato M, Tachihara K, Umezawa Y. Split Luciferase as an Optical Probe for Detecting Protein-Protein Interactions in Mammalian Cells Based on Protein Splicing. Anal Chem. 2001;73:2516–2521. doi: 10.1021/ac0013296. [DOI] [PubMed] [Google Scholar]
- 15.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat Biotechnol. 2002;20:619–22. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 16.Spotts JM, Dolmetsch RE, Greenberg ME. Time-lapse imaging of a dynamic phosphorylation-dependent protein-protein interaction in mammalian cells. PNAS. 2002;99:15142–15147. doi: 10.1073/pnas.232565699. [DOI] [PMC free article] [PubMed] [Google Scholar]