Abstract
In an unprecedented finding, Davis et al. [Davis, R. E., Miller, S., Herrnstadt, C., Ghosh, S. S., Fahy, E., Shinobu, L. A., Galasko, D., Thal, L. J., Beal, M. F., Howell, N. & Parker, W. D., Jr. (1997) Proc. Natl. Acad. Sci. USA 94, 4526–4531] used an unusual DNA isolation method to show that healthy adults harbor a specific population of mutated mitochondrial cytochrome c oxidase (COX) genes that coexist with normal mtDNAs. They reported that this heteroplasmic population was present at a level of 10–15% in the blood of normal individuals and at a significantly higher level (20–30%) in patients with sporadic Alzheimer’s disease. We provide compelling evidence that the DNA isolation method employed resulted in the coamplification of authentic mtDNA-encoded COX genes together with highly similar COX-like sequences embedded in nuclear DNA (“mtDNA pseudogenes”). We conclude that the observed heteroplasmy is an artifact.
Keywords: cytochrome c oxidase, respiratory chain, oxidative phosphorylation, mitochondria
Three nuclear genes have been identified as the cause of Alzheimer’s disease (AD) in families with autosomal dominant transmission of early-onset disease: presenilin 1, presenilin 2, and amyloid precursor protein (1–4). The E4 variant of apolipoprotein E has been associated with increased risk of familial and sporadic late-onset AD (5).
Mitochondria have also been implicated in the pathogenesis of AD (6, 7). Mitochondria are unique, because they are under dual genetic control and contain the oxidative-phosphorylation enzyme pathway, the major energy producing system of the cell. The vast majority of mitochondrial proteins are derived from genes encoded by nuclear DNA (nDNA), but 13 mitochondrial polypeptides, all of which are components of the oxidative-phosphorylation pathway, are encoded in mtDNA, a 16,569-bp circular molecule (8) that is maternally inherited (9). Normal individuals typically contain a single mtDNA genotype, a condition known as homoplasmy. However, patients with mitochondrial diseases due to mutations in mtDNA (both point mutations and large-scale rearrangements) often harbor two populations of mtDNA, wild-type and mutated, a condition known as heteroplasmy (10).
Cytochrome c oxidase (COX) is the terminal enzyme of the respiratory chain. It catalyzes the oxidation of cytochrome c, reduces oxygen, and translocates protons from the mitochondrial matrix. COX is composed of 13 subunits, of which 3 (COX I, II, and III) are encoded by mtDNA and the remainder are encoded by nDNA. Deficiencies in COX activity have been identified in brain and in platelets of AD patients (11–14), but the pathogenic significance of these findings is unclear.
Davis et al. (15) identified six heteroplasmic mutations—three in COX I (G6366A, C6483T, and A7146G; notation of ref. 8) and three in COX II (C7650T, C7868T, and A8021G), but none in COX III—in all AD patients and in all controls studied in a screening of DNA isolated from “platelet-enriched pellets” subjected to a unique “boiling” method (15). The COX genes in the “. . . AD cases exhibited statistically significant increases in mutational burden at each of the six nucleotide sites relative to age-matched and other controls” (15). An average of 20–30% mutation was observed in AD patients, compared with 10–15% in controls.
If true, these findings would have profound implications regarding the nature of mtDNA maintenance and transmission in human populations and would have great significance for the understanding, diagnosis, and treatment of sporadic AD. We therefore attempted to confirm and to clarify further the observed mtDNA heteroplasmy in general, and the six COX mutations in particular.
MATERIALS AND METHODS
Subjects.
Venous blood was obtained in vacuum tubes with EDTA from four patients with AD and four controls similar in age, gender, and ethnic group, residing in the Washington Heights-Inwood community of northern Manhattan (New York City), who had previously participated in a study of genetic risk factors for AD (16). Patients and their next of kin, as well as the controls, provided informed consent. The blood was analyzed blindly in our laboratory.
Cell Culture.
We cultured human osteosarcoma-derived cell lines containing mtDNA (ρ+ cells) and lacking mtDNA (ρo cells) (provided by E. Shoubridge, Montreal Neurological Institute, Montreal, Canada) as described (17). The absence of mtDNA in these cells was confirmed by both PCR and Southern blot analyses (data not shown).
Extraction of DNA.
To reproduce the results of Davis and colleagues (15), we replicated their DNA extraction protocol essentially as described (15, 18). Two buffy coat pellets were extracted from each subject’s blood sample. One buffy coat sample was processed according to the methods of Davis et al. (15) and Fahy et al. (18). The pellet was resuspended in 0.2 ml of sterile water and placed in a boiling water bath for 10 min before placing the sample on ice. The sample was centrifuged at 14,000 rpm in a microcentrifuge for 2 min. The supernatant containing DNA was transferred to a sterile tube.
The second buffy coat sample was used to isolate DNA by a “standard” method, specifically, proteinase K digestion in SDS solution followed by phenol/chloroform purification and ethanol precipitation (19). We also used the standard method to isolate DNA from the ρ+ and ρo cells and from the post-boiling precipitate, after removing a small aliquot for the microscopic studies described below. DNA concentrations were measured by using optical density readings at 260 nm.
PCR Amplifications.
We used oligonucleotide PCR primers with sequences identical to those used by Davis and colleagues (15). Primers for the nDNA-encoded phosphoglycerate kinase (PGK) gene were 17F (GGTGTGCAGCCCTGAGTTCT) in intron 8 and 18B (AGCTAATGCCAAGTGGAGATGC) in exon 11 (20).
For PCR amplification of patient and control DNA, we used 0.5–1.0 μg of total DNA as template, based on the A260 measurements of the boiled supernatant, as recommended (15). For PCR amplification of DNA isolated from the post-boiling pellet and from ρ+ and ρo cell DNA, we used approximately 100 ng of DNA. Each 100-μl PCR contained all four dNTPs (each at 200 μM), 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, and 2.5 units of Taq polymerase (Boehringer Mannheim). PCR conditions were denaturation at 94°C for 1 min, annealing at 55°C for 1 min (for COX I) or 60°C for 1 min (for COX II and III), and extension for 72°C for 2 min, for 25 cycles in a Perkin–Elmer thermocycler 480 (Perkin–Elmer) and were the same as those used by Davis and colleagues (15, 18).
DNA Sequence Analysis.
The PCR products were gel-purified from 1% low melting point agarose (Boehringer Mannheim). The bands were excised and eluted from the gel by using a QIAquick gel extraction kit (Qiagen, Chatsworth, CA). Sequence reactions using PCR-amplified COX I and COX II from ρ+ and ρo cell DNA were performed with the fmol DNA cycle sequencing system (Promega).
Restriction Fragment Length Polymorphism (RFLP) Analysis.
DNA was amplified with the COX II primers with [α-32P]dATP added to the last cycle (21). The fragments were digested with HpaII and electrophoresed through a 12% nondenaturing polyacrylamide gel. The relative amounts of the digestion products were quantitated by scanning the vacuum-dried gel in a phosphorimager (Bio-Rad, model GM363) using md-image quant software version 3.22 (Molecular Dynamics).
Southern Blot Hybridization Analysis.
We digested 1 μg of patient total-blood DNA, 40% of the patient buffy-coat DNA released after 10 min of boiling, and 1 μg of DNA from the pelleted cell debris obtained after boiling, with PvuII, which linearizes normal mtDNA by cutting the circular molecule at one site. DNA was electrophoresed through 0.8% agarose and blotted onto a nylon membrane (Zetaprobe, Bio-Rad) (19). Nearly full-length mtDNA (15.6 kb), generated by a long PCR (22), was used as the template for random primer labeling. We also hybridized the membrane with a nuclear 18S ribosomal DNA probe to estimate the relative proportions of mtDNA and nDNA (23).
Morphological Studies.
The pellet obtained after boiling the buffy coat was resuspended in PBS and treated with MitoTracker CMX ROS (Molecular Probes) at a final concentration of 0.5 μM. This compound is taken up specifically by mitochondria and forms a fluorescent conjugate that can be visualized with a standard rhodamine filter.
RESULTS
PCR Amplifications.
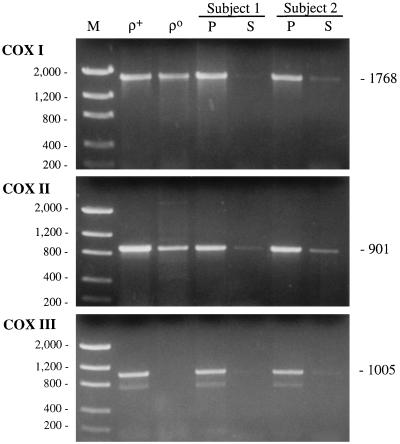
We used the methods of Davis et al. (15) to examine mtDNA heteroplasmy in the COX I, II, and III gene regions in blood from AD patients and controls. We also examined DNA from ρ+ and ρo osteosarcoma-derived cells, which served as a positive and a negative control, respectively. We were able to PCR-amplify all three COX gene regions from patient and control DNA present in supernatants from boiled buffy-coat cells, albeit in low yields, and from the ρ+ DNA (Fig. 1). Unexpectedly, we also were able to amplify PCR fragments from the ρo DNA with the COX I and II primers, although not with the COX III primers (Fig. 1). We amplified all three COX fragments from DNA extracted from total blood (data not shown) and, surprisingly, from the pelleted cell debris as well (Fig. 1).
Figure 1.
PCR amplification of ρ+ and ρo DNA and of DNA from the supernatant (S) and the pelleted cell debris (P) after 10 min of boiling of the buffy coat of an AD patient (subject 1) and a control (subject 2), using primers corresponding to the three COX genes. The predicted sizes of the PCR products, in bp, are at right. M, DNA Mass Ladder markers (GIBCO/BRL) with sizes, in bp, at left.
DNA Sequencing.
DNA sequencing of the PCR fragments amplified with the COX I and COX II primers from the ρ+ DNA revealed the normal Cambridge mtDNA sequence (8), except for a single neutral polymorphism in COX I (T6221C). By contrast, the corresponding amplification products from ρo DNA diverged from the Cambridge sequence at 19 nucleotide positions in “COX I” and at 13 nucleotide positions in “COX II” (Table 1). All six mtDNA mutations identified by Davis and colleagues (15) in COX I and COX II were present in the ρo DNA PCR products (Table 1).
Table 1.
Polymorphisms in ρ0-derived PCR fragments
| COX I
|
COX II
|
|||
|---|---|---|---|---|
| A5984G | G6383A* | C6935T* | C7650T*† | G8065A*† |
| G5985C | C6410T* | C6938T† | T7705C*† | C8080T*† |
| G6023A*† | C6483T*† | A7146G*† | C7810T* | C8140T |
| C6242T* | T6512C | C7256T*† | C7868T*† | G8153A |
| A6266C*† | C6542T*† | G7316A† | C7891T* | T8167C*† |
| A6299G | C6569A*† | G7912A*† | C8203T*† | |
| G6366A*† | T6641C | A8021G*† | ||
Only unambiguous nucleotide differences found between the “Cambridge” sequence and the sequence of uncloned PCR products amplified from total DNA isolated from ρ0 cells are shown. Polymorphisms in boldface type were reported by Davis et al. (15).
Polymorphism is present at the analogous nucleotide position in chimpanzee mtDNA (Genbank accession no. D38113).
Polymorphism is present at the analogous nucleotide position in gorilla mtDNA (Genbank accession no. X93347).
RFLP Analysis.
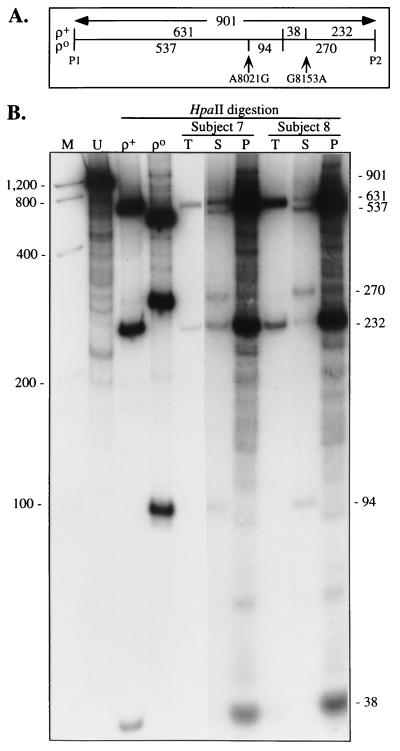
Two of these mutations, both in “COX II,” generate HpaII polymorphisms: the A → G transition at nucleotide 8,021 creates a new HpaII restriction site, and the G → A transition at nucleotide 8,153 destroys a HpaII site (Fig. 2A). Digestion of the 901-bp COX II PCR product amplified from ρ+ cells with HpaII produced DNA fragments that corresponded to the expected pattern predicted by the Cambridge mtDNA sequence (8), i.e., 631, 232, and 38 bp (Fig. 2B). Digestion of the 901-bp COX II PCR product amplified from ρo cells with HpaII produced DNA fragments that corresponded to a different pattern, but one that was consistent with the presence of the A8021G (gain-of-site) and G8153A (loss-of-site) polymorphisms identified in the sequence analysis (Table 1), i.e., 537, 270, and 94 bp (Fig. 2B).
Figure 2.
PCR/RFLP analysis of the COX II region. (A) Schematic of the HpaII digestion pattern of the COX II region amplified from ρ+- and ρo-derived DNA by using the COX II primers (P1 and P2). The HpaII sites (interior vertical lines), the location of the two HpaII polymorphisms, and the predicted sizes of the HpaII-digested fragments, in bp, are shown. (B) Autoradiogram of HpaII-digested 0.9-kb PCR products of ρ+ and ρo DNA and of total cellular DNA (T), DNA from the supernatant (S), and DNA from the pelleted cell debris (P) after 10 min of boiling of the buffy coat from the blood of an AD patient (subject 7) and a control (subject 8), by using primers corresponding to the COX II gene. Sizes of expected fragments, in bp, are at right. U, uncut PCR product from ρ+ DNA. Other notations are as in Fig. 1. The photo is a composite of two exposures of the same gel.
Digestion with HpaII of the 0.9-kb COX II PCR products amplified from the boiled supernatants of the buffy coats from the blood of four AD patients and four controls revealed two DNA populations, one similar to ρ+-derived DNA (i.e., normal mtDNA) and the other similar to ρo-derived DNA (i.e., consistent with the presence of the A8021G and G8153A mutations). Typical results for two of the eight subjects are shown in Fig. 2B. There were no obvious differences in the percentage of the mutated mtDNA measured in three patients (44 ± 4%) and three controls (44 ± 19%). The HpaII digestion patterns of the pelleted cell debris from all eight samples were consistent with normal mtDNA (Fig. 2).
Time Course of Boiling.
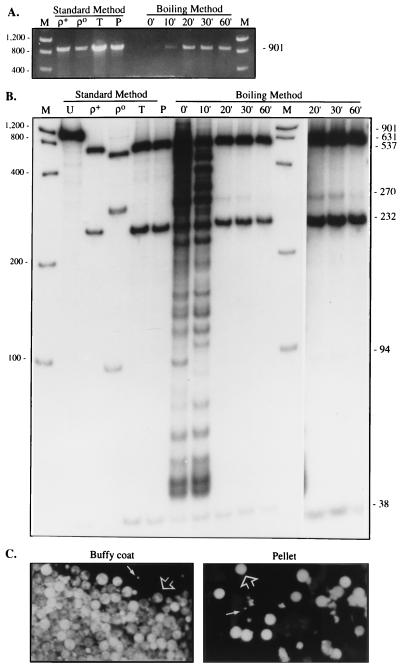
As noted above, the yield of amplified PCR product from the three COX genes by using the DNA obtained after 10 min of boiling was surprisingly low (Figs. 1 and 3). However, the yield was much greater when the cells were boiled for at least 20 min (Fig. 3A). Moreover, the DNA released after 20 min revealed a HpaII digestion pattern more similar to the normal mtDNA pattern than that revealed after 10 min, and the pattern after 60 min of boiling was essentially 100% wild type (Fig. 3B).
Figure 3.
Time course of boiling. (A) The buffy coat pellet from subject 1 was resuspended in water, boiled for the indicated times, amplified with the COX II primers, and electrophoresed through an agarose gel; equal amounts of PCR product were loaded in each lane. DNA from ρ+ and ρo cells, and the total cellular DNA (T) and pelleted buffy-coat DNA after 10 min of boiling (P), all isolated by the standard method, are also shown. (B) The samples shown in A were digested with HpaII and electrophoresed through a nondenaturing polyacrylamide gel. Note the decline of the 270-bp and 94-bp fragments at the later time points (more clearly visible in the original autoradiogram, and in the darker exposure of the 20-min, 30-min, and 60-min results shown in the last three lanes). Other notations are as in Fig. 2. (C) Micrographs of resuspended cells from the buffy coat prior to boiling (the cells were clumped and difficult to disaggregate) and of the resuspended pellet after 10 min of boiling, after staining with MitoTracker. Note the mixed population of lymphocytes (large arrows) and platelets (small arrows). (×400.)
Morphology.
Staining with MitoTracker of the cell debris collected after 10 min of boiling revealed many positively stained intact cells (mainly platelets and white cells), indicating that mitochondria were present in those cells (Fig. 3C).
Southern Blot Analysis.
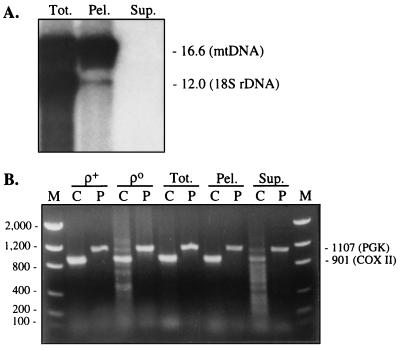
We performed Southern blot analysis to see whether the presumed reduced amounts of mtDNA in the boiled supernatant could be quantitated. Total cellular DNA, DNA released into the supernatant after 10 min of boiling, and DNA isolated from the pelleted cell debris from subject 7 were digested with PvuII and electrophoresed through agarose. The digestion products were transferred to a nylon membrane and were hybridized first with a nearly full-length mtDNA probe and then with an nDNA-encoded 18S rDNA probe. There was a much higher amount of mtDNA, as estimated by the ratio of mtDNA to nDNA (23), in the pelleted cell debris as compared with the original amount present in total cellular DNA (Fig. 4A). On the other hand, we were unable to detect any hybridizing species from the supernatant, even after exposing the blot for long times, and even after performing the experiment with a 6-fold greater amount of supernatant (from subject 1; data not shown). The failure to see a signal in the supernatant lane was not due to the absence of DNA in the sample, because PCR of the supernatant produced signals for the mtDNA-encoded COX II gene and for the nDNA-encoded phosphoglycerate kinase gene (Fig. 4B). Thus, these results imply that the supernatant contained amounts of both mtDNA and nDNA below the level of detection of the Southern blot analysis (i.e., estimated at <1% of either mtDNA or nDNA present in the original cellular DNA), despite digestion of an amount of supernatant-derived DNA (as measured by A260) that should have been equivalent to that in the other samples and that should have been sufficient to produce detectable bands. The use of A260 measurements to quantitate DNA in the supernatants may have been misleading, as the ratio of A260 to A280 in these samples was well above 2.0 (Table 2), implying that the supernatant contained impurities, such as free nucleotides and RNA. Thus, using the A260 value alone may have resulted in overestimating the actual amount of DNA present.
Figure 4.
(A) Southern blot hybridization analysis of DNA from subject 7. Total cellular DNA (Tot.), and the DNA from the supernatant (Sup.) and the pelleted cell debris (Pel.) after 10 min of boiling of the buffy coat were digested with PvuII, electrophoresed through agarose, transferred to a nylon membrane, and hybridized with probes to reveal mtDNA and 18S rDNA (predicted sizes, in kb, at right). The blot was overexposed to reveal the nDNA signal in the pellet. (B) PCR amplification of the samples shown in A, as well as of DNA from ρ+ and ρo cells, using primers to amplify the mtDNA-encoded COX II gene (C) and the nDNA-encoded PGK gene (P). Note the strong signal for PGK compared with that for COX II in the supernatant samples. Predicted sizes of the PCR products, in bp, are at right.
Table 2.
A260/A280 measurements
| Material | Subject 1 | Subject 2 |
|---|---|---|
| Total cellular DNA | 1.7 | 1.8 |
| Supernatant (0-min boil) | 1.3 | 1.2 |
| Supernatant (10-min boil) | 2.4 | 2.8 |
| Supernatant (20-min boil) | 2.4 | 2.4 |
| Supernatant (30-min boil) | 2.3 | 2.2 |
| Supernatant (60-min boil) | 2.2 | 2.0 |
| Pellet after 10-min boil | 1.6 | 1.5 |
A260/A280 for ρ+ DNA, 1.7; for ρ0 DNA, 1.8.
Mixing Experiment.
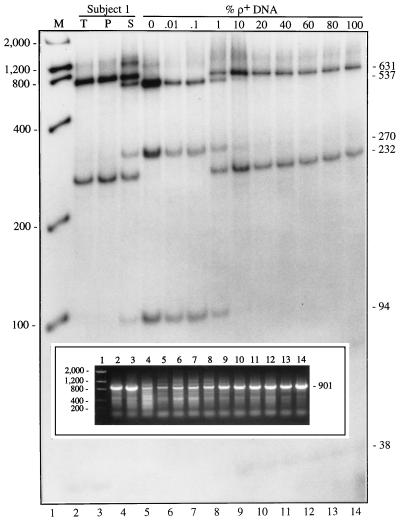
Since the COX II primers recommended by Davis et al. (15) amplify two types of PCR products from the buffy-coat boiled supernatant, we postulated that the PCR primers distribute themselves quantitatively on their target sites in proportion to the amount of mtDNA and nDNA present. We therefore mixed ρ+ DNA with ρo DNA in various proportions (ranging from 0 to 100% ρ+ DNA) and subjected each sample to PCR/RFLP analysis, to determine the amount of ρo DNA required to produce the observed heteroplasmy. At proportions of ρ+ DNA at or below 0.1%, only the ρo-type pattern was observed (Fig. 5), and conversely, at proportions at or above 10%, only the normal (ρ+-type) pattern was observed. However, at proportions of ρ+ DNA between 0.1% and 10%, we were able to detect heteroplasmy. The heteroplasmy was particularly evident when the proportion was about 1% (Fig. 5) and matched closely the heteroplasmic pattern obtained from the boiled supernatant from subject 1 (Fig. 5).
Figure 5.
Mixing experiment. DNA from ρ+ and ρo cells were mixed in the indicated proportions [reported as the percentage of ρ+ DNA present in the mixture, i.e., ρ+/(ρ+ + ρo) × 100], amplified with the COX II primers (see agarose gel shown in Inset), and the 901-bp fragment was digested with HpaII, and electrophoresed through a nondenaturing polyacrylamide gel. Lane numbers at bottom correspond to those in the Inset. Other notations are as in Fig. 2.
DISCUSSION
Davis and colleagues (15) identified six mtDNA point mutations in COX genes that “segregate with Alzheimer’s disease” and that “… define a unique mtDNA molecule or set of molecules that coexists with the wild-type mtDNA molecules in both AD and control cases in blood cells”. They claimed that “The ratio of this highly mutated molecule relative to the wild-type mitochondrial genome is elevated significantly in clinically defined cases of AD but not in age-matched, cognitively normal controls, patients with other neurologic diseases, or patients with NIDDM” (15). To identify these putative mtDNA mutations, Davis and colleagues (15) extracted “total cellular DNA” from “platelet-enriched pellets” that were resuspended in water and placed in a boiling water bath for 10 minutes; they deemed this to be “a critical step in the isolation of DNA for our analyses.” They hypothesized (15) that “standard SDS/proteinase K, phenol/chloroform treatments resulted in the quantitative loss of mutant mitochondrial genes.”
This finding of a significant degree of mtDNA heteroplasmy (10–30%) in the normal human population is unprecedented. In fact, based on the the results reported herein, we think that such heteroplasmy does not exist, in spite of the fact that we were able to reproduce the qualitative findings of Davis et al. (15). Rather, we believe that their data are consistent with the possibility that the “heteroplasmic” population of mtDNAs was actually derived from an artifactual PCR amplification of “mtDNA-like” sequences embedded in nDNA (“mtDNA pseudogenes”). We think that the boiling method preferentially releases nDNA relative to mtDNA, thereby enhancing the ability of PCR to amplify such pseudogenes, resulting in an erroneous interpretation of mtDNA heteroplasmy.
PCR amplification of DNA isolated from cells completely devoid of endogenous mtDNA (ρo cells) revealed the existence of nDNA sequences that are highly similar to the mtDNA-encoded COX I and II subunits and that are almost certainly nuclear pseudogenes. We deem it significant that the pseudogenes that we amplified from ρo cell DNA by using the primers recommended by Davis et al. (15) contained all six nucleotide changes that were identified by them as AD-associated mtDNA mutations. Equally significant, our attempts to amplify ρo cell DNA with the recommended COX III primers consistently failed to amplify COX III, the one mtDNA-encoded COX subunit that Davis and colleagues (15) found not to be associated with mtDNA mutations.
RFLP analysis with HpaII demonstrated the existence of two populations of COX II genes amplified from the patient and control DNAs extracted by 10 min of boiling. One population had a digestion pattern consistent with the normal Cambridge mtDNA sequence, but the other revealed a pattern indistinguishable from that obtained from digestion of the ρo-derived COX II pseudogenes. Thus, our results are compatible with a pseudoheteroplasmic state due to the simultaneous coamplification of both nDNA and mtDNA in all samples amplified from DNA extracted by the boiling method.
The coamplification of nDNA and mtDNA is likely to be the result of a poor yield of authentic mtDNA in the 10-min boiling protocol. Although we used similar amounts of template DNA (quantitated by A260), the DNA isolated by the boiling method produced far less PCR product than did the DNA extracted by the standard method. These results suggest that impurities in the DNA isolated by the boiling method interfered with the DNA quantitation, with the PCR amplification, or both.
In addition to low absolute yields of mtDNA, our results suggest that boiling the cells for 10 min released a disproportionately low amount of mtDNA relative to nDNA, because to produce an artifactual heteroplasmic state of 10–30% “mutation,” the number of nDNA pseudogene copies released by the boiling method must have been of the same order of magnitude as the number of released mtDNA molecules. Both the Southern blot analysis (Fig. 4A) and the mixing experiment (Fig. 5) support these contentions and imply that only about 1% of the total mtDNA was released into the supernatant after 10 min of boiling. Moreover, when we boiled the buffy coats for greater lengths of time (Fig. 3), we obtained greater yields of PCR product that, in turn, contained more of the authentic COX II gene (and less of the pseudo-COX II genes), again suggesting that 10 minutes of boiling were insufficient to lyse the mitochondria or to release significant quantities of mtDNA. In agreement with this concept, staining with MitoTracker of the cell debris pellet, normally discarded after boiling, revealed an abundance of positively staining cells, implying that significant numbers of mitochondria survived the 10-min boiling protocol, and PCR/RFLP analysis of the DNA isolated from this debris demonstrated that it contained almost exclusively wild-type mtDNA. The presence of relatively high levels of mtDNA in the pellet could thus account for the relative paucity of mtDNA molecules present in the supernatant.
The presence of mtDNA-like sequences in the human nuclear genome has been well documented (24, 25). From screening of human cDNA libraries, Fukuda and colleagues (26) estimated that hundreds of mtDNA-like fragments are present in the human nuclear genome. nDNA fragments similar to portions of the COX I and COX III genes have been sequenced (25, 27), including a HeLa genomic library clone that was highly similar to the COX I gene [92.3% between nucleotides 6,553 and 7,302 (27)] and that contained not only the A7146G change identified by Davis and colleagues as an AD-associated mtDNA mutation (15), but also three of the other five polymorphisms (C6569A, C6935T, and C7256T) identified by us in this region (Table 1).
The vast majority of mtDNA pseudogenes found in nDNA today probably entered the nucleus prior to the mammalian radiation, with only a minority arising after the relatively recent divergence of hominids from other primates. However, the most easily recognizable pseudogenes (and, not coincidentally, the sequences that can be most easily amplified by PCR) are those that entered the nuclear genome most recently in evolution. Not unexpectedly, when we sequenced PCR-amplified human COX-like fragments from human ρo DNA, we, like Davis et al. (15), noticed a striking similarity to nonhuman primate mtDNAs. In fact, 25 of the 32 COX polymorphisms listed in Table 1, including all six of the “AD-associated mutations,” are present in authentic mtDNA from chimpanzees, gorillas, or both. It was therefore surprising that Davis et al. (15) asserted that these six mutations are associated with COX deficiency, especially in light of the fact that human cells containing exclusively chimp or gorilla mtDNAs (28) had normal COX biochemical activity (A. Barrientos and C. T. Moraes, personal communication).
There is other, admittedly indirect, support for our contention that the COX I- and COX II-like PCR products identified by Davis et al. (15) are pseudogenes: there is no evidence that these sequences are transcribed into mRNA. Specifically, a blast search (29) of the Expressed Sequence Tag database (dbest) for cDNA sequences corresponding to the COX I and COX II mRNAs failed to identify a single cDNA (of 197 COX I- and COX II-positive “hits” examined in cDNAs synthesized from 15 independent libraries) containing any of the six putative AD-associated mutations.
In summary, we have identified nucleus-embedded human pseudogenes of the mtDNA-encoded COX I and II genes that contain all six of the purported AD-associated mutations reported by Davis and colleagues (15). We have presented evidence that the boiling method used by Davis and colleagues (15) releases nDNA preferentially and believe that this artifact led them to misidentify nucleus-embedded pseudogenes as authentic heteroplasmic mtDNA mutations.
We cannot exclude the possibility that AD patients may have mitochondria that are more resistant to lysis by boiling or, conversely, that AD nuclei are more susceptible to lysis by boiling. This might account for the higher proportion of the “mutations” in AD compared with control samples found by Davis and colleagues (15); nevertheless, because the “mutations” originate from nuclear pseudogenes, they are unlikely to be of functional significance.
Acknowledgments
We thank C. Bruno for providing the phosphoglycerate kinase gene primers; C. P. Krishnamurthy, E. Limon, G. Manfredi, J. Sadlock, and Y.-Y. Tang for technical assistance; and P. Magalhães and S. Lieberman for helpful discussions. This work was supported by grants from the National Institutes of Health (NS11766, NS01617, NS28828, AG07232, AG10963, AG08702, and RR00645), the Muscular Dystrophy Association, the Columbia-Presbyterian Medical Center Irving Scholars Program, and the Charles S. Robertson Memorial Gift for Alzheimer’s Disease Research from the Banbury Fund.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: AD, Alzheimer’s disease; nDNA, nuclear DNA; RFLP, restriction fragment length polymorphism.
References
- 1.Goate A, Chartier-Harlin M-C, Mullan M, Brown J, Crawford F, et al. Nature (London) 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 2.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, et al. Nature (London) 1995;375:754–760. [Google Scholar]
- 3.Levy-Lahad E, Bird T D. Ann Neurol. 1996;40:829–840. doi: 10.1002/ana.410400604. [DOI] [PubMed] [Google Scholar]
- 4.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingell W H, Yu C-E, Jondro P D, Schmidt S D, Wand K, Crowley A C, Fu Y H, Fuenette SY, Galas D, Nemens E, Wijsmann E M, Bird TD, Schellenberg GD, Tanzi R E. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 5.Stritmatter W J, Saunders A M, Schmechel D, Pericak-Vance M, Enghild J, Salvensen G S, Roses A D. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sims N R, Finegan J M, Blass J P, Bowen D M, Neary D. Brain Res. 1987;436:30–38. doi: 10.1016/0006-8993(87)91553-8. [DOI] [PubMed] [Google Scholar]
- 7.Hoyer S. Prog Neuro-Psychopharmacol & Biol Psychiat. 1993;17:199–228. doi: 10.1016/0278-5846(93)90043-r. [DOI] [PubMed] [Google Scholar]
- 8.Anderson S, Bankier A T, Barrell B G, de Bruijn M H L, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, Schreier P H, Smith A J H, Staden R, Young I G. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 9.Giles R E, Blanc H, Cann R M, Wallace D C. Proc Natl Acad Sci USA. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schon E A, Bonilla E, DiMauro S. J Bioenerg Biomembr. 1997;29:131–149. doi: 10.1023/a:1022685929755. [DOI] [PubMed] [Google Scholar]
- 11.Kish S J, Bergeron C, Rajput A, Dozic S, Mastrogiacomo F, Chang L-J, Wilson J M, DiStefano L M, Nobrega J N. J Neurochem. 1992;59:776–779. doi: 10.1111/j.1471-4159.1992.tb09439.x. [DOI] [PubMed] [Google Scholar]
- 12.Mutisya E M, Bowling A C, Beal M F. J Neurochem. 1994;63:2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 13.Parker W D, Parks J, Filley C M, Kleinschmidt-DeMasters B K. Neurology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 14.Parker W D, Parks J K. Neurology. 1995;45:482–486. doi: 10.1212/wnl.45.3.482. [DOI] [PubMed] [Google Scholar]
- 15.Davis R E, Miller S, Herrnstadt C, Ghosh S S, Fahy E, Shinobu L A, Galasko D, Thal L J, Beal M F, Howell N, Parker W D., Jr Proc Natl Acad Sci USA. 1997;94:4526–4531. doi: 10.1073/pnas.94.9.4526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Tang M X, Maestre G, Tsai W Y, Liu X H, Feng L, Chung W Y, Chun M, Schofield P, Stern Y, Tycko B, Mayeux R. Am J Hum Genet. 1996;58:574–584. [PMC free article] [PubMed] [Google Scholar]
- 17.King M P, Attardi G. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 18.Fahy E, Nazarbaghi R, Zomorrodi M, Herrnstadt C, Parker W D, Davis R E, Ghosh S S. Nucleic Acids Res. 1997;25:3102–3109. doi: 10.1093/nar/25.15.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeviani M, Moraes C T, Nakase H, Bonilla E, Schon E A, DiMauro S. Neurology. 1988;38:1339–1346. doi: 10.1212/wnl.38.9.1339. [DOI] [PubMed] [Google Scholar]
- 20.Michelson A M, Blake C C F, Evans S T, Orkin S H. Proc Natl Acad Sci USA. 1985;82:6965–6969. doi: 10.1073/pnas.82.20.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moraes C T, Ricci E, Bonilla E, DiMauro S, Schon E A. Am J Hum Genet. 1992;50:934–949. [PMC free article] [PubMed] [Google Scholar]
- 22.Fromenty B, Manfredi G, Sadlock J, Zhang L, King M P, Schon E A. Biochim Biophys Acta. 1996;1308:222–230. doi: 10.1016/0167-4781(96)00110-8. [DOI] [PubMed] [Google Scholar]
- 23.Moraes C T, Shanske S, Tritschler H J, Aprille J R, Andreetta F, Bonilla E, Schon E A, DiMauro S. Am J Hum Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuzuki T, Nomiyama H, Setoyama C, Maeda S, Shimada K. Gene. 1983;25:223–229. doi: 10.1016/0378-1119(83)90226-3. [DOI] [PubMed] [Google Scholar]
- 25.Shay J W, Werbin H. Mutat Res. 1992;275:227–235. doi: 10.1016/0921-8734(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda M, Wakasugi S, Tsuzuki T, Nomiyama H, Shimada K. J Mol Biol. 1985;186:257–266. doi: 10.1016/0022-2836(85)90102-0. [DOI] [PubMed] [Google Scholar]
- 27.Kamimura N, Ishii S, Liandon M, Shay J W. J Mol Biol. 1989;210:703–707. doi: 10.1016/0022-2836(89)90103-4. [DOI] [PubMed] [Google Scholar]
- 28.Kenyon L, Moraes C T. Proc Natl Acad Sci USA. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, D. J. L. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]