Abstract
Phosphorus is an essential nutrient for all living organisms. Under conditions of inorganic phosphate starvation, genes from the Pho regulon are induced allowing microorganisms to use phosphonates as a source of phosphorus. The phnO gene was previously annotated as a transcriptional regulator of unknown function due to sequence homology with members of the GCN5-related N-acyltransferase family (GNAT). PhnO can now be functionally annotated as an aminoalkylphosphonic acid N-acetyltransferase which is able to acetylate a range of aminoalkylphosphonic acids. Studies revealed that PhnO proceeds via an ordered, sequential kinetic mechanism with acetyl-CoA binding first followed by aminoalkylphosphonate. Attack by the amine on the thioester of AcCoA generates the tetrahedral intermediate that collapses to generate the products. The enzyme also requires a divalent metal ion for activity, which is the first example of this requirement for a GNAT family member.
Keywords: Phosphonate metabolism, Enzyme kinetics, Chemical Mechanism, N-acetyltransferases
Phosphorus plays an essential role in the physiology and biochemistry of all living organisms. It is therefore not surprising that microorganisms have evolved sophisticated systems to acquire phosphorus from sources other than inorganic orthophosphate. Under phosphate starvation conditions, when other more readily utilizable forms of phosphate are not present, microorganisms have evolved to utilize more reduced organophosphorus sources, including phosphonates (1). Phosphonates are a class of compounds which contain a carbon-phosphorus (C-P) bond that is exceptionally stable and resistant to chemical hydrolysis, thermal decomposition and photolysis (2). Phosphonates are a naturally occurring class of compound which are present in a number of organisms; e.g. Tetrahymena which possesses up to 30% of its membrane lipids in the form of phosphonolipids (3).
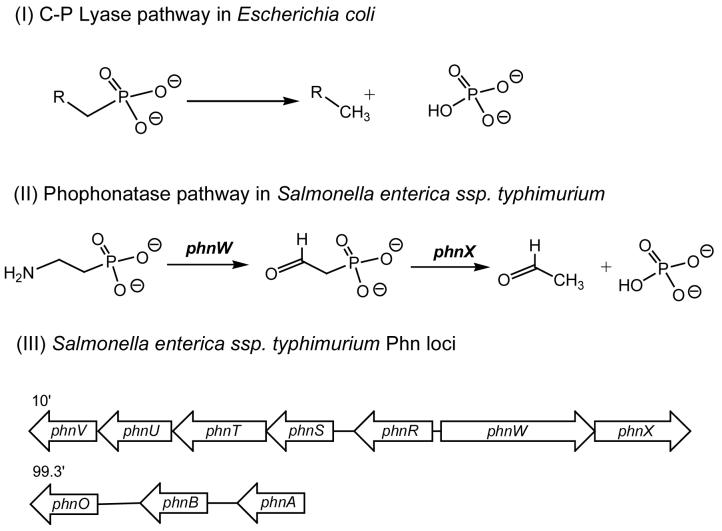
Changing the source of phosphorus from orthophosphate to organophosphorus compounds is a tightly regulated process. Under phosphate starvation conditions, genes from the Pho (phosphate starvation) regulon are induced (1). In Gram-negative organisms, like Salmonella enterica serovar Typhimurium, the pho genes are regulated by a two-component system, which consists of PhoB and PhoR; PhoB binds directly to the pho gene promoter whilst PhoR is a Pi sensory histidine protein kinase (4). Two alternative pathways exist for cleavage of the C-P bond: the C-P lyase and the phosphonatase pathways (Scheme 1; (5). Escherichia coli carries genes for the reductive C-P lyase pathway (6), whilst S. enterica carries genes for the phosphonatase pathway (7). The C-P lyase pathway has a much broader substrate specificity and is able to cleave alkylphosphonates as well as aminoalkylphosphonates (8, 9). In comparison, the phosphonatase pathway has much narrower substrate specificity and acts only on 2-aminoethylphosphonate (10). Mutagenic studies and sequence analysis of the Pho regulon in Escherichia coli identified a number of genes involved in phosphonate catabolism designated phnC to phnP (11, 12). PhnC, PhnD, and PhnE were proposed to constitute a phosphonate transporter while PhnG, PhnH, PhnI, PhnJ, PhnK, PhnL, and PhnM were required for catalysis and were proposed to form a membrane-associated carbon-phosphorus (C-P) lyase. PhnN and PhnP were not absolutely required for phosphonate catabolism and were therefore proposed to be accessory proteins for the C-P lyase. PhnF and PhnO were non-essential and were proposed to play regulatory roles because of sequence similarities to other regulatory proteins (11).
Scheme 1.
Pathways for phosphonate catabolism. (I) The C-P lyase pathway acts on alkylphosphonates as well as aminoalkylphosphonates. (II) The phosphonatase pathway that uses 2-aminoethylphosphonate and cleaves the C-P bond in a two-step transamination and hydrolysis process. (III) The S. enterica phn gene locus; phnR encodes a putative aminoalkylphosphonate transport repressor, phnS-V encode putative aminoalkylphosphonate transport components, phnW encodes a 2-aminoethylphosphonate transaminase, phnX encodes a 2-phosphonoacetaldehyde hydrolase, phnA encodes a putative phosphonoacetate hydrolase, phnB encodes a putative phosphonoacetate transporter, phnO encodes an aminoalkylphosphonic N-acetyltransferase.
In contrast to the E. coli Pho regulon which consists of at least 31 co-regulated genes, the S. enterica Pho regulon is smaller consisting of only 21 genes (7). The S. enterica Phn locus differs from the E. coli Phn locus in its size, layout and composition, primarily due to their differing mechanisms of phosphonate catabolism. The S. enterica Phn locus is composed of seven genes, designated phnR to phnX although another cluster containing three genes designated phnA, phnB and phnO is putatively involved in phosphonate catabolism (Scheme 1; (7, 13). Unlike the C-P lyase pathway, the phosphonatase pathway has been extensively mechanistically characterized (7, 10, 14, 15). The initial step in the catabolism of 2-aminoethylphosphonate in S. enterica is a PLP-dependent transamination, carried out by 2-aminoethylphosphonate transaminase (the phnW gene product;(10). This is followed by a metal-dependent hydrolysis of the C-P bond catalyzed by 2-phosphonoacetaldehyde hydrolase (the phnX gene product;(16, 17). Interestingly the phnO gene is present in both E. coli and S. enterica, even though they have very different mechanisms of phosphonate catabolism (6, 7). Gene knockout studies have shown that the E. coli phnO gene product was not required for the catabolism of phosphonates and was presumed to play a regulatory role (11). Furthermore, phnO showed significant sequence homology to the GCN5 related N-acetyltransferases family (GNAT), of which some members such as the histone acetyltransferases play a regulatory role (18).
In this paper, we describe the cloning, expression, and purification of PhnO from S. enterica serovar Typhimurium LT2 and define its functionality as an aminoalkylphosphonic acid N-acetyltransferase. We have determined the steady-state kinetic parameters, defined the divalent metal ion dependency and explored the steady-state kinetic mechanism. General acid catalysis is used by the enzyme, and this is likely to be the rate-limiting step as assessed by solvent deuterium kinetic isotope effects.
MATERIAL AND METHODS
All chemicals, acetyl-coenzyme A (CoA), and aminoalkylphosphonic acids were purchased from Sigma-Aldrich Chemical Co or Fisher Scientific. pET-17b(-) plasmid was purchased from Novagen. All restriction enzymes and T4 DNA ligase were obtained from New England Biolabs. Escherichia coli strain BL21 (DE3) star cells, PCR primers and the pCR-Blunt plasmid kit was obtained from Invitrogen. Pfu DNA polymerase was purchased from Stratagene. DesulfoCoA was synthesized as previously described (19). All divalent metals used in this study were prepared from their respective chloride salt.
General methods
Solution pH values were measured at 25°C with an Accumet model 20 pH meter and Accumet combination electrode standardized at pH 7.0 and 4.0 or 10.0. Protein purification was performed at 4°C using a fast protein liquid chromatography system (Amersham-Pharmacia Biotech). Spectrophotometric assays were performed using a UVIKON XL double beam UV-vis spectrophotometer (BIO-TEK Instruments). All buffers were passed through Chelex 100 resin (Bio Rad) to remove any divalent metal ions present.
Cloning, expression, and purification of PhnO
The S. enterica serovar Typhimurium LT2 gene phnO was amplified by PCR using the primers 5-TTTTTCATATGCCAGTCTGTGAATTACGCCA-3 and 5-TTTTTTGAATTCTCACAATGCTTTCGTAAACC-3 from the plasmid template pET-28a(-):phnO (generously provided by Sophie Magnet, Albert Einstein College of Medicine), incorporating NdeI and EcoRI restriction endonuclease sites (underlined). The amplified DNA product was ligated into a pCR-Blunt plasmid and transformed into One Shot TOP10 cells. Plasmid DNA isolated from these cells was then digested with NdeI and EcoRI, and the purified insert was ligated into purified plasmid pET-17b(-) previously linearized with the same restriction enzymes, yielding an expression plasmid for S.enterica phnO.
The plasmid pET-17b(-):phnO was then isolated, sequenced, and used to transform E. coli BL21 (DE3) star cells. Transformed cells were grown overnight in 50 ml of LB broth containing 100 μg/ml ampicillin. One-liter cultures were then inoculated, the cells were grown at 37°C to an A600 of 0.8. Cells and were then induced by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and left to grow for a further 16 h at 37°C. Soluble expression of the protein was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
All protein purification steps were carried out at 4°C. The cell pellet (30 g) was resuspended in 50 ml of buffer A (Buffer A; 20 mM CHES, pH 8.75), containing two tablets of Complete protease inhibitor cocktail (Roche). The cells were disrupted on ice by sonication using a Branson Sonifier 450. The suspension was then centrifuged (10,000 g for 30 min) to remove cell debris. The supernatant was filtered using a 0.2 μm Millipore syringe filter and applied to a 140 ml Fast Flow Q-Sepharose anion-exchange column pre-equilibrated with buffer A. The proteins were eluted with a linear, one liter gradient of NaCl (0 to 1 M NaCl) in buffer A volumes at 1 ml/min. Protein was detected with an on-line detector monitoring A280, and column fractions were collected and analyzed by SDS-PAGE. Fractions containing the ca. 16 kDa protein were pooled and (NH4)2SO4 was added to a final concentration of 1.5 M. After centrifugation to remove insoluble material, the supernatant was applied to a 140 ml Phenyl Sepharose column pre-equilibrated with buffer C (20 mM CHES, pH 8.75, 1.5 M (NH4)2SO4). Proteins were eluted with a linear, one liter 1.5 - 0 M (NH4)2SO4 gradient at 1 ml/min. The active fractions were pooled and dialyzed extensively against buffer A. The sample was then applied to a 25 ml Mono-Q column anion-exchange column pre-equilibrated with buffer A, and proteins were eluted with a linear 750 ml gradient of NaCl (0 to 1 M NaCl) in buffer A at 1 ml/min. The active fractions were pooled and dialyzed four times against 4 liters of buffer D (Buffer D; 20 mM HEPES 10 mM EDTA, 100 mM (NH4)2SO4, pH 8.25) and concentrated using a YM10 Amicon ultrafiltration membrane to a final concentration of 20 mg/ml and stored in 50% glycerol at -20°C.
Determination of protein concentration
The enzyme concentration was determined from ε 280 nm = 17780 M-1 cm-1 for native PhnO, and turnover numbers are based on an enzyme monomer. The concentration of enzyme was also determined using the bicinchoninic acid protein assay (Pierce) with bovine serum albumin as a standard, which agreed favorably with the value obtained by A280.
Measurement of Enzyme Activity
Initial velocities for the reaction of PhnO were determined using 4,4′-dithiodipyridine (DTDP) to continuously detect the formation of the product CoA at 324 nm (thiopyridone: ε = 19,800 M-1 cm-1) at 25°C A typical reaction mix contained 100 mM HEPES, pH 7.5, 100 mM (NH4)2SO4, 200 μM DTDP, 1 mM AcCoA, and 2 mM aminoalkylphosphonate in a final volume of 1 ml. Reactions were initiated by the addition of enzyme, typically 2 nM final concentration. 100 μM Ni2+ was typically included because a divalent metal is essential for activity.
Initial Velocity Experiments
Initial velocity kinetic data were fitted using Sigma Plot 2000. The substrate specificity of PhnO was determined at pH 7.5 at 10 different concentrations of the variable substrate and a fixed, saturating concentration of the second substrate and at saturating concentrations of Ni2+ [100 μM]. Kinetic constants for acetyl-CoA and other CoA derivatives were determined at fixed saturating concentrations of (S)-1-aminoethylphosphonic acid (S-1AEP) [3 mM] and Ni2+, whilst kinetic constants for the aminoalkylphosphonates were determined using fixed, saturating concentrations of acetyl-CoA [1 mM] and Ni2+. Kinetic constants for divalent metal ions were determined at fixed saturating concentrations of S-1AEP and acetyl-CoA. Individual substrate saturation kinetic data were fitted to Eq. 1
| (Eq. 1) |
where V is the maximal velocity, A is the substrate concentration, and K is the Michaelis-Menten constant (Km). Initial velocity patterns were obtained by measuring the initial rate at five concentrations of each substrate. Eq. 2 was used to fit the intersecting initial velocity pattern,
| (Eq. 2) |
where A and B are the concentrations of the substrates, KA and KB are the Michaelis-Menten constants for the substrates and Kia is the inhibition constant for substrate A.
Dead-end inhibition Studies
Dead-end inhibition patterns were determined by measuring initial velocities (<10% completion) at variable concentrations of one reactant, the second reactant concentration fixed at its Km value, a fixed, saturating concentration of Ni2+ and the inhibitor at several concentrations. Eq. 3 and 4 were used to fit linear, competitive and linear noncompetitive inhibition data, respectively,
| (Eq. 3) |
| (Eq. 4) |
where I is the inhibitor concentration and Kis and Kii are the slope and intercept inhibition constants, respectively.
Dependence of PhnO activity on pH
The pH dependence of the kinetic parameters exhibited by PhnO were determined using S-1AEP as the variable substrate. Acetyltransferase activity was monitored from pH 6.75 to 8.5 every ∼ 0.5 pH unit using the following buffers: PIPES (pH 6.75-7.35) and HEPES (pH 7.15-8.5). The resulting kinetic data were fitted to Eq.1 to obtain the kinetic parameters; kcat and kcat/Km. Profiles were generated by plotting the log of kcat or kcat/Km versus the pH and fitted using Eq. 7 and 8,
| (Eq. 7) |
| (Eq. 8) |
where y is kcat or kcat/Km, C is the pH-independent value of kcat or kcat/Km, H is [H+] and K represents the observed dissociation constant(s) for the ionizing group(s).
Solvent Kinetic Isotope Effects
The solvent kinetic isotope effects on kcat and kcat/Km were determined by measuring the initial velocities using saturating concentrations of acetyl-CoA and Ni2+ whilst varying the concentration of S-1AEP in either H2O or 90% D2O at pH 8.15. Solvent deuterium kinetic isotope effects were fitted to Eq. 9, where EV/K and EV are the isotope effects on kcat/Km -1 and kcat -1, respectively and Fi represents the fraction of isotope
| (Eq. 9) |
RESULTS AND DISCUSSION
Cloning, expression and purification of PhnO
To obtain large quantities of PhnO for mechanistic studies, the phnO gene was cloned into plasmid pET-17b(-) and expressed in E. coli BL21 (DE3) star cells. PhnO was purified using a combination of anion exchange/hydrophobic interaction column chromatographies. These methods were sufficient to achieve a catalytically active protein that was greater than 95% homogeneous (as judged by SDS-PAGE). Approximately 200 mg of purified enzyme was obtained from 30 g of cell paste. Protein electrospray ionization-mass spectrometry was performed on the purified protein, revealing a single species with a molecular mass of 16,499 Da, compared to 16,632 Da expected for full length PhnO, indicating that the N-terminal N-formyl-methionine has been posttranslationally removed. Dynamic light scattering was performed on the protein sample revealing that the protein existed as dimer (data not shown).
Substrate Specificity of PhnO
The substrate specificity of S. .enterica PhnO for acyl-CoA derivatives, summarized in Table 1 was determined at fixed saturating concentrations of S-1AEP and Ni2+. On the basis of kcat/Km values and the probability that acetyl-CoA was the likely physiologically relevant substrate, acetyl-CoA was used as the acyl donor in all subsequent experiments. The kinetic parameters of various aminoalkylphosphonic acids determined at fixed saturating concentrations of acetyl-CoA and Ni2+ are also summarized in Table 1. The kinetic behavior of the aminoalkylphosphonic acid followed two clear patterns. Increasing the alkyl chain from ethyl to butyl resulted in approximately a 70-fold increase in the Km values and 13-fold decrease in kcat values, indicating a preference for shorter aminoalkylphosphonic acids. The kinetic data also suggested that the position of the amine group was important with a preference for the 1 position. The kcat/Km value for S-1AEP was 15-fold higher than 2-aminoethyl phosphonic acid, primarily being an effect on the Km value. The selectivity for the phosphonic acid group was also investigated. Replacement with a carboxyl group, yielding d-alanine, yielded a very poor substrate for which saturation could not be obtained. For d-alanine, only an observed kcat/Km of 3.0 × 101 M-1 sec could be calculated, some 2600-fold lower than the kcat/Km observed for S-1AEP. Mimics of the phosphonate group were also investigated with aminoalkylsulfinic (SO2H) and aminoalkylsulfonic (SO3H) acids. Approximately a five-fold decrease of kcat/Km values were observed with respect to their corresponding phosphonic acid (2-aminoethyl phosphonic acid) and this is primarily a Km effect.
Table 1.
Kinetic parameters for PhnO with various acyl-CoAs and aminoalkylphosphonates. A) Values for acyl-CoA’s were determined at a fixed, saturating concentration of (S)-1-aminoethylphosphonic acid. B) Values for aminoalkylphosphonates, and analogs, were determined at a fixed, saturating concentration of AcCoA. NS: not a substrate
| Compound | Km (mM) | kcat (s-1) | kcat/Km (M-1 s-1) |
|---|---|---|---|
| A | |||
| Acetyl-CoA | 0.06 ± 0.01 | 13.1 ± 0.1 | (2.2 ± 0.3) × 105 |
| Propionyl-CoA | 0.07 ± 0.01 | 14.7 ± 0.7 | (2.1 ± 0.3) × 105 |
| Malonyl-CoA | 0.05 ± 0.01 | 0.4 ± 0.1 | (8.0 ± 0.6) × 103 |
| B | |||
| Aminomethyl phosphonic acid | 1.7 ± 0.1 | 7.0 ± 0.1 | (4.1 ± 0.2) × 103 |
| 2-Aminoethyl phosphonic acid | 1.8 ± 0.1 | 9.0 ± 0.2 | (5.0 ± 0.2) × 103 |
| 3-Aminopropyl phosphonic acid | 4.0 ± 0.4 | 5.4 ± 0.1 | (1.4 ± 0.1) × 103 |
| 4-Aminobutylphosphonic acid | 13.0 ± 1.0 | 0.5 ± 0.1 | (3.8 ± 0.4) × 101 |
| (S)-1-Aminoethylphosphonic acid | 0.17 ± 0.01 | 13.3 ± 0.1 | (7.8 ±0.4) × 104 |
| (R)-1-Aminoethylphosphonic acid | NS | ||
| 1-Aminopropyl phosphonic acid | 4.1 ± 0.2 | 7.4 ± 0.1 | (1.8 ± 0.1) × 103 |
| 1-Aminobutyl phosphonic acid | 12.5 ± 0.8 | 1 ± 0.1 | (8.0 ± 0.5) × 101 |
| D-Alanine | (3.0 ± 0.3) × 101 | ||
| 2-Aminoethanesulfonic acid | 18.9 ± 0.9 | 14.8 ± 0.2 | (7.8 ± 0.3) × 102 |
| 2-Aminoethanesulfinic acid | 16.4 ± 0.7 | 20 ± 0.3 | (1.2 ± 0.1) × 103 |
Metal Ion Requirments
Various divalent metal ions were examined as activators for PhnO. Other enzymes involved in phosphonate catabolism have shown a metal ion dependency, most notably 2-phosphonoacetaldehyde hydrolase (phosphonatase), which requires magnesium for coordination of the phosphonate (17). To investigate whether a tightly-bound metal ion was playing a similar role to that observed in phosphonatase, following the final Mono Q column, PhnO was dialyzed extensively against buffer D in the presence and absence of 10 mM EDTA. Following dialysis with EDTA, PhnO was assayed in the absence of any divalent metal ions. The observed kcat/Km for S-1AEP decreased dramatically after EDTA treatment to 7.2 × 102 M-1 sec-1, this being primarily being a Km effect (Table 2). In contrast when PhnO was extensively dialyzed against buffer alone, the observed kcat/Km value was 4.6 × 104 M-1 sec-1, or half the value determined in the presence of saturating divalent metal ion. This suggests that the enzyme retains half an equivalent of tightly bound divalent metal ion throughout the purification. The metal ion specificity of EDTA-treated PhnO was examined as a function of both the metal ion at fixed, saturating concentrations of acetyl-CoA and S-1AEP and the amino phosphonate at fixed concentrations of acetyl-CoA and divalent metal ions (Table 2). The resulting hyperbolic curves (data not shown) were fitted to Eq. 1 to obtain a maximal kcat and Km values and in the case of the metal ions, the parameter Kact, which is defined as the concentration of divalent metal ion required to half saturate the rate. The S. enterica PhnO exhibited a preference for Ni2+ followed by Mn2+ and very poor activity with Mg2+, as evaluated by the kcat/Kact values, however it is unclear what the physiologically relevant metal ion activator is and further studies are required.
Table 2.
Kinetic parameters for PhnO with various divalent metal ions. A) Kinetic parameters for S-1AEP were determined at a fixed, saturating concentration of AcCoA and the indicated metal ion. B) Kinetic parameters for divalent metal ion activation at saturating concentrations of S-1AEP and AcCoA
| A | KmS-1AEP (mM) | kcat (s-1) | kcat/km (M-1 s-1) |
|---|---|---|---|
| Mn2+ | 0.30 ± 0.02 | 20.6 ± 0.2 | (6.9 ± 0.4) × 104 |
| Ni2+ | 0.17 ± 0.01 | 13.1 ± 0.1 | (8.1 ± 0.3) × 104 |
| Co2+ | 0.28 ± 0.06 | 10.0 ± 0.4 | (3.6 ± 0.7) × 104 |
| Mg2+ | 2.00 ± 0.06 | 17.0 ± 0.3 | (8.5 ± 0.3) × 103 |
| EDTA treated | 18.00 ± 0.76 | 13.0 ± 0.3 | (7.2 ± 0.3) × 102 |
| Non EDTA treated | 0.20 ± 0.02 | 9.2 ± 0.2 | (4.6 ± 0.4) × 104 |
| B | Kact (μM) | kcat (s-1) | kcat/Kact (M-1 s-1) |
| Mn2+ | 6.8 ± 0.6 | 18.7 ± 1.1 | (2.7 ± 0.3) × 106 |
| Ni2+ | 0.6 ± 0.1 | 13.0 ± 0.3 | (2.2 ± 0.3) × 107 |
| Co2+ | 1.3 ± 0.4 | 8.9 ± 1.1 | (6.8 ± 2.2) × 106 |
| Cu2+ | 15.9 ± 2.4 | 4.5 ± 0.2 | (2.8 ± 0.4) × 105 |
| Mg2+ | 850 ± 63 | 16.8 ± 0.2 | (2.0 ± 0.1) × 104 |
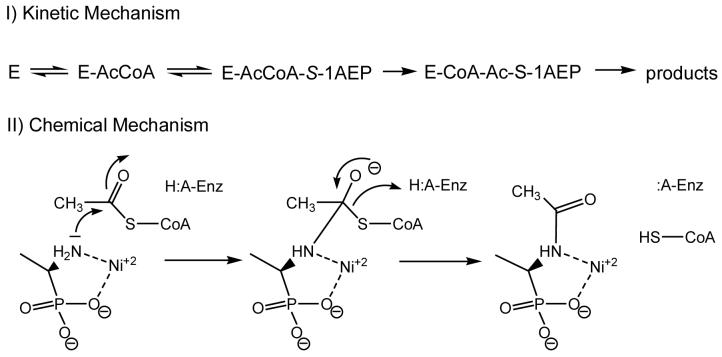
Kinetic Mechanism
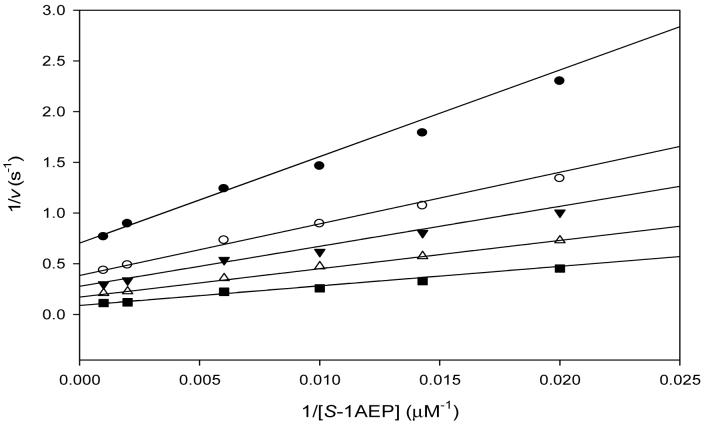
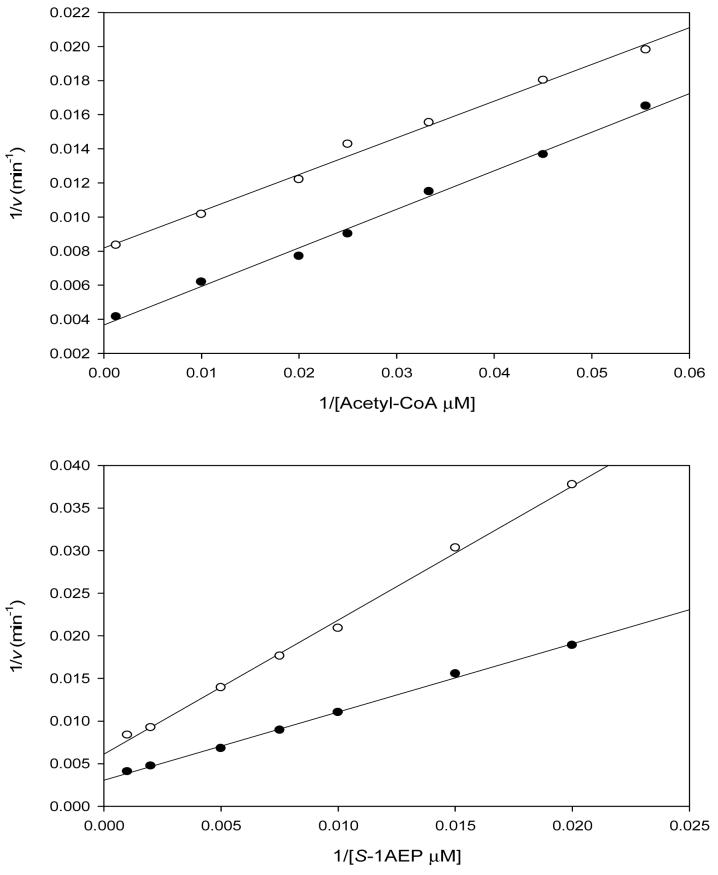
Reactions catalyzed by GNAT acetyltransferases are known to proceed through two distinct mechanisms (18). Only the ESA1 histone acetyltransferase has been shown to proceed via a ping-pong mechanism, with the transfer of the acetyl group from acetyl-CoA to a cysteinyl residue of the enzyme and the subsequent transfer to the ε-amino group of the histone lysine residue (20). The other is a sequential mechanism, where a ternary complex of enzyme, acetyl-CoA, and substrate forms and the acetyl group of acetyl-CoA is directly transferred to a substrate. Reactions catalyzed by all other GNAT superfamily members studied so far, proceed through such a sequential kinetic mechanism (18). The initial velocity patterns were determined using S-1AEP and acetyl-CoA at five different concentrations, respectively, and at a fixed, saturating concentration of Ni2+. The resultant double-reciprocal plot was intersecting, diagnostic of a sequential kinetic mechanism in which both substrates must bind to the enzyme before chemistry can take place (Figure 1). Dead-end inhibition experiments were carried out using desulfocoenzyme A versus either acetyl-CoA or S-1AEP. Desulfocoenzyme A is a coenzyme A analogue that lacks the terminal sulfhydryl of CoA and is a dead-end inhibitor since it forms a non-productive complex with PhnO and the aminophosphonic acid. Desulfocoenzyme A exhibited linear, competitive inhibition versus acetyl-CoA (Kis: 79 ± 3 μM) and linear, noncompetitive inhibition versus S-1AEP (Kis: 219 ± 44 μM; Kii: 204 ±: 33 μM). These dead-end inhibition patterns are only compatible with the ordered binding of acetyl-CoA followed by S-1AEP, as has been observed with other GNAT acetyltransferases (18).
Figure 1.
Double reciprocal plot of initial rate data at varying S-1AEP concentrations and fixed concentrations of acetyl-CoA at 10 μM (♀), 20 μM (○), 30 μM (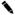 ), 60 μM (→) and 250 μM (†). The pattern of intersecting lines is indicative of a sequential kinetic mechanism.
), 60 μM (→) and 250 μM (†). The pattern of intersecting lines is indicative of a sequential kinetic mechanism.
Dependence of pH
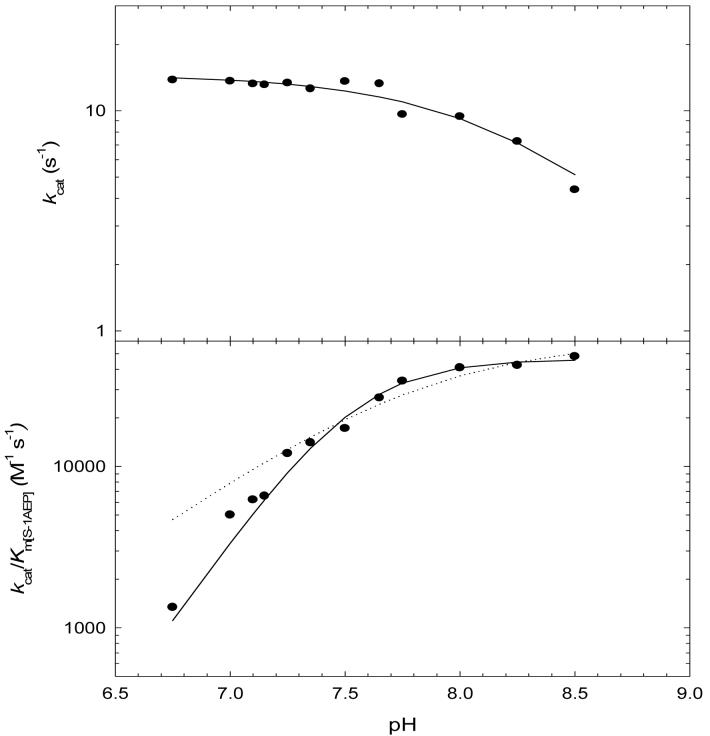
pH studies were performed to investigate the ionization behavior of groups responsible for catalysis and binding. The pH depencency of kcat and kcat/KmS-1AEP were investigated at saturating concentrations of acetyl-CoA and Ni2+. The kcat pH profile revealed the presence a single ionizable group exhibiting a pK value of 8.2 (± 0.1). We ascribed this group to an enzymic group that functions as a general acid involved in protonation of the initially formed thiolate after tetrahedral intermediate collapse. The kcat/KmS-1AEP profile reveals two groups with similar pK values of 7.5 (± 0.1) (Figure 2). The two similar pKa’s are likely to be ionizable groups from the aminoalkylphosphonate substrate, most likely the amine and the phosphonoxygen groups (both have pKa values around 7; (21)), both of which are unprotonated when S-1AEP binds to the E-acetyl-CoA complex. From these data a chemical mechanism can be proposed where after ordered substrate binding, a tetrahedral intermediate is formed followed by intermediate collapse and protonation of the CoA thiolate (Scheme 2). The exact role of the metal ion is unclear, but could bind to the enzyme and aid in the binding of the aminoalkylphosphonate by coordinating the oxygen’s of the phosphonate group in a similar fashion to which 2-phosphonoacetaldehyde hydrolase binds to Mg2+ (17). However, we favor a role in which the metal binds to the enzyme, and the aminoalkylphosphonate contributes both the amine and one of the phosphonoxygens atoms as metal ligands, which is supported by the absence of any general base catalysis observed in the kcat pH profile. Preliminary isothermal titration calorimetry data suggest that metals can bind to the free enzyme (data not shown). This could position the amine group of the aminoalkylphosphonate for acetyltransfer and reduce the pK of the amine, making it more nucleophilic for attack on the thioester (such metal complexes of aminoalkylphosphonates have previous been described (22)).
Figure 2.
Dependence of kcat and kcat/KS-1AEP on pH at saturating levels of acetyl-CoA and Ni2+. The dotted and solid lines are fits to Eqs 7 and 8, respectively. The kcat pH profile revealed the presence a single ionizable group with a pK value of 8.2 (± 0.1). The kcat/KS-1AEP profile was fit to Eq. 8, which describes the dependence on 2 groups with similar pK values of 7.5 (± 0.1) and Eq. 7, which describes the dependence on 1 group (dotted line).
Scheme 2.
Proposed kinetic and chemical mechanisms for aminophosphonate N-acetylation by PhnO
Solvent Kinetic Isotope Effects
Solvent kinetic isotope effects were determined by measuring initial velocities in both H2O and 90% D2O. Both acetyl-CoA and S-1AEP were investigated as the variable substrate at seven different concentrations, with reactions performed under fixed, saturating concentrations of the other substrate and Ni2+. These experiments were performed at pH 8.15 where both kcat and kcat/Km are relatively independent of pH. When acetyl-CoA was the variable substrate, a D2Okcat/Kacetyl-CoA value of unity, and a D2Okcat value of 2.2 (± 0.1) were observed, implying that acetyl-CoA binds first, and its dissociation is prevented by S-1AEP binding. When S-1AEP was the variable substrate, equivalent solvent kinetic isotope effects of 2.2 (± 0.1) were observed on D2Okcat and D2Okcat/KS-1AEP suggesting that S-1AEP is not a “sticky substrate” (Figure 3). These data are in agreement with the dead-end studies that suggested an ordered binding of acetyl-CoA followed by the aminoalkylphosphonate.
Figure 3.
Reciprocal plots of the solvent kinetic isotope effect determined for PhnO using acetyl-CoA (top panel) or S-1AEP (bottom panel) as the variable substrate, in the presence of saturating concentrations of Ni2+. The symbols are the experimentally determined values in H2O (♀)or 90% D2O ( ), while the lines are fits of the data to Eq. 9.)
), while the lines are fits of the data to Eq. 9.)
A proton inventory experiment was performed by varying the atom fraction of D2O at saturating concentrations of both S-1AEP and acetyl-CoA using Ni2+ as the metal activator. Each data point was determined in triplicate, yielding a linear relation between the rate and mole fraction of deuterium (data not shown). The equivalent values of D2Okcat and D2Okcat/KS-1AEP and the linear proton inventory suggest that a single proton is transferred in a step that is at least partially rate-limiting in the chemical reaction catalyzed by PhnO. On the basis of the kcat pH profile, the most likely single proton transfer is from the enzymic general acid to the thiolate of CoA, formed upon decomposition of the tetrahedral intermediate. In the structures of a number of GNAT-substrate complexes, a suitably positioned tyrosine residue has been ascribed as the general acid (18).
CONCLUDING REMARKS
Although we have demonstrated that PhnO functions as an aminoalkylphosphonic N-acetyltransferase, a number of questions still exist. PhnO acetylates a range of aminoalkylphosphonates, including the physiologically relevant 2-aminoethylphosphonic acid. The chemical and kinetic mechanisms proposed are similar to those proposed for other GNAT acetyltransferases, with the sequential binding of acetyl-CoA followed by the binding of the acyl acceptor (the aminoalkylphosphonate), with subsequent formation of the tetrahedral intermediate and product release (23). However, the divalent metal ion activation that was observed is, to date, unique for a GNAT acetyltransferase. It seems likely that the divalent metal ion activation has evolved to play a specific role in aminoalkylphosphonate binding similar to that observed in other enzymes involved in phosphonate catabolism (phosphonatase;(17). This would also serve to reduce the pK value of the amine and assist in catalysis. The physiological relevance of the PhnO catalyzed reaction is unclear, as acetylation of the amine group would prevent its transamination and subsequent cleavage of the C-P bond. We propose two physiological roles for aminoalkylphosphonate acetylation: storage and protection. When amounts of aminoalkylphosphonates are available in the environment that exceed the organisms phosphate requirements, these phosphonates can be stored in an acetylated form and later used after hydrolysis by a deacetylase. S-1AEP is known to have antibacterial properties (24). S-1AEP is a structural analogue of d-alanine, a component of the muramyl-pentapeptide involved in the formation of Gram-negative cross-linked peptidoglycan. S-1AEP is a slow onset inhibitor of the S. enterica d-alanine:d-alanine ligase, exhibiting a Ki value of 0.5 mM (25). S-1AEP is also a slow onset inhibitor of the Streptococcus faecalis alanine racemase (26). Thus, PhnO, by acetylating S-1AEP, would protect against the deleterious effects of S-1AEP as a result of the inhibition of these two key enzymes in peptidoglycan biosynthesis.
Acknowledgement
We thank Sophie Magnet (AECOM) for the generous donation of the S. enterica pET-28a(-):phnO onstruct. The authors wish to thank Argyrides Argyrou for thoughtful discussions.
This work was supported by NIH Grant AI33696 and AI60899.
ABBREVATIONS
- AcCoA
acetyl-Coenzyme A
- CHES
2-(Cyclohexylamino)ethanesulfonic acid
- CoA
Coenzyme A
- DTDP
4,4′-dithiodipyridine
- EDTA
(Ethylenedinitrilo)tetraacetic acid
- GNAT
GCN5-related N-acyltransferase
- HEPES
N-[2-hydroxyethyl] piperazine-N’-[2-ethanesulfonic acid]
- IPTG
Isopropyl-thio-β-d-galactopyranoside
- LB
Luria Broth
- PCR
polymerase chain reaction
- PIPES
Piperazine-1,4-bis(2-ethanesulfonic acid)
- S-1AEP
(S)-1-aminoethylphosphonic acid
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TEA
triethanolamine
REFERENCES
- 1.Vershinina OA, Znamenskaia LV. The Pho regulons of bacteria. Mikrobiologiia. 2002;71:581–95. [PubMed] [Google Scholar]
- 2.Kononova SV, Nesmeyanova MA. Phosphonates and their degradation by microorganisms. Biochemistry (Mosc) 2002;67:184–95. doi: 10.1023/a:1014409929875. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy KE, Thompson GA., Jr. Phosphonolipids: localization in surface membranes of Tetrahymena. Science. 1970;168:989–91. doi: 10.1126/science.168.3934.989. [DOI] [PubMed] [Google Scholar]
- 4.Conlin CA, Tan SL, Hu H, Segar T. The apeE gene of Salmonella enterica serovar Typhimurium is induced by phosphate limitation and regulated by phoBR. J. Bacteriol. 2001;183:1784–6. doi: 10.1128/JB.183.5.1784-1786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanner BL. Molecular genetics of carbon-phosphorus bond cleavage in bacteria. Biodegradation. 1994;5:175–84. doi: 10.1007/BF00696458. [DOI] [PubMed] [Google Scholar]
- 6.Metcalf WW, Wanner BL. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene. 1993;129:27–32. doi: 10.1016/0378-1119(93)90692-v. [DOI] [PubMed] [Google Scholar]
- 7.Jiang W, Metcalf WW, Lee KS, Wanner BL. Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J. Bacteriol. 1995;177:6411–21. doi: 10.1128/jb.177.22.6411-6421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wackett LP, Shames SL, Venditti CP, Walsh CT. Bacterial carbon-phosphorus lyase: products, rates, and regulation of phosphonic and phosphinic acid metabolism. J. Bacteriol. 1987;169:710–7. doi: 10.1128/jb.169.2.710-717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White AK, Metcalf WW. Two C-P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 2004;186:4730–9. doi: 10.1128/JB.186.14.4730-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim AD, Baker AS, Dunaway-Mariano D, Metcalf WW, Wanner BL, Martin BM. The 2-aminoethylphosphonate-specific transaminase of the 2-aminoethylphosphonate degradation pathway. J. Bacteriol. 2002;184:4134–40. doi: 10.1128/JB.184.15.4134-4140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metcalf WW, Wanner BL. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA’ elements. J. Bacteriol. 1993;175:3430–42. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalf WW, Wanner BL. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 1991;173:587–600. doi: 10.1128/jb.173.2.587-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulakova AN, Kulakov LA, Akulenko NV, Ksenzenko VN, Hamilton JT, Quinn JP. Structural and functional analysis of the phosphonoacetate hydrolase (phnA) gene region in Pseudomonas fluorescens 23F. J. Bacteriol. 2001;183:3268–75. doi: 10.1128/JB.183.11.3268-3275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumora C, Marche M, Doignon F, Aigle M, Cassaigne A, Crouzet M. First characterization of the phosphonoacetaldehyde hydrolase gene of Pseudomonas aeruginosa. Gene. 1997;197:405–12. doi: 10.1016/s0378-1119(97)00185-6. [DOI] [PubMed] [Google Scholar]
- 15.Baker AS, Ciocci MJ, Metcalf WW, Kim J, Babbitt PC, Wanner BL, Martin BM, Dunaway-Mariano D. Insights into the mechanism of catalysis by the P-C bond-cleaving enzyme phosphonoacetaldehyde hydrolase derived from gene sequence analysis and mutagenesis. Biochemistry. 1998;37:9305–15. doi: 10.1021/bi972677d. [DOI] [PubMed] [Google Scholar]
- 16.Olsen DB, Hepburn TW, Lee SL, Martin BM, Mariano PS, Dunaway-Mariano D. Investigation of the substrate binding and catalytic groups of the P-C bond cleaving enzyme, phosphonoacetaldehyde hydrolase. Arch. Biochem. Biophys. 1992;296:144–51. doi: 10.1016/0003-9861(92)90556-c. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Morais MC, Dai J, Zhang W, Dunaway-Mariano D, Allen KN. Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily. Biochemistry. 2004;43:4990–7. doi: 10.1021/bi036309n. [DOI] [PubMed] [Google Scholar]
- 18.Vetting MW, LP S. d. C., Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–26. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Chase JF, Middleton B, Tubbs PK. A coenzyme A analogue, desulpho-coA; preparation and effects on various enzymes. Biochem. Biophys. Res. Commun. 1966;23:208–13. doi: 10.1016/0006-291x(66)90529-8. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 2002;9:862–9. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 21.Wozniak M, Nicole J, Tridot G. N°701 - Complexes du cuivre (II) et des acides α-aminoalkylphosphoniques: correlation entre acidites, facteurs steriques et pouvoir complexant. Bull. Soc. Chem. 1972;11:4445–4452. [Google Scholar]
- 22.Song B, Chen D, Bastian M, Martin BR, Sigel H. 155. Metal-Ion-Coordination Properties of a Viral Inhibitor, a Pyrophosphate Analogue, and a Herbicide Metabolite, a Glycinate Analogue: The Solution Properties of the Potentially Five-Membered Chelates Derived from Phosphonoformic acid and (Aminoethyl)phosphonic acid. Helvetica Chem. Acta. 1994;77:1738–1756. [Google Scholar]
- 23.Draker KA, Northrop DB, Wright GD. Kinetic mechanism of the GCN5-related chromosomal aminoglycoside acetyltransferase AAC(6′)-Ii from Enterococcus faecium: evidence of dimer subunit cooperativity. Biochemistry. 2003;42:6565–74. doi: 10.1021/bi034148h. [DOI] [PubMed] [Google Scholar]
- 24.Badet B, Inagaki K, Soda K, Walsh CT. Time-dependent inhibition of Bacillus stearothermophilus alanine racemase by (1-aminoethyl)phosphonate isomers by isomerization to noncovalent slowly dissociating enzyme-(1-aminoethyl)phosphonate complexes. Biochemistry. 1986;25:3275–82. doi: 10.1021/bi00359a029. [DOI] [PubMed] [Google Scholar]
- 25.Duncan K, Walsh CT. ATP-dependent inactivation and slow binding inhibition of Salmonella typhimurium d-alanine: d -alanine ligase (ADP) by (aminoalkyl)phosphinate and aminophosphonate analogues of d -alanine. Biochemistry. 1988;27:3709–14. doi: 10.1021/bi00410a028. [DOI] [PubMed] [Google Scholar]
- 26.Badet B, Walsh C. Purification of an alanine racemase from Streptococcus faecalis and analysis of its inactivation by (1-aminoethyl)phosphonic acid enantiomers. Biochemistry. 1985;24:1333–41. doi: 10.1021/bi00327a010. [DOI] [PubMed] [Google Scholar]