Abstract
The voltage-gated Ca2+ channels that effect tonic release of neurotransmitter from hair cells have unusual pharmacological properties: unlike most presynaptic Ca2+ channels, they are sensitive to dihydropyridines and therefore are L-type. To characterize these Ca2+ channels, we investigated the expression of L-type α1 subunits in hair cells of the chicken’s cochlea. In PCRs with five different pairs of degenerate primers, we always obtained α1D products, but only once an α1C product and never an α1S product. A full-length α1D mRNA sequence was assembled from overlapping PCR products; the predicted amino acid sequence of the α1D subunit was about 90% identical to those of the mammalian α1D subunits. In situ hybridization confirmed that the α1D mRNA is present in hair cells. By using a quantitative PCR assay, we determined that the α1D mRNA is 100–500 times more abundant than the α1C mRNA. We conclude that most, if not all, voltage-gated Ca2+ channels in hair cells contain an α1D subunit. Furthermore, we propose that the α1D subunit plays a hitherto undocumented role at tonic synapses.
By controlling synaptic transmission and electrical tuning, the voltage-gated Ca2+ channels of hair cells play a key role in auditory signaling (reviewed in ref. 1). These channels open when mechanical stimulation of the hair bundle depolarizes the cell membrane. The rising local Ca2+ concentration increases the tonic release of neurotransmitter, which stimulates afferent neurons (reviewed in ref. 2). The entering Ca2+ also opens Ca2+-activated K+ channels that are clustered with the Ca2+ channels and repolarize the cell membrane. The interplay of these ion channels can electrically tune each cell to a characteristic frequency (reviewed in refs. 3 and 4).
At most synapses, transmitter release depends on N-type or P-type Ca2+ channels, which are blocked by ω-conotoxin GIVA and ω-agatoxin IVA, respectively (reviewed in ref. 5). In hair cells, however, the Ca2+ channels have different pharmacological properties: they are sensitive to dihydropyridines and therefore are L-type channels (6–9). In only a few other cell types, such as cultured sensory neurons (10) and retinal bipolar cells (11), have L-type channels been shown to effect neurotransmitter release.
The drug sensitivity and permeability of a voltage-gated Ca2+ channel depend on its type of α1 subunit (reviewed in ref. 12). The pore-forming α1 protein is 160–240 kDa in size, with a cytoplasmic amino terminus, four homologous repeats (I–IV) of six transmembrane segments (S1-S6) each, and a cytoplasmic carboxyl terminus. L-type channels contain the product of the α1C (cardiac), the α1D (neuroendocrine), or the α1S (skeletal muscle) gene. To characterize the unusual L-type Ca2+ channels that control synaptic transmission, we sought to determine which of these three α1 genes are expressed in hair cells of the chicken’s cochlea.
MATERIALS AND METHODS
Histology.
White Leghorn chickens (Gallus gallus) were asphyxiated with CO2 and decapitated. The temporal bones with intact cochleae were excised and fixed overnight at 4°C with 0.75% (wt/vol) paraformaldehyde and 2.5% (vol/vol) glutaraldehyde in a buffer solution containing 70 mM sodium phosphate (pH 7.4), 75 mM sucrose, and 0.9 mM CaCl2. After two rinses in the buffer solution, the cochleae were carefully dissected from the bone, fixed with 1% (wt/vol) OsO4 in buffer solution, dehydrated successively with ethanol and propylene oxide, and embedded in epoxy resin consisting of EMbed 812, Araldite 6005, dodecenyl succinic anhydride, and 2,4,6-tris(dimethylaminomethyl)phenol (25:20:60:1 by volume; Electron Microscopy Sciences, Fort Washington, PA). Semi-thin sections were cut 1 μm thick.
RNA and DNA Isolation.
Chickens 1 week to 2 months old were killed as above. Under a dissecting microscope, layers of bone were shaved from the skull with a scalpel until the entire length of each cochlea was exposed. The cochlear duct was grasped at the lagena with fine forceps, lifted from the temporal bone while leaving most of the ganglion behind, and placed into low-divalent cation solution (9). After the tegmentum vasculosum had been removed with fine forceps, the basilar papilla and tectorial membrane were scraped out with a hypodermic needle and frozen in liquid nitrogen. The brain was also dissected and frozen. Total RNA was isolated from these tissue samples with a solution of phenol and guanidinium isothiocyanate (Trizol, Life Technologies, Gaithersburg, MD). Genomic chicken DNA was purified by lysing blood-cell nuclei in an acidic solution of guanidine hydrochloride (13).
cDNA Synthesis and PCR Amplification.
First-strand cDNA was synthesized with reverse transcriptase (Superscript II, Life Technologies). A 20-μl reaction typically contained total RNA from 10 basilar papillae or 1 μg total RNA from other tissues, 20 units of recombinant RNase inhibitor (RNasin, Promega), and random hexamers or oligo(dT) as primers. The reaction mixture was incubated first at 44°C for 1 h, then at 95°C for 10 min, diluted to 500 μl with a solution containing 10 mM Tris⋅HCl (pH 8.0) and 1 mM EDTA, and stored at −20°C.
PCRs were conducted in 20- to 50-μl volumes and typically contained 1–2 μl diluted cDNA, 50 mM KCl, 10 mM Tris⋅HCl (pH 9.0 at 25°C), 0.1% (vol/vol) polyethylene glycol-p-isooctylphenyl ether (Triton X-100), 2 mM MgCl2, 1 μM of each oligonucleotide primer, 0.1 mM each dATP, dCTP, dGTP, and dTTP, and 0.03 units Taq DNA polymerase (Promega). An initial denaturation step at 94°C for 2 min was followed by 30 cycles at 94°C for 1 min, 46–50°C with degenerate primers or 55°C with nondegenerate primers for 1 min, and 72°C for 2 min, and a final extension step at 72°C for 5 min.
The cDNA clones from the basilar papilla (Fig. 2) were obtained with the following primer pairs (5′ to 3′; N = any base; R = A or G; Y = C or T; D = A, G, or T; H =A, C, or T): pSE193/194, CCGGATGTGAGTGTCATGTT, TGGTGCTGCTTGCATAGTTC; pSE42/43-1, GAARCCNTTYGAYATHTTYA, CYTCCATNGTDATRCAYTG; pSE9/39-1, GGNAARATGCAYAARACNTG, CAAGCAACGAGAAGATTATG; pSE12/13-1, TTTYAAYCGNTTYGAYTGYTT, CCNACRAADATRTTCATCAT; pSE4/6-1, GTNCARCTNTTYAARGGNAA, CCRAANACYTGCATNCCDAT; pSE1/3-1, GAYSCNTGGAAYGTNTTYGA, NARRTARTCRAARTTRTCCAT; pSE14/15-1, TTYCARACNTTYCCNCARGCNGT, AAYTTNCCNACNGTNACYTCRTC; pSE48/154-1, TGATGAACCAGAGGAAAACAA, TGAACGCGTGGAACGACTT; and pSE141/142-1, ATGCARCARCARATHATGGC, GTDATRCADATCATYTCRTC.
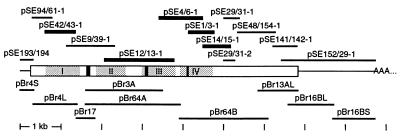
Figure 2.
Cloning of α1D cDNAs from the chicken’s basilar papilla and brain. Horizontal bars indicate the extent of each partial cDNA and are aligned with a diagram of the α1D mRNA in the middle. pSE… , PCR products from the basilar papilla; pBr… , clones from a brain cDNA library; rectangle, the ORF; AAA… , poly(A) tail; gray areas, homologous repeats I–IV; and black areas, the I-II-loop insert, the alternative IIIS2 segment, and the IVS2–3 insert in the basilar papilla. Thicker bars indicate products of PCRs with degenerate primers that were designed to amplify cDNAs of any L-type α1 subunit.
The termini of the α1D mRNA in the basilar papilla were obtained by rapid amplification of cDNA ends (14). For the 5′ end, cDNA was synthesized with the α1D-specific primer CAGGACAATTTGTAAACTGGG and tailed with dCTP. Consecutive PCRs with the nested α1D-specific primers ATATGCATTGGGGTGTAATAATAAT and CCATATGCTATAATCTTCAAAAATGT yielded only products that extended from exon 2 into intron 1, such as clone pSE94/61-1. The 5′-most clone from the basilar papilla, pSE193/194, was obtained only after the 5′ end of the α1D mRNA had been isolated from a brain cDNA library (see below). For the 3′ end, cDNA was synthesized with an anchored oligo(dT) primer. Consecutive PCRs with the nested α1D-specific primers GAGAGCAGGAATATTATAGTGGAGA and CCTGGTACACAGACGACCCT yielded clone pSE152/29-1; reactions with the nested primers GGCCTGCAAGAGGTTAGT and GCTTTGGTTAGGACTGCTC produced clones pSE29/31-1 and -2, which diverge from the composite sequence after exons 42 and 41, respectively (Fig. 4).
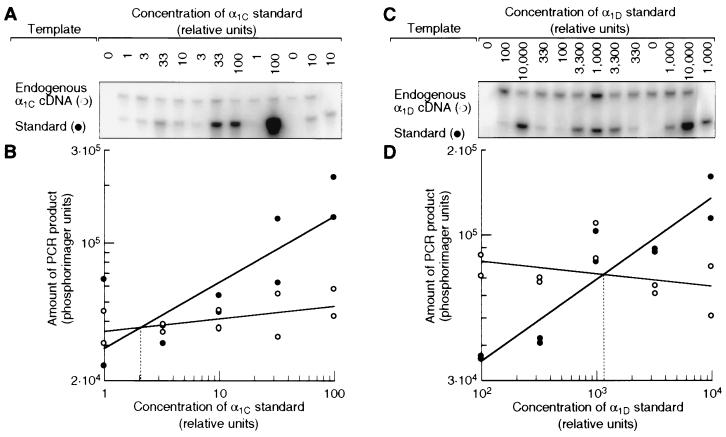
Figure 4.
Relative abundances of α1C and α1D mRNAs in the basilar papilla. (A) Southern blot of quantitative PCR assay. Aliquots of cDNA from the basilar papilla were spiked with serial dilutions of an α1C template as an internal standard. The PCR product of this standard was shorter than that of the endogenous α1C cDNA but accumulated with the same efficiency (data not shown). Both templates were amplified together with a primer pair specific for the α1C subunit, and the products were detected by hybridization with a radiolabeled oligonucleotide. The result from one of two independent experiments is shown. The control PCRs for the first and last lanes contained no cDNA from the basilar papilla. (B) Amounts of PCR products from endogenous α1C cDNA (○) and from α1C standard (•) plotted against the initial standard concentration. Straight lines fitted to each data set intersect at the standard concentration that gave rise to the same amount of PCR product as the endogenous α1C cDNA (dotted line). This standard concentration therefore was equal to the concentration of endogenous α1C cDNA in the sample. (C and D) Same as A and B, but with primers and standards for the α1D cDNA. The relative unit of concentration represents the same number of template molecules for both standards.
Library Screening.
From a library of chicken-brain cDNA in the phage vector λgt11 (CLONTECH), we isolated clones pBr17, pBr3A, pBr13AL, and pBr16BL/S (Fig. 2) by plaque hybridization with the radiolabeled α1D cDNAs from the basilar papilla. Screening subpools of this library by PCRs yielded the clones pBr4S/L and pBr64A/B. To obtain additional clones from the 3′ end of the α1D mRNA, we screened by plaque hybridization with the clones pBr64B, pBr13AL, and pSE152/29-1 two libraries in the phage vector HybriZAP (Stratagene) of cDNAs from cochleae at embryonic days 14–19 and from basilar papillae at 1–2 weeks after hatching.
DNA Sequencing.
PCR products were ligated into the plasmid vectors pCRII (Invitrogen) or pGEM-T Easy (Promega). Inserts in λgt11 were excised with the restriction enzyme EcoRI and ligated into the plasmid vector pBluescript II SK(+) (Stratagene). All cDNA clones were analyzed by manual or, in a few cases, automated sequencing. All templates were sequenced completely on both strands by primer walking; 27 single-base discrepancies—probably amplification errors or sequence polymorphisms—were resolved by sequencing at least three independent clones.
In Situ Hybridization.
Cochleae were fixed in periodate-lysine-paraformaldehyde (15), cryoprotected with 30% (wt/vol) sucrose in a 0.1-M sodium-phosphate solution at pH 7.4, and cryosectioned at 20-μm thickness. In situ hybridizations were conducted with digoxigenin-labeled RNA probes (16). No staining was observed in control hybridizations without probe or with a sense probe.
Quantitative PCR.
To prepare a standard template for the α1C mRNA, we first deleted 34 bp from cDNA clone pSE1/3-3 by replacing base pairs 313–363 with ACGCATAGAGAAGCACT; this clone had been obtained from the basilar papilla with the same primers as pSE1/3-1 but contained 0.6 kb of α1C sequence. For the α1D mRNA, we similarly deleted 29 bp from cDNA clone pSE9/39-1 by replacing base pairs 1919–1964. The α1 sequences in these deletion clones were then amplified in PCRs with the same pair of vector primers, one of which was radiolabeled. The products were purified from agarose gels and quantitated relative to each other both by scintillation counting and by gel electrophoresis and detection with a phosphor-storage screen (PhosphorImager, Molecular Dynamics). The values obtained with either method differed by less than a factor of 2.
To quantitate the endogenous α1C and α1D mRNA, we synthesized cDNA with random-hexamer primers from total RNA of the basilar papilla and the brain; we assumed that the isolation and reverse transcription were equally efficient for both mRNAs. Aliquots of each cDNA were spiked with serial dilutions of either standard; the appropriate dilutions were established for each combination in pilot experiments. PCRs were conducted as above, except that the number of amplification cycles was reduced to 25 to avoid saturation, with the primers CAGGTGTTTGGTAAAATTGCAC and GGGGTCGCACTTCTTGTCT (α1C) or TGTAGAGCTGCAGTAAAATCTGTC and CAGGTGAACAAAGCCAGAAGAA (α1D). From the endogenous α1C and α1D cDNAs, these primer pairs amplified 293-bp and 156-bp products, respectively, both from regions where alternative splicing has not been observed.
The PCR products were analyzed by Southern blotting. They were fractionated in a 6% (wt/vol) polyacrylamide gel containing 8 M urea (NOVEX, San Diego), transferred electrophoretically onto a positively charged nylon membrane, and hybridized (ExpressHyb, CLONTECH) at 50°C for 3 h with the radiolabeled internal oligonucleotides TACCACAGAAATCAACCGCA (α1C) or ACCAAGACAATAACAAGCCAA (α1D). The membrane was washed thrice at 37°C for 10 min with a solution containing 4× SSC (1× SSC is 0.15 M NaCl and 15 mM sodium citrate) and 0.1% (wt/vol) SDS and twice at 37°C for 15 min with a solution containing 2× SSC and 0.1% (wt/vol) SDS. To detect the PCR products, a storage phosphor screen was exposed to the membrane and scanned. The PCR products were quantitated by summing the pixel values for each band of the scanned image with iplab gel software (Signal Analytics, Vienna, VA). The entire quantitative PCR experiment was performed twice with independently prepared standards and cDNAs.
RESULTS
Expression of L-type α1 Genes in the Basilar Papilla.
We isolated RNA and synthesized cDNA from the basilar papilla (Fig. 1A), the sensory epithelium that runs the length of the cochlea and contains about 104 hair cells and twice as many supporting cells (17, 18). Because α1 sequences from birds were not available, we aligned all published full-length α1C (19–25), α1D (26–30), and α1S (31–33) sequences and designed five pairs of degenerate oligonucleotide primers on the basis of the most conserved amino acid stretches. In PCRs with these different primer pairs and cDNA from the basilar papilla, we always obtained products that were most similar to the mammalian α1D sequences, but never an α1S and only once an α1C product. To demonstrate that these primer pairs did not exclusively amplify α1D products, we conducted PCRs with one pair and cDNA from the chicken’s brain and obtained products not only from α1C mRNA, but even from the more distantly related, non-L-type α1A (P/Q-type) and α1B (N-type) mRNAs.
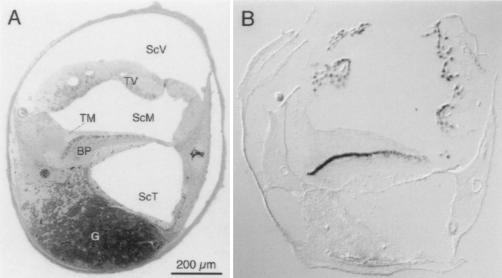
Figure 1.
Expression of the α1D subunit in cochlear hair cells. (A) Structure of the chicken’s cochlea. A semi-thin transverse section was taken near the middle of the organ and stained with toluidine blue. The hair cells are the dark, oblong cells at the upper margin of the basilar papilla (BP), the homolog of the mammalian organ of Corti; tall hair cells are toward the left and short hair cells are toward the right. ScV, scala vestibuli; ScM, scala media; ScT, scala tympani; TV, tegmentum vasculosum; TM, tectorial membrane; G, cochlear ganglion. (B) Localization of α1D mRNA by in situ hybridization. Digoxigenin-labeled antisense RNA derived from clone pBr64B was hybridized to a cryosection from a position similar to that in A and detected with anti-digoxigenin antibodies in a color reaction.
To ascertain the cellular origin of the α1D PCR products, we conducted in situ hybridizations. On transverse cryosections through the cochlea, an antisense RNA probe for the α1D mRNA bound to hair cells in the basilar papilla and to the tegmentum vasculosum, the vascular structure responsible for maintaining the cochlea’s ionic milieu (Fig. 1B). This result confirmed that the α1D gene is expressed in hair cells of the basilar papilla.
Full Length α1D Sequences from the Basilar Papilla and the Brain.
We first closed the gaps in the sequence from the basilar papilla by conducting additional PCRs (Fig. 2). We also screened cDNA libraries from the embryonic cochlea and from the chick’s basilar papilla, but found no sequences different from the PCR products. Among 2⋅106 clones from each library, we detected 23 and 24 α1D clones, respectively. Because hair cells represent about one-third of the cells in the basilar papilla, we estimate the abundance of the α1D mRNA in hair cells to be about 1 in 104. To detect possible amplification errors, we then used all the PCR products to isolate α1D clones from a brain cDNA library (Fig. 2). We chose this organ because a functional α1D cDNA has been cloned from the human brain (28). From these two sets of overlapping, partial clones, we assembled full-length α1D mRNA sequences for the basilar papilla and brain that both terminate after a consensus polyadenylation signal (34); without their poly(A) tails, these are 8719 bases and 8611 bases long, respectively. These composite sequences are identical except for three alternatively spliced exons (35).
Three lines of evidence suggested that translation starts at the fourth AUG (nucleotides 253–255) from the 5′ ends of these α1D sequences. First, the sequence around this codon matches the consensus of vertebrate translation initiation sites (36). Second, the longest ORFs of about 6.5 kb begin only with this codon. Third, sequencing of genomic DNA around the 5′ end indicated that the first 295 bases are identical in the primary transcript and the mRNA (data not shown).
Comparison of α1D Subunits from Chicken and from Mammals.
The ORFs encode proteins of 2,190 aa in the basilar papilla (Fig. 3) and 2,154 aa in the brain. They are 88–92% identical to the sequences of the mammalian α1D proteins, but only 70–71% identical to the α1C proteins and 65–66% identical to the α1S proteins. All four glutamate residues that are conserved in the presumptive pore-lining regions of voltage-gated Ca2+ channels and that confer Ca2+ selectivity (37) are present in the chicken sequence (residues 359, 726, 1122, and 1435). The region of highest similarity, a stretch of 227 aa that begins in the sixth transmembrane segment of the fourth repeat (IVS6), is identical in the human and the chicken; in L-type α1 subunits, this region contains conserved binding sites for dihydropyridines (38–40) and phenylalkylamines (41). Two other regions that are necessary for dihydropyridine binding, transmembrane segments IIIS5 and IIIS6 (38, 39, 42), are identical at all but one residue. Aside from the alternative exons mentioned above, the biggest difference is the absence in the chicken sequence of a run of six or seven methionine residues found at the amino termini of all mammalian α1D subunits. Together, these results indicate that the cDNAs from the basilar papilla and brain are products of the chicken ortholog of the mammalian L-type α1D gene.
Figure 3.
Comparison of α1D protein sequences from the chicken and human. The chicken’s sequence (Top) was predicted from the composite of the cDNAs from the basilar papilla; the sequence predicted from the chicken’s brain cDNAs is identical except for the I-II-loop insert (exon 9a), the alternative IIIS2 segment (exon 22a), and the IVS2–3 insert (exon 30a), which are printed in reverse contrast. The sequence from the human brain (28) is aligned underneath; only residues that differ from the chicken’s are given, and periods indicate gaps. The sequence of the alternative IIIS2 segment isolated from the chicken’s brain (exon 22) is identical to that of the human IIIS2 segment; the sequence of an alternative IVS3 segment isolated from the chicken’s brain (exon 31a, replacing amino acids 1294–1321) is HYFTDAWNTFDALIVVGSVVDIAITEVN. Arrows indicate where additional splice variants from the basilar papilla diverge at the ends of exons 41, 42, and 45. Horizontal lines above the alignment mark the putative transmembrane segments IS1-IVS6, the consensus binding site for the channel’s β subunit (65), and the Ca2+-binding EF-hand motif (66). (For a detailed description of the alternative exons, see ref. 35.)
Predominance of the α1D mRNA in the Basilar Papilla.
To verify that the amount of α1C mRNA in the basilar papilla is negligible, we developed a quantitative PCR assay. The relative concentrations of α1C and α1D mRNA were measured indirectly by comparing each with a precisely quantitated internal standard. We thus determined that there is 100–500 times as much α1D as α1C mRNA in the basilar papilla (Fig. 4). In the brain, by contrast, the α1D mRNA is only 2–5 times as abundant (data not shown). α1D mRNA therefore is the predominant, if not the only, L-type species in the basilar papilla.
DISCUSSION
Our results indicate that the predominant α1 subunit of L-type Ca2+ channels in hair cells of the chicken’s cochlea is the product of the ortholog of the mammalian α1D gene. First, the protein sequence predicted from our cloned cDNA is more than 90% identical to those of the functional human and rat α1D subunits (28, 30) and contains such hallmarks of L-type Ca2+ channels as the glutamate residues necessary for Ca2+ selectivity and the binding site for dihydropyridines. Because functional expression of the α1D subunit requires auxiliary subunits (28, 30), we have not yet attempted to demonstrate that the composite cDNA encodes an L-type Ca2+ channel. Second, α1D mRNA and the cognate protein were detected with specific probes in the basilar papilla by RNA blotting and protein immunoblotting and were of sizes consistent with the composite cDNA sequence (35). Furthermore, the mRNA was localized within hair cells by in situ hybridization. Because of this localization, we conclude that the results of the PCR and blotting experiments with material from the basilar papilla reflect the properties of the α1D subunit in hair cells. The α1D subunit, like the α1C subunit, has previously been detected in the mouse cochlea by the PCR (43) but not localized to specific cell types. Third, by using the PCR, the most sensitive method available, we could detect the other two L-type subunits, α1C and α1S, barely or not at all in the basilar papilla. Even though the α1D mRNA is the predominant L-type mRNA in hair cells, its estimated abundance of about 1 in 104 places it among the cell’s scarce mRNAs (44).
These results do not exclude the presence of other α1 subunits in hair cells. Even though we employed several different methods, it is possible that all of our assays were biased against the detection of α1C and α1S mRNAs, that there are other L-type α1 genes besides α1C, α1D, and α1S, or that the mRNA levels do not reflect the abundance of the proteins. For example, an mRNA encoding a non-L-type α1 subunit has been found in the sensory epithelium of the rainbow trout’s sacculus.‡ Moreover, low-conductance channels have been detected in single-channel recordings from hair cells of the chicken’s cochlea (45), and putative N-type channels have been described in hair cells of the frog’s sacculus (46). In hair cells of the chicken’s cochlea, however, at least 95% of the Ca2+ conductance is sensitive to dihydropyridines and can be described by a single activation process (9). This electrophysiological study and the molecular characterization reported here suggest that subunits other than the α1D are present in only a small fraction of the hair cell’s Ca2+ channels.
Our conclusion is consistent with a comparison of the physiological properties of the hair cell’s Ca2+ channel—low activation threshold, rapid activation, and lack of Ca2+-dependent inactivation—to those of the three L-type α1 subunits. The α1S subunit is expressed only in skeletal muscle (47), in which Ca2+-channel activation is about 100-fold slower than in hair cells (48). Both the α1C and the α1D subunit occur in a variety of organs, including the brain. Channels containing the former activate at a more positive potential, around −20 mV, and inactivate more rapidly during prolonged depolarization (49). In contrast, heterologously expressed channels containing α1D subunits activate at potentials positive to −40 mV and do not inactivate for at least 0.7 s (28). The α1D subunit is also expressed in hippocampal CA3 pyramidal neurons (50–52) that contain L-type Ca2+ conductances with unusually low activation thresholds (53).
The α1D subunit generally is termed “neuroendocrine” (54) because of its broad distribution in the brain (50–52) and its localization and prevalence in the insulin-secreting β cells of the pancreas (27, 55). In addition, it has been found in Müller cells of the retina (56), in sympathetic neurons (57), in the kidney (58), and in the ovary (59). In most neurons, α1D protein is distributed diffusely in the cell membrane of cell bodies and proximal dendrites (50), suggesting a role in general Ca2+ signaling. Our results do not demonstrate directly that neurotransmitter release in hair cells is controlled by Ca2+ channels containing the α1D subunit. However, the predominance of the α1D subunit in hair cells, as demonstrated by our results, and the consistent properties of the hair cell’s Ca2+ conductance (9) implicate the α1D subunit in tonic neurotransmitter release.
Tonic synapses often are found in sensory pathways and are distinguished from spiking synapses by continuous transmitter release that can track a protracted stimulus (60). Because of its physiological properties, the α1D subunit has been postulated to be present at the tonic synapse of cone photoreceptors (61). The α1D subunit’s lack of Ca2+-dependent inactivation (28, 30) may make it more suitable than the rapidly inactivating α1C subunit for a role in tonic transmitter release. There are, however, important differences between hair cells and photoreceptors. The resting potentials of the former are more negative, the frequencies of auditory stimuli are much higher, and phase information is more important in the auditory system. The demands on the electrophysiological properties of the Ca2+ channels, therefore, differ between these sensory cells, and we do not necessarily expect the channels’ molecular structures to be identical.
The presence of α1D protein in the basilar papilla (35) is consistent with the proposed role of the L-type Ca2+ channel in tonic transmitter release. Unambiguous localization of the α1D protein in the hair cell’s presynaptic membrane, however, will require immunoelectron microscopy with subunit-specific antibodies. In hair cells from the bullfrog’s sacculus, the Ca2+ channels are clustered at presynaptic active zones (6, 62) to localize the Ca2+ entry. In hair cells from the chicken’s basilar papilla, we also expect the distribution of the Ca2+ channels to vary along and across the basilar papilla. First, the magnitude of the Ca2+ current increases from the apex, where the hair cells are tuned to low frequencies, to the base, where the cells respond to high frequencies; second, both the Ca2+ current and the number of presynaptic dense bodies are larger in tall hair cells, which receive mostly afferent innervation, than in short hair cells, which receive mostly efferent innervation (63).§ A molecular mechanism for Ca2+-channel clustering is suggested by the last four amino acids of the α1D subunit, ITSL. Similar motifs, such as (E, S)(S, T)XV and SS(S, T)L, occur at the carboxyl termini of membrane-associated proteins that are clustered by binding to scaffolding proteins with PDZ domains (64).
Acknowledgments
We thank Dr. S. Heller for the cochlear libraries and Drs. P. Farnham and J. Imredy, as well as members of our research group, for comments on drafts of the manuscript. Begun at University of Texas Southwestern Medical Center, this research was supported by National Institutes of Health Grant DC00317. R.K. was an Associate and A.J.H. is an Investigator of Howard Hughes Medical Institute.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF027602–AF027610).
Sheikhali, S. A. Karadaghy, A. A. & Drescher, D. G., 20th Annual Midwinter Research Meeting, Assoc. Res. Otolaryngol., Des Moines, IA, February 2–6, 1997, abstr. 163.
Michaels, R. L., Martinez, C. A. & Fuchs, P. A., 19th Annual Midwinter Research Meeting, Assoc. Res. Otolaryngol., Des Moines, IA, February 4–8, 1996, abstr. 500.
References
- 1.Hudspeth A J. Nature (London) 1989;341:397–404. doi: 10.1038/341397a0. [DOI] [PubMed] [Google Scholar]
- 2.Fuchs P A. Curr Opin Neurobiol. 1996;6:514–519. doi: 10.1016/s0959-4388(96)80058-4. [DOI] [PubMed] [Google Scholar]
- 3.Fettiplace R. Trends Neurosci. 1987;10:421–425. [Google Scholar]
- 4.Rosenblatt K P, Sun Z-P, Heller S, Hudspeth A J. Neuron. 1997;19:1061–1075. doi: 10.1016/s0896-6273(00)80397-9. [DOI] [PubMed] [Google Scholar]
- 5.Reuter H. Curr Opin Neurobiol. 1996;6:331–337. doi: 10.1016/s0959-4388(96)80116-4. [DOI] [PubMed] [Google Scholar]
- 6.Roberts W M, Jacobs R A, Hudspeth A J. J Neurosci. 1990;10:3664–3684. doi: 10.1523/JNEUROSCI.10-11-03664.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Art J J, Fettiplace R, Wu Y-C. J Physiol. 1993;470:109–126. doi: 10.1113/jphysiol.1993.sp019850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Nenov A, Norris C H, Bobbin R P. Hearing Res. 1995;86:25–33. doi: 10.1016/0378-5955(95)00050-e. [DOI] [PubMed] [Google Scholar]
- 9.Zidanic M, Fuchs P A. Biophys J. 1995;68:1323–1336. doi: 10.1016/S0006-3495(95)80305-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perney T M, Hirning L D, Leeman S E, Miller R J. Proc Natl Acad Sci USA. 1986;83:6656–6659. doi: 10.1073/pnas.83.17.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tachibana M, Okada T, Arimura T, Kobayashi K, Piccolino M. J Neurosci. 1993;13:2898–2909. doi: 10.1523/JNEUROSCI.13-07-02898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofmann F, Biel M, Flockerzi V. Annu Rev Neurosci. 1994;17:399–418. doi: 10.1146/annurev.ne.17.030194.002151. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Frohman M A. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 15.McLean I W, Nakane P K. J Histochem Cytochem. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- 16.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka K, Smith C A. Am J Anat. 1978;153:251–271. doi: 10.1002/aja.1001530206. [DOI] [PubMed] [Google Scholar]
- 18.Tilney L G, Tilney M S, Saunders J S, DeRosier D J. Dev Biol. 1986;116:100–118. doi: 10.1016/0012-1606(86)90047-3. [DOI] [PubMed] [Google Scholar]
- 19.Mikami A, Imoto K, Tanabe T, Niidome T, Mori Y, Takeshima H, Narumiya S, Numa S. Nature (London) 1989;340:230–233. doi: 10.1038/340230a0. [DOI] [PubMed] [Google Scholar]
- 20.Biel M, Ruth P, Bosse E, Hullin R, Stühmer W, Flockerzi V, Hofmann F. FEBS Lett. 1990;269:409–412. doi: 10.1016/0014-5793(90)81205-3. [DOI] [PubMed] [Google Scholar]
- 21.Koch W J, Ellinor P T, Schwartz A. J Biol Chem. 1990;265:17786–17791. [PubMed] [Google Scholar]
- 22.Snutch T P, Tomlinson W J, Leonard J P, Gilbert M M. Neuron. 1991;7:45–57. doi: 10.1016/0896-6273(91)90073-9. [DOI] [PubMed] [Google Scholar]
- 23.Ma W-J, Holz R W, Uhler M D. J Biol Chem. 1992;267:22728–22732. [PubMed] [Google Scholar]
- 24.Soldatov N M. Proc Natl Acad Sci USA. 1992;89:4628–4632. doi: 10.1073/pnas.89.10.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz D, Mikala G, Yatani A, Engle D B, Iles D E, Segers B, Sinke R J, Weghuis D O, Klöckner U, Wakamori M, Wang J-J, Melvin D, Varadi G, Schwartz A. Proc Natl Acad Sci USA. 1993;90:6228–6232. doi: 10.1073/pnas.90.13.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hui A, Ellinor P T, Krizanova O, Wang J-J, Diebold R J, Schwartz A. Neuron. 1991;7:35–44. doi: 10.1016/0896-6273(91)90072-8. [DOI] [PubMed] [Google Scholar]
- 27.Seino S, Chen L, Seino M, Blondel O, Takeda J, Johnson J H, Bell G I. Proc Natl Acad Sci USA. 1992;89:584–588. doi: 10.1073/pnas.89.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams M E, Feldman D H, McCue A F, Brenner R, Velicelebi G, Ellis S B, Harpold M M. Neuron. 1992;8:71–84. doi: 10.1016/0896-6273(92)90109-q. [DOI] [PubMed] [Google Scholar]
- 29.Yaney G C, Wheeler M B, Wei X, Perez-Reyes E, Birnbaumer L, Boyd A E, III, Moss L G. Mol Endocrinol. 1992;6:2143–2152. doi: 10.1210/mend.6.12.1337146. [DOI] [PubMed] [Google Scholar]
- 30.Ihara Y, Yamada Y, Fujii Y, Gonoi T, Yano H, Yasuda K, Inagaki N, Seino Y, Seino S. Mol Endocrinol. 1995;9:121–130. doi: 10.1210/mend.9.1.7760845. [DOI] [PubMed] [Google Scholar]
- 31.Tanabe T, Takeshima H, Mikami A, Flockerzi V, Takahashi H, Kangawa K, Kojima M, Matsuo H, Hirose T, Numa S. Nature (London) 1987;328:313–318. doi: 10.1038/328313a0. [DOI] [PubMed] [Google Scholar]
- 32.Ellis S B, Williams M E, Ways N R, Brenner R, Sharp A H, Leung A T, Campbell K P, McKenna E, Koch W J, Hui A, Schwartz A, Harpold M M. Science. 1988;241:1661–1664. doi: 10.1126/science.2458626. [DOI] [PubMed] [Google Scholar]
- 33.Grabner M, Friedrich K, Knaus H-G, Striessnig J, Scheffauer F, Staudinger R, Koch W J, Schwartz A, Glossmann H. Proc Natl Acad Sci USA. 1991;88:727–731. doi: 10.1073/pnas.88.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proudfoot N. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 35.Kollmar R, Fak J, Montgomery L G, Hudspeth A J. Proc Natl Acad Sci USA. 1997;94:14889–14893. doi: 10.1073/pnas.94.26.14889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellinor P T, Yang J, Sather W A, Zhang J-F, Tsien R W. Neuron. 1995;15:1121–1132. doi: 10.1016/0896-6273(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 38.Grabner M, Wang Z, Hering S, Striessnig J, Glossmann H. Neuron. 1996;16:207–218. doi: 10.1016/s0896-6273(00)80037-9. [DOI] [PubMed] [Google Scholar]
- 39.Peterson B Z, Tanada T N, Catterall W A. J Biol Chem. 1996;271:5293–5296. doi: 10.1074/jbc.271.10.5293. [DOI] [PubMed] [Google Scholar]
- 40.Schuster A, Lacinová L, Klugbauer N, Ito H, Birnbaumer L, Hofmann F. EMBO J. 1996;15:2365–2370. [PMC free article] [PubMed] [Google Scholar]
- 41.Döring F, Degtiar V E, Grabner M, Striessnig J, Hering S, Glossman H. J Biol Chem. 1996;271:11745–11749. doi: 10.1074/jbc.271.20.11745. [DOI] [PubMed] [Google Scholar]
- 42.He M, Bodi I, Mikala G, Schwartz A. J Biol Chem. 1997;272:2629–2633. doi: 10.1074/jbc.272.5.2629. [DOI] [PubMed] [Google Scholar]
- 43.Green G E, Khan K M, Beisel K W, Drescher M J, Hatfield J S, Drescher D G. J Neurochem. 1996;67:37–45. doi: 10.1046/j.1471-4159.1996.67010037.x. [DOI] [PubMed] [Google Scholar]
- 44.Davidson E H, Britten R J. Science. 1979;204:1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- 45.Kimitsuki T, Nakagawa T, Hisashi K, Komune S, Komiyama S. Acta Otolaryngol (Stockholm) 1994;114:144–148. doi: 10.3109/00016489409126033. [DOI] [PubMed] [Google Scholar]
- 46.Su Z-L, Jiang S-C, Gu R, Yang W-P. Hear Res. 1995;87:62–68. doi: 10.1016/0378-5955(95)00079-j. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Reyes E, Schneider T. Drug Dev Res. 1994;33:295–318. [Google Scholar]
- 48.McDonald T F, Pelzer S, Trautwein W, Pelzer D J. Physiol Rev. 1994;74:365–507. doi: 10.1152/physrev.1994.74.2.365. [DOI] [PubMed] [Google Scholar]
- 49.Tomlinson W J, Stea A, Bourinet E, Charnet P, Nargeot J, Snutch T P. Neuropharmacology. 1993;32:1117–1126. doi: 10.1016/0028-3908(93)90006-o. [DOI] [PubMed] [Google Scholar]
- 50.Hell J W, Westenbroek R E, Warner C, Ahlijanian M K, Prystay W, Gilbert M M, Snutch T P, Catterall W A. J Cell Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka O, Sakagami H, Kondo H. Brain Res Mol Brain Res. 1995;30:1–16. doi: 10.1016/0169-328x(94)00265-g. [DOI] [PubMed] [Google Scholar]
- 52.Ludwig A, Flockerzi V, Hofmann F. J Neurosci. 1997;17:1339–1349. doi: 10.1523/JNEUROSCI.17-04-01339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avery R B, Johnston D. J Neurosci. 1996;16:5567–5582. doi: 10.1523/JNEUROSCI.16-18-05567.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birnbaumer L, Campbell K P, Catterall W A, Harpold M M, Hofmann F, Horne W A, Mori Y, Schwartz A, Snutch T P, Tanabe T, Tsien R W. Neuron. 1994;13:505–506. doi: 10.1016/0896-6273(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 55.Iwashima Y, Pugh W, Depaoli A M, Takeda J, Seino S, Bell G I, Polonsky K S. Diabetes. 1993;42:948–955. doi: 10.2337/diab.42.7.948. [DOI] [PubMed] [Google Scholar]
- 56.Puro D G, Hwang J-J, Kwon O-J, Chin H. Brain Res Mol Brain Res. 1996;37:41–48. doi: 10.1016/0169-328x(96)80478-5. [DOI] [PubMed] [Google Scholar]
- 57.Lin Z, Harris C, Lipscombe D. J Mol Neurosci. 1996;7:257–267. doi: 10.1007/BF02737063. [DOI] [PubMed] [Google Scholar]
- 58.Yu A S, Hebert S C, Brenner B M, Lytton J. Proc Natl Acad Sci USA. 1992;89:10494–10498. doi: 10.1073/pnas.89.21.10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Reyes E, Wei X, Castellano A, Birnbaumer L. J Biol Chem. 1990;265:20430–20436. [PubMed] [Google Scholar]
- 60.Juusola M, French A S, Uusitalo R O, Weckström M. Trends Neurosci. 1996;19:292–297. doi: 10.1016/S0166-2236(96)10028-X. [DOI] [PubMed] [Google Scholar]
- 61.Wilkinson M F, Barnes S. J Gen Physiol. 1996;107:621–630. doi: 10.1085/jgp.107.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Issa N P, Hudspeth A J. Proc Natl Acad Sci USA. 1994;91:7578–7582. doi: 10.1073/pnas.91.16.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer F P. Hear Res. 1992;61:167–178. doi: 10.1016/0378-5955(92)90048-r. [DOI] [PubMed] [Google Scholar]
- 64.Kornau H-C, Seeburg P H, Kennedy M B. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 65.Pragnell M, De Waard M, Mori Y, Tanabe T, Snutch T P, Campbell K P. Nature (London) 1994;368:67–70. doi: 10.1038/368067a0. [DOI] [PubMed] [Google Scholar]
- 66.de Leon M, Wang Y, Jones L, Perez-Reyes E, Wei X, Soong T W, Snutch T P, Yue D T. Science. 1995;270:1502–1506. doi: 10.1126/science.270.5241.1502. [DOI] [PubMed] [Google Scholar]